Abstract
Endoreduplication is the process where a cell replicates its genome without mitosis and cytokinesis, often followed by cell differentiation. This alternative cell cycle results in various levels of endoploidy, reaching 4× or higher one haploid set of chromosomes. Endoreduplication is found in animals and is widespread in plants, where it plays a major role in cellular differentiation and plant development. Here, we show that variation in endoreduplication between Arabidopsis thaliana accessions Columbia-0 and Kashmir is controlled by two major quantitative trait loci, ENDO-1 and ENDO-2. A local candidate gene association analysis in a set of 87 accessions, combined with expression analysis, identified CYCD5;1 as the most likely candidate gene underlying ENDO-2, operating as a rate-determining factor of endoreduplication. In accordance, both the overexpression and silencing of CYCD5;1 were effective in changing DNA ploidy levels, confirming CYCD5;1 to be a previously undescribed quantitative trait gene underlying endoreduplication in Arabidopsis.
Keywords: change-of-function allele, endopolyploidy, regulatory haplotype
Endopolyploidy, defined as the occurrence of different DNA ploidy levels within an organism, is a common feature in seed plants. Endopolyploidy in plants is most often generated by endoreduplication, a biological process that allows extra rounds of genome duplication to occur without mitosis (1). Endoreduplicating cells increase their nuclear DNA content and size, thereby reducing the overall production cost per tissue of cell walls and cytoplasm, and facilitating faster growth compared with diploid tissue (2). Though apoptotic cell death is the mammalian response against cellular stress and DNA damage (3), endoreduplication in plants leads to differentiation and prohibits cells from reentry into the cell cycle, basically preventing transmission of deleterious mutations (4, 5). Indeed, endoreduplication is found more abundantly among angiosperms that grow under environmentally challenging conditions, suggesting an evolutionary advantage for endoreduplication (6).
Reverse genetics experiments have demonstrated that genes controlling the mitotic cell cycle control the plant endocycle as well (1). The endoreduplication onset is achieved by a decrease in cyclin-dependent kinase (CDK) activity obtained through different interconnected mechanisms, including the interaction of CDKs with small inhibitory proteins (7–9) and inhibitory kinases (10), and the selective destruction of mitotic cyclins (11–13). Surprisingly, little is known about the molecular mechanisms controlling endoreduplication kinetics. CDKA;1 transcription in endoreduplicating tissues, combined with reduced endoreduplication levels in loss-of-function mutants, pinpoint CDKA;1 as a key regulator (14–17). In addition, oscillations in CDKA;1 activity trigger consecutive endocycles (18). Nevertheless, it remains unclear which cyclins control CDKA;1 activity during endoreduplication.
Endoreduplication levels in Arabidopsis thaliana accessions vary in degree (19) and, therefore, are likely controlled by the interaction of environmental factors and multiple genetic loci, most probably, although not exclusively, cell cycle related. Despite the numerous mapping populations available for Arabidopsis (http://www.inra.fr/vast/RILs.htm), and the fact that conventional quantitative trait loci (QTL) linkage mapping is an effective tool for the identification of genetic loci underlying natural variation, only one attempt to map QTL for endoreduplication has been reported so far (20). Recently, genome-wide association (GWA) studies have received increased attention for the identification of QTL in plants, and in Arabidopsis in particular (21), as an alternative to, or in combination with, linkage mapping approaches (22–24). Association approaches provide much higher mapping resolution than linkage mapping, but population structure can be a strong confounding factor, resulting in inflated false-positive associations (24). Recently developed GWA models (25, 26) that control for population structure showed successful in detecting plant QTL for flowering time (22–24), glucosinolates (27, 28), and 107 variable A. thaliana phenotypes (29). Candidate gene association studies are an extension to GWA, focusing the association analysis exclusively on a selection of genes with known or potential functions in the trait of interest, instead of anonymous genome wide markers. The candidate gene association approach has the potential to enrich the number of meaningful trait associations, and has proven to be successful in the identification of genes for trait variation in wild and cultivated maize (30–32), pine (33), and Arabidopsis (23, 34).
