Abstract
Transferring lipid antigens from membranes into CD1 antigen-presenting proteins represents a major molecular hurdle necessary for T-cell recognition. Saposins facilitate this process, but the mechanisms used are not well understood. We found that saposin B forms soluble saposin protein–lipid complexes detected by native gel electrophoresis that can directly load CD1 proteins. Because saposin B must bind lipids directly to function, we found it could not accommodate long acyl chain containing lipids. In contrast, saposin C facilitates CD1 lipid loading in a different way. It uses a stable, membrane-associated topology and was capable of loading lipid antigens without forming soluble saposin–lipid antigen complexes. These findings reveal how saposins use different strategies to facilitate transfer of structurally diverse lipid antigens.
Keywords: Natural Killer T cell, tuberculosis, immunogenicity, lipid binding protein
Antigen presentation molecules have a fundamental role in host defense and impact innate and adaptive immune responses. Whereas the MHC class I and II proteins present protein-derived peptide antigens to T cells, CD1 molecules present lipid antigens to T cells with invariant and diverse T-cell receptors (1). Although the molecular mechanisms governing MHC class I and II peptide loading are well appreciated, the mechanisms of CD1 lipid loading remain poorly understood. In the case of MHC class I molecules, proteasome-derived peptides are loaded onto MHC class I molecules by proteins of the peptide loading complex whose components include calnexin, calreticulin, ERp57, tapasin, and TAP (2). MHC class II molecules are loaded in endosomal compartments, where HLA-DM facilitates the exchange of the class II associated invariant peptide for high-affinity peptides derived from lysosomal processing of exogenous proteins (3, 4). The capacity of the MHC class I and II processing and antigen loading mechanisms are major factors determining which peptides are antigenic. In contrast to MHC-presented peptides, CD1-restricted lipid antigens are amphipathic molecules that are typically embedded in cellular membranes or micelles (5) and thus may be largely inaccessible to luminal proteins. Given the nonpolar component of lipids, mobilization of membrane-derived lipids across an aqueous endocytic environment likely requires extraction from membranes and, in most cases, transport across aqueous biological buffers into the lipid-binding grooves of CD1 molecules. These related processes are predicted to have unfavorable dissolution energetics and is unlikely to occur efficiently without the assistance of lipid-transfer proteins or other molecular mediators (6, 7).
Recent studies have shown that saposins, a family of small, nonenzymatic lipid-binding proteins, facilitate CD1 lipid loading in endosomal compartments (6, 8–10). To a relative extent, all saposins enhance CD1 lipid loading in cellular and in vitro assays, but they appear to have a differential capacity to load particular lipids into individual CD1 isoforms. In one study, saposin B (SapB) was shown to facilitate α-galactosylceramide (αGalCer) loading onto human CD1d molecules more efficiently than other saposin isoforms (9). In cellular studies, saposin C (SapC), but not other saposins, restored CD1b presentation of long-chain mycobacterial antigens in saposin-deficient antigen presenting cells (8). Furthermore, direct visualization of lipid-loaded mouse CD1d molecules (mCD1d) by native isoelectric focusing revealed that saposin A (SapA) and SapC were more efficient at facilitating the transfer of lipids onto mCD1d molecules than SapB or saposin D (SapD) (10). It is not clear if any interactions occur between CD1 molecules, saposins and lipid antigens that may favor preferential lipid loading “partnerships” among saposins, specific lipids, and CD1 isoforms. Given the extensive structural and biophysical diversity of CD1-presented lipid antigens, more than one lipid loading mechanism may be necessary to effectively load CD1 molecules with acyl chains ranging from C8 to C90 (1, 11).
Here, we define two fundamentally distinct saposin loading mechanisms that allow for short and long acyl-chain lipid antigens to be loaded. We show that SapC loads CD1 molecules in a process that occurs at the membrane surface containing the lipid antigen to be transferred. Instead, SapB binds lipids to form soluble complexes separate from the membrane that contained the lipid antigen and can transport lipid to recipient CD1d molecules. These differences in topology and lipid loading mechanisms allow CD1 molecules to load lipid antigens having widely different structures.
Results and Discussion
SapC, but Not SapB, Makes Stable Contacts with Lipid Bilayers.