The objective of this study was to identify the quantitative trait genes (QTGs) underlying natural variation of endoreduplication in A. thaliana, using traditional linkage mapping complemented with a candidate gene association analysis. We phenotyped 82 recombinant inbred lines (RILs) derived from a cross of Columbia carrying a glabrous1 allele and Kashmir-1 (Col-gl1 × Kas-1), and mapped two large QTL for endoreduplication. Because the genetic network underlying the cell cycle in A. thaliana is very well characterized and known to be involved in endoreduplication, we nominated the mitotic cell cycle genes underlying the identified QTL as candidate genes for local association mapping in a set 87 A. thaliana accessions. Statistical and genetic evidence for the gene-trait association was obtained for CYCD5;1, suggesting CYCD5;1 as a QTG for variation in endoreduplication. Both overexpression and silencing of CYCD5;1 effectively changed the rate of DNA ploidy accumulation, demonstrating its role in endoreduplication kinetics. These results shed light on the unique role of the CYCD5;1 protein, for which a function in endoreduplication was not assigned before.
Results
Endoreduplication Analysis.
To quantify variation in endoreduplication, we evaluated a set of 87 accessions for variation in DNA ploidy levels and the endoreduplication index (EI) (6), calculated as the weighted number of endoreduplication cycles per nucleus [EI = (0 × %2C) + (1 × %4C) + (2 × %8C) + (3 × %16C) + (4 × %32C)]. The endoploidy profile was determined by flow cytometric analysis on nuclei isolated from the first leaf pair at growth stage 1.06 (35). The accessions varied considerably in the extent of EI (Fig. S1A and Table S1), most notably at higher DNA ploidy levels (Fig. S1 B–E and Table S1). Broad sense heritability (H2)—the proportion of phenotypic variation attributed to genetic effects—was 0.52 for EI and ranged from 0.43 to 0.72 for the individual DNA ploidy levels.
QTL Analysis.
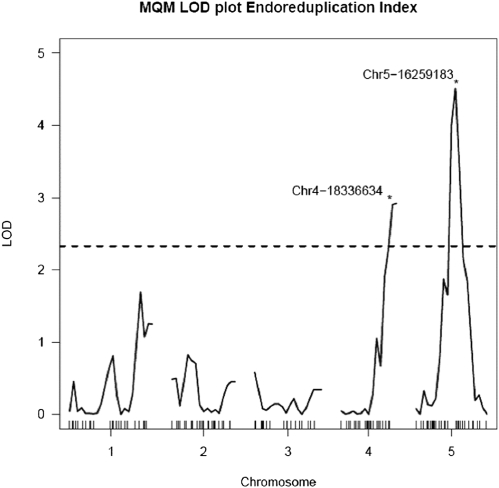
To identify QTL underlying the variation for EI, we accessed an existing F6 RIL mapping population derived from a Col-gl1 × Kas-1 cross (36). The choice for the Col-gl1 × Kas-1 cross was motivated by the large difference measured in EI between Kas-2, having the highest EI value, and Col-0, having an EI level close to the median, in the screen of 87 accessions (Fig. S1A and Table S1). EI values measured in Kas-1 and Kas-2 are comparable, and the glabrous (gl1) mutation, located on chromosome 3, did not affect the EI of Col-gl1 compared with Col-0 (Table S1 and Figs. S1A and S2A). We examined DNA ploidy levels and calculated the EI on 82 RILs (Fig. S2), followed by a multiple QTL linkage method using 119 markers. The logarithm of the odds (LOD) significance threshold (P = 0.05) for the detection of QTL cosegregating with variation in EI was 2.33. Two genome-wide significant QTL for EI were identified on chromosomes 5 (ENDO-1; marker 16259183; LOD = 4.51) and 4 (ENDO-2; marker 18336634; LOD = 2.92; Fig. 1). ENDO-1 accounted for 19.2%, and ENDO-2 for 11.8%, of the EI variance. Both QTL did not exhibit epistasis, suggesting that each QTL contributed additively to EI (Fig. S3). At both QTL, the Kas-1 allele inferred an increase in EI. For the individual DNA ploidy levels, ENDO-1 coincided with a QTL for %8C (LOD = 5.18; Table S2), explaining 23.2% of %8C variance. ENDO-2 coincided with a %16C QTL (LOD = 3.75; Table S2), accounting for 16.7% of %16C variation, and overlapped a QTL for %4C (marker chr4-14936282; LOD = 5.77).
Fig. 1.
QTL likelihood map for EI in the Col-gl1 × Kas-1 F6 RIL population. The x axis corresponds to the genetic map in centimorgans, with tick lines showing the relative position of genetic markers per linkage group; the y axis corresponds to the LOD value as calculated by MQM. The dashed line indicates the 5% significance LOD threshold = 2.33. Two significant QTL were located on two chromosomes: ENDO-1 at marker 16259183 (LOD = 4.51) located on chromosome 5, and ENDO-2 at marker 18336634 (LOD = 2.92) located on chromosome 4.
Reverse genetics experiments have demonstrated that genes controlling the mitotic cell cycle, such as cyclins, most likely control the endocycle as well (7, 8, 11, 15, 17, 37–40). Hence, we focused on 61 previously described cell cycle genes (41) as primary candidate genes for natural variation in endoreduplication, and nominated those residing in a 2-Mb [∼10 centimorgans (cM)] interval surrounding the QTL peaks. The ENDO-1 QTL contained CYCA3;1 (AT5G43080), whereas ENDO-2 contained CYCB2;2 (AT4G35620), CYCB1;1 (AT4G37490), and CYCD5;1 (AT4G37630).
Candidate Gene Association Mapping.
Next, we resequenced the four candidate genes at four 600-bp amplicons covering promoter, 5′-UTR, exonic, intronic, and 3′-UTR regions for 87 accessions. We selected relevant tag sequence polymorphisms (SPs) for each gene [minor allelic frequency (MAF) > 0.04] to moderate the number of SPs while preserving genetic information. Rather than testing for each individual tag SP association with EI, and hence running into multiple testing issues, we applied a haplotype–EI association analysis using all tag SP data simultaneously (i.e., haplotype) by fitting a semiparametric regression model using the least-squares kernel machine procedure (42, 43). This mixed-model approach also allows for population structure adjustment (24–26), and the kernel used incorporates a weight that upweights tag SPs with a rare MAF and downweights tag SPs with more common MAF. For ENDO-1, no significant haplotype–EI association was identified for CYCA3;1 (P = 0.67). Across the ENDO-2 QTL, we identified a significant haplotype–EI association for CYCD5;1 (P = 8 × 10−4) and CYCB2;2 (P = 0.03), but not for CYCB1;1 (P = 0.88). The CYCD5;1 haplotypes also associated strongly with %4C (P = 0.003), %16C (P = 2 × 10−7), and, to a lesser extent %8C (P = 0.01), reflecting our observations from the QTL analysis, i.e., colocalization of QTL for EI, %4C and %16C but not for %8C. In contrast, CYCB2;2 haplotype variation associated with %4C (P = 0.003) and %8C (P = 0.009), but not with %16C (P = 0.11). Further investigation of the CYCD5;1-EI association using G-estimation identified an 8-bp insertion (INDEL17681135; Fig. 2A) in the CYCD5;1 3′-UTR region of the Kas-2 haplotype (frequency = 9.2%), contributing the most (P = 2 × 10−4) to the CYCD5;1 haplotype–EI association, accounting for 20.0% of the EI variance. In the case of CYCB2;2, a single tag SP (SNP16901259) residing in the promotor region of the gene (Fig. 2B) was identified as contributing the most (P = 0.006) to the CYCB2;2 haplotype–EI association, explaining 8.4% of the EI variance.
Fig. 2.
The haplotype structure maps and LD plots for (A) CYCD5;1 and (B) CYCB2;2. In the haplotype structure map, each column represents a polymorphic site with minor alleles in yellow, major alleles in blue, and missing data in gray. Accessions (represented by rows) are clustered by haplotype. The tag SPs most strongly contributing to the significant CYCD5;1 and CYCB2;2 haplotype–EI association (respectively, INDEL17681135 and SNP16901259) are indicated by vertical rectangles. The LD plot reflects r2 estimates for each pair of polymorphic sites visualized by a color matrix, with red indicating strongest LD between a pair of markers.
An analysis of the full-length CYCD5;1 sequence showed no sequence polymorphisms between Col-0 and Kas-1 at the coding level (http://signal.salk.edu/atg1001/3.0/gebrowser.php). In contrast, CYCB2;2 showed two nonsynonymous (NS) substitutions between Col-0 and Kas-1 (amino acids 135 and 147), flanking the CDKA binding site of CYCB2;2 (44). However, analysis of 261 A. thaliana alleles at CYCB2;2 revealed low linkage disequilibrium (LD; r2 = 0.050 and 0.055, respectively) between SNP16901259 and the haplotypes comprising the two NS substitutions. We concluded, therefore, that both analyses suggest that any quantitative effect from CYCD5;1 and CYCB2;2 on EI is regulatory in origin.