To examine SapB and SapC membrane interactions, we used a sucrose gradient separation approach. Small unilamellar liposomes composed of 70% mol phosphatidylcholine, 15% mol bis(monoacylglycero)phosphate and 15% mol cholesterol were generated and incubated with SapB or SapC at pH 5.0. After incubation, samples were transferred to a three-step sucrose gradient buffered to pH 5.0 and centrifuged to separate saposins bound to liposomes from those unable to stably associate with liposome vesicles. After centrifugation, gradient fractions were collected, analyzed by SDS/PAGE, and visualized by silver staining. Liposomes centrifuged in the absence of saposins were almost entirely detected in low-density sucrose fractions at the top of the gradient, whereas high-density sucrose fractions lacked detectable liposome content (Fig. S1). Conversely, in the absence of liposomes, the SapC protein was detected only in high-density sucrose fractions at the bottom of the gradient that were shown to be devoid of lipid content (Fig. 1A, Top). These data indicate that SapC and liposomes, when centrifuged separately, sediment to opposite ends of the sucrose gradient. SapC and liposomes were then incubated together at pH 5.0 and centrifuged on the sucrose gradient at this pH. In contrast to the SapC sedimentation profile observed in the absence of liposomes, incubation of SapC and liposomes together at pH 5.0 showed a markedly different sedimentation profile with a considerable fraction of SapC being detected in low-density, liposome-positive sucrose fractions at the top of the gradient (Fig. 1A, Middle). This suggests that, under acidic conditions, SapC stably associates with liposome membranes. de Alba et al. showed that SapC has a negatively charged protein surface at neutral pH. Under acidic conditions (approximately pH 5.0), negatively charged amino acid residues on the SapC surface are partially protonated, thereby neutralizing negatively charged carboxyl groups that normally repel the negative charge on anionic phospholipid bilayers (12). It is then expected that SapC binding to lipid bilayers is a reversible process whereby the release of SapC from the bilayer is triggered by deprotonation of acidic amino acid residues at neutral or increased pH. To demonstrate pH-dependent interactions between SapC and the liposomal bilayer, SapC and liposomes were incubated at pH 5 and then transferred to a sucrose gradient buffered to neutral pH. Following gradient centrifugation and analysis by SDS/PAGE, SapC was mostly detected in high-density sucrose fractions previously shown to be devoid of liposome content (Fig. 1A, Bottom). These findings imply that, although SapC has the capacity to make stable contacts with liposomal membranes at acidic pH, SapC is released from the liposomal bilayer at neutral pH. Importantly, the sedimentation profile of SapB incubated with liposomes was entirely different from that observed for SapC. Here, SapB was detected only in liposome-negative sucrose gradient fractions (Fig. 1B), and its sedimentation profile was unchanged in the presence or absence of liposomes; suggesting that, unlike SapC, SapB is not capable of establishing stable interactions with the liposomal bilayer under the same experimental conditions.
Fig. 1.
SapC, but not SapB, makes stable contacts with lipid bilayer. (A) SapC was incubated with liposomes for 60 min in sodium citrate buffer, pH 5.0. Liposomes and SapC samples were transferred to a sucrose gradient buffered to pH 5 or pH 7 where indicated, and centrifuged at 200,000 × g. Gradient fractions were collected, separated by SDS/PAGE, and visualized by silver staining. (B) SapB was incubated with liposomes, transferred to a sucrose gradient and visualized as described in A. “Top” and “bottom” indicate low and high-density sucrose gradient fractions, respectively. Triangle (Bottom) indicates increasing sucrose density.
SapC Interacts with CD1c-Fc in Liposome-Proximal Topology.
The cosedimentation of SapC in liposome fractions on sucrose gradients suggested that SapC might have the capacity to interact with CD1 while associating with the lipid bilayer, thereby situating CD1 adjacent to accessible lipid substrates embedded in the bilayer. To evaluate this possibility, SapC, CD1c-Fc fusion protein and liposomes were coincubated under acidic conditions. Then, samples were transferred to the sucrose gradient buffered to pH 5 and centrifuged to separate liposome-bound protein components from those present in the soluble phase. Following sucrose gradient fractionation and analysis by SDS/PAGE, a detectable fraction of CD1c-Fc fusion protein was noted in liposome