Haplotype Expression Analysis.
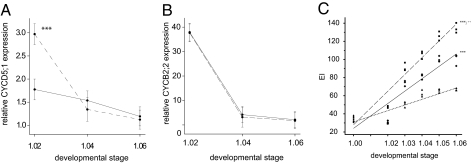
To test the regulatory variant hypothesis, we measured CYCD5;1 and CYCB2;2 gene expression following a leaf development course in Col-0 and Kas-2. As early as leaf developmental stage 1.02, CYCD5;1 expression was significantly higher (P < 0.001) in Kas-2 than in Col-0 (Fig. 3A). This difference, however, disappeared over later growth stages. In contrast, we found no evidence for differential expression of CYCB2;2 (P = 0.99) between Col-0 and Kas-2 (Fig. 3B). Together, these data suggested that increased expression of CYCD5;1, but not of CYCB2;2, early on in the development of leaf tissue, instigates increased DNA duplication in Kas-2.
Fig. 3.
Progression of relative (A) CYCD5;1 and (B) CYCB2;2 expression through first leaf pair development in Col-0 and Kas-2. Mean values ± SE (n = 3) (y axis) for first leaf pair of Col-0 (solid line) and Kas-2 (dashed line) sampled at a series of leaf developmental stages (x axis). ***P < 0.001 pairwise contrast tested after ANOVA. The developmental stages are shown along the x axis. (C) Endoreduplication levels through first leaf pair development in an amiRNA line (▲), Col-0 (◆), and Kas-2 (■). A simple linear regression model was fitted for each genotype (amiRNA line: dotted line; Col-0: solid line; and Kas-2: long dash line). An analysis of parallelism was performed to test for differences: *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the amiRNA-CYCD5;1 line using Student t test); ++P < 0.01 (compared with Col-0 using Student t test).
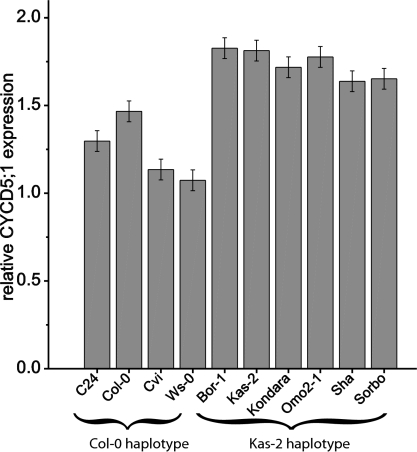
To further substantiate the association between CYCD5;1 expression levels and variation in endoreduplication, CYCD5;1 expression was measured in the first leaf pair at stage 1.02 in a set of accessions having either the 8-bp insertion (Kas-2 haplotype; Bor-1, Kas-2, Kondara, Omo2-1, Shahdara, Sorbo) or not (Col-0 haplotype; C24, Col-0, Cvi, Ws-1). Expression of CYCD5;1 transcripts was 35% higher (P < 0.001) in accessions having the Kas-2 haplotype compared with accessions having the Col-0 haplotype (Fig. 4). According to the Arabidopsis Information Resource release no. 10 annotation (http://www.arabidopsis.org/), CYCD5;1 expresses two transcript variants, of which only the long variant contains the 8-bp insertion. Using quantitative RT-PCR, we specifically measured the abundance of the long transcript across the same set of 10 accessions, but found no significantly higher expression (P = 0.181) in the Kas-2 compared with the Col-0 haplotype group (Fig. S4). The amplification levels of the long CYCD5;1 transcript variant have threshold cycle (Ct) values three- to fourfold higher than those of both transcripts together, suggesting that the proportion of the long transcript variants in the total transcript is ∼10% and, hence, of minor significance. Together, these data further support CYCD5;1 as the QTG underlying the ENDO-2 QTL for endoreduplication, and indicate that the joint expression of both CYCD5;1 transcript variants are important.
Fig. 4.
CYCD5;1 expression variation per CYCD5;1 haplotype group. Means ± SE (n = 6) for CYCD5;1 expression measured in first leaf pair sampled at developmental stage 1.02 of natural accessions carrying either the Col-0 haplotype (C24, Col-0, Cvi, Ws) or the Kas-2 haplotype [Bor-1, Kas-2, Kondara, Omo2-1, Shahdara (Sha), and Sorbo] at the CYCD5;1 locus.
Transgenic Experiments.
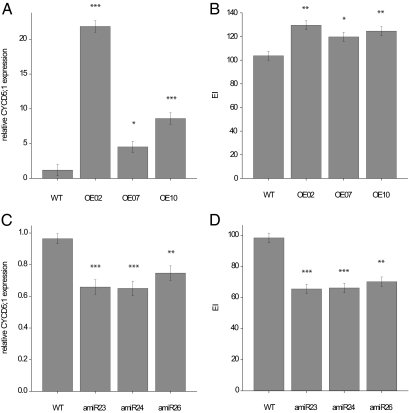
We next used a transgenic strategy to confirm that CYCD5;1 expression variation contributes to variation in endoreduplication. We measured DNA ploidy levels and a number of cell parameters in three independent transgenic lines (OE2, OE7, and OE10) constitutively overexpressing CYCD5;1 in a Col-0 genetic background. The three OE lines, having a 3- to 18-fold up-regulation (P < 0.05) of CYCD5;1 expression compared with Col-0 WT (Fig. 5A), showed EI levels increased by ∼20% (P < 0.05; Fig. 5B). Analysis of the effect of CYCD5;1 overexpression on leaf cell number and cell size showed a doubling of pavement cell number (P < 0.001; Fig. S5B) and a ∼40% (P < 0.001; Fig. S5A) reduction in cell size in OE2 and OE10 compared with WT, which is likely the result of a higher mitotic activity rather than endocycle activity. The smaller cell size observation is in disagreement with previous observations that endoreduplication is positively correlated with cell size (39, 45–47). A possible explanation for this discrepancy is that cell number and cell size jointly control total leaf size, where the increase in one parameter can to some extent be compensated by the reduction of the other parameter.
Fig. 5.
Analysis of CYCD5;1 expression and EI measured in first leaf pair sampled at developmental stage 1.06. Means ± SE for (A) CYCD5;1 expression (n = 3) and (B) EI (n = 3) measured in three independent transgenic lines constitutively overexpressing CYCD5;1 under control of the cauliflower mosaic virus 35S promoter in a Col-0 genetic background (OE2, OE7, and OE10) and Col-0 (WT); means ± SE for (C) CYCD5;1 expression (n = 3) and (D) EI (n = 3) measured in three amiRNA lines (amiR23, amiR24, amiR26) and Col-0 (WT). *P < 0.05; **P < 0.01; ***P < 0.001 (compared with WT after ANOVA).
Given the role of CYCD5;1 overexpression on EI, we also investigated the effect of CYCD5;1 silencing on endoreduplication in three independent transgenic artificial microRNA (amiRNA)-CYCD5;1 lines in a Col-0 genetic background. A reduction to ∼70% (P < 0.001) of the WT CYCD5;1 expression levels (Fig. 5C) increased %2C and %4C ploidy levels (P < 0.001 and P < 0.05, respectively), while decreasing %8C ploidy levels (P < 0.001) (Fig. S6 B–D). Collectively this resulted in a reduction to 67–71% of the WT EI (P < 0.001) (Fig. 5D). We further observed a decrease (P < 0.001) in cell number (Fig. S5D), which is again more likely the result of less mitotic than endocycle activity upon silencing of CYCD5;1. Cell sizes in the amiRNA lines were comparable (P = 0.231) to those of the WT (Fig. S5C).
CYCD5;1 Controls Endoreduplication Kinetics.
To test whether CYCD5;1 controls the kinetics of endoreduplication over time, we measured DNA ploidy levels of the first leaf pair following a detailed development course in Col-0, Kas-2, and a transgenic amiRNA-CYCD5;1 line, and performed an analysis of parallelism by fitting a simple linear regression with the three genotypes as groups. The endoreduplication levels were significantly (P < 0.001) slowed down in the amiRNA-CYCD5;1 line compared with Col-0 and Kas-2, whereas the DNA content increased significantly (P < 0.004) faster in Kas-2 compared with Col-0 (Fig. 3C). These data suggested a role for CYCD5;1 as a rate-determining factor of endoreduplication during the endoreduplication process. These observations were further substantiated by promoter β-glucuronidase (GUS) analysis. At the 1.02 developmental stage, CYCB1;1 promoter activity, marking mitotic activity, was exclusively detected at the basal part of the first leaf pair (Fig. S7), corresponding with the observation that at the proliferation-to-expansion transition of the leaf, cell division ceases along a longitudinal gradient from the leaf tip to the base (48, 49). In contrast, CYCD5;1 promoter activity could be observed throughout the complete leaf blade, marking both dividing and endoreduplicating cells (Fig. S7).
Discussion
Nearly 30 genes and functional polymorphisms underlying natural variation in plant development and physiology have been identified in Arabidopsis, of which only a few had not been found previously in mutant screens (50). The genes identified are mainly involved in the timing of germination and flowering, plant growth and morphology, primary metabolism, and mineral accumulation. Although the cell cycle machinery displays a variety in natural allelic variation with signatures of natural selection (51), so far, natural alleles of cell cycle genes underlying variation in cell cycle-related processes, such as cell differentiation, cell proliferation, mitotic arrest under stress, and endoreduplication, have not been identified yet. In this study, we describe two QTL with a moderate effect on endoreduplication in A. thaliana, one of which could be identified as CYCD5;1 using candidate gene association mapping. The failure to identify the causal gene underlying the QTL ENDO-1 might have been compromised by an incomplete list of candidate genes. Beside more extensive linkage mapping and/or GWA mapping, identifying the QTG underlying the QTL ENDO-1 could also profit from recently published lists of novel genes suggested to be (indirectly) involved in the mitosis, obtained by, e.g., protein interaction data generated by tandem affinity technology (52).
Nucleotide polymorphisms underlying QTL shed a light on the nature of mutations that generate natural variation. A large proportion of natural alleles carry loss-of-function mutations, which are often produced by indel or structural nonsense mutations (53). A second type of common allele is a change-of-function allele produced by a missense or splice-site mutation, altering protein structure and function, or by regulatory mutations affecting spatiotemporal expression. Lack of CYCD5;1 coding variants between Col-0 and Kas-2, and the higher CYCD5;1 expression in the accessions carrying the Kas-2 allele, clearly suggest a change-of-function allele carrying regulatory mutations. The 8-bp insertion in the 3′-UTR genomic region of the CYCD5;1 could be easily hypothesized to either have a regulatory effect on CYCD5;1 expression or to be in very strong LD with a nearby regulatory polymorphism.
So far, CDKA;1 was pinpointed as being essential for endoreduplication in Arabidopsis (15, 16). The observed decrease in cell number and EI in the amiRNA-CYCD5;1 lines suggests CYCD5;1 to be a rate-determining factor for DNA replication. This previously undescribed role for CYCD5;1 is further supported by the association between CYCD5;1 transcript level and the DNA ploidy level across different accessions. In addition, CYCD5;1 and CDKA;1 were found to interact in vivo (51), suggesting that the CYCD5;1/CDKA;1 complex determines the pace of DNA replication through the phosphorylation of yet-to-be-identified substrates.
Here, we identified CYCD5;1 as a QTG that controls endoploidy in nature by modulating the progression of successive endocycles during leaf development. The partial confounding of the population structure with the genetic variation for CYCD5;1 suggests an adaptive mechanism to an environmental gradient that results in differential endopolyploidy and organ growth over a broad geographic range. With only eight accessions possessing the CYCD5;1 allele effecting increased endoreduplication, the identification of an obvious adaptive response to an environmental gradient was not evident. Therefore, there is every reason to believe that GWA studies for endoreduplication involving larger samples will be more fruitful in identifying a potential selective environmental gradient for variation in endoreduplication. Uncovering the QTGs and, ultimately, the nucleotide polymorphisms that underlie adaptation to environmental gradients will lead to a better understanding of the mutation types and gene functions that constitute the bulk of natural phenotypic variation.
Materials and Methods
A. thaliana genotypes, growth conditions, and experimental design; measurement of DNA ploidy levels and EI calculation; cell imaging and cell size measurements; sequencing data, SP detection and tag SP selection; statistical and genetic analysis of data; and map construction and QTL mapping are described in SI Materials and Methods.
Linkage and QTL Mapping.
The genotypes for the 82 RILs at each of the 119 markers can be found at http://naturalvariation.org/KasCol. Genetic maps for each linkage group were constructed using JoinMap 4.0 (54). A multiple QTL mapping (MQM) approach was followed to identify QTL. First, unconditioned QTL mapping was conducted using the MQM scan of R/QTL (55) to identify putative QTL. Next, forward stepwise cofactor selection was performed to select a marker as cofactor at each suggestive (LOD > 2; P < 0.10; 1,000 permutations) QTL region identified. Third, a MQM model including the selected cofactors was fitted to the data to optimize LOD scores and minimize QTL intervals. Mapping was conducted with an interval size of 5 cM.
Association Mapping.
We applied a two-stage association analysis (56). First, endoreduplication data were analyzed on the basis of the linear model yijk = μ + Gi + Rk + GRik + eijk, where yijk is the phenotypic observation of the jth sample of the ith accession of the kth replicate; Gi, Rk, and GRik represent the fixed genotype, replicate, and genotype × replicate effect, respectively; and eijk represent the error effect. Second, the obtained adjusted entry means were analyzed by a semiparametric linear mixed model (42, 43). Given Gis as the genotype of phenotype i at tag SP s (s = 1, … , S) coded as the number of copies of the minor allele, and Gi = (Gi1, …, GiS). The semiparametric model is subsequently given by (random terms underlined) yi = μ + h(Gi) + genotypei + ei, where μ is the intercept, genotypei is the random factor to account for population structure, ei the random error term, and h(Gi) is the joint effect of all S tag SP genotypes, i.e., the haplotype. All random effects are assumed to be zero-mean normally distributed. This model accounts for population structure by including the genome-wide estimates of genetic similarities to correct for genetic relatedness. The genetic similarities were calculated as the proportion of shared haplotypes for each pair of individuals at 5,000 SNPs (1,000 SNPs per chromosome) randomly selected from the 250,000 SNP data set (29).
We used G-estimation (57), a propensity score-based estimation technique from the field of causal inference (58), to detect association between an individual tag SP and the trait while considering all possible interactions among all tag SP genotypes. The semiparametric linear mixed model and the generalized linear mixed model used in the G-estimation procedure were fitted by a restricted maximum likelihood approach as implemented in SAS (59). Further information about the semiparametric linear mixed model and the G-estimation procedure can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wilson Ardiles-Diaz, Raimundo Villarroel, and Hilde Van den Daele for excellent technical assistance in resequencing, sequence data processing, endoreduplication, and promoter-GUS analyses. We thank F. van Eeuwijk for comments on the manuscript. This work was supported by Ghent University, Bijzonder Onderzoeksfonds Methusalem Project BOF08/01M00408; the Agency for Innovation by Science and Technology in Flanders predoctoral fellowships (to R.S., R.K., and S.D.); and European Union Human Resources and Mobility for an Early Stage Training Grant MEST-CT-2004-514632 predoctoral fellowship (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120811109/-/DCSupplemental.
References
- 1.De Veylder L, Larkin JC, Schnittger A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011;16:624–634. doi: 10.1016/j.tplants.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Barlow P. Patterned cell determination in a plant tissue: The secondary phloem of trees. Bioessays. 2005;27:533–541. doi: 10.1002/bies.20214. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 4.Radziejwoski A, et al. Atypical E2F activity coordinates PHR1 photolyase gene transcription with endoreduplication onset. EMBO J. 2011;30:355–363. doi: 10.1038/emboj.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi S, et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barow M, Meister A. Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ. 2003;26:571–584. [Google Scholar]
- 7.Churchman ML, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verkest A, et al. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinl C, et al. Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell. 2005;17:1704–1722. doi: 10.1105/tpc.104.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J. 2007;51:642–655. doi: 10.1111/j.1365-313X.2007.03167.x. [DOI] [PubMed] [Google Scholar]
- 11.Boudolf V, et al. CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 2009;150:1482–1493. doi: 10.1104/pp.109.140269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasili R, et al. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics. 2010;185:257–268. doi: 10.1534/genetics.109.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieu-Rivet E, Gévaudant F, Cheniclet C, Hernould M, Chevalier C. The Anaphase Promoting Complex activator CCS52A, a key factor for fruit growth and endoreduplication in Tomato. Plant Signal Behav. 2010;5:985–987. doi: 10.4161/psb.5.8.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiva-Neto JT, et al. A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell. 2004;16:1854–1869. doi: 10.1105/tpc.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dissmeyer N, et al. T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell. 2007;19:972–985. doi: 10.1105/tpc.107.050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dissmeyer N, et al. Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell. 2009;21:3641–3654. doi: 10.1105/tpc.109.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bramsiepe J, et al. Endoreplication controls cell fate maintenance. PLoS Genet. 2010;6:e1000996. doi: 10.1371/journal.pgen.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roodbarkelari F, et al. Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc Natl Acad Sci USA. 2010;107:15275–15280. doi: 10.1073/pnas.1006941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inzé D. Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 2002;129:854–864. doi: 10.1104/pp.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massonnet C, et al. New insights into the control of endoreduplication: Endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol. 2011;157:2044–2055. doi: 10.1104/pp.111.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- 22.Brachi B, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrenreich IM, et al. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009;183:325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA. A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics. 2007;175:879–889. doi: 10.1534/genetics.105.054932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 27.Chan EKF, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 2011;9:e1001125. doi: 10.1371/journal.pbio.1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan EKF, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185:991–1007. doi: 10.1534/genetics.109.108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber A, et al. Major regulatory genes in maize contribute to standing variation in teosinte (Zea mays ssp. parviglumis) Genetics. 2007;177:2349–2359. doi: 10.1534/genetics.107.080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber AL, et al. The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): New evidence from association mapping. Genetics. 2008;180:1221–1232. doi: 10.1534/genetics.108.090134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson LM, et al. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell. 2004;16:2719–2733. doi: 10.1105/tpc.104.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Martínez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB. Association genetics in Pinus taeda L. I. Wood property traits. Genetics. 2007;175:399–409. doi: 10.1534/genetics.106.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenreich IM, Stafford PA, Purugganan MD. The genetic architecture of shoot branching in Arabidopsis thaliana: A comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics. 2007;176:1223–1236. doi: 10.1534/genetics.107.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyes DC, et al. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson IW, Schiff CL, Hughes DE, Somerville SC. Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics. 2001;158:1301–1309. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 38.Lammens T, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlieghe K, et al. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol. 2005;15:59–63. doi: 10.1016/j.cub.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 40.Boudolf V, et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandepoele K, et al. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu D, Lin X, Ghosh D. Semiparametric regression of multidimensional genetic pathway data: Least-squares kernel machines and linear mixed models. Biometrics. 2007;63:1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renaudin J-P, et al. Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- 45.Castellano MdelM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell. 2004;16:2380–2393. doi: 10.1105/tpc.104.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabatini S, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 47.Sugimoto-Shirasu K, Roberts K. “Big it up”: Endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Beemster GTS, et al. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 2005;138:734–743. doi: 10.1104/pp.104.053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andriankaja M, et al. Exit from proliferation during leaf development in Arabidopsis thaliana: A not so gradual process. Dev Cell. 2012;22:64–78. doi: 10.1016/j.devcel.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Alonso-Blanco C, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sterken R, et al. A population genomics study of the Arabidopsis core cell cycle genes shows the signature of natural selection. Plant Cell. 2009;21:2987–2998. doi: 10.1105/tpc.109.067017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Leene J, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alonso-Blanco C, et al. Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol. 2005;139:1304–1312. doi: 10.1104/pp.105.068510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Ooijen JW. JoinMap 4. Software for the Calculation of Genetic Linkage Maps on Experimental Populations. Wageningen, The Netherlands: Kyazma; 2006. [Google Scholar]
- 55.Broman KW, Wu H, Sen Ś, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 56.Stich B, et al. Comparison of mixed-model approaches for association mapping. Genetics. 2008;178:1745–1754. doi: 10.1534/genetics.107.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robins JM, Blevins D, Ritter G, Wulfsohn M. G-estimation of the effect of prophylaxis therapy for Pneumocystis carinii pneumonia on the survival of AIDS patients. Epidemiology. 1992;3:319–336. doi: 10.1097/00001648-199207000-00007. [DOI] [PubMed] [Google Scholar]
- 58.Rosenbaum PR, Rubin DB. Constructing a control group using multivariate matched sampling methods that incorporate the propensity score. Am Stat. 1985;39:33–38. [Google Scholar]
- 59.SAS Institute Inc . SAS/STAT User's Guide Version 9.2. Cary, NC: SAS Institute; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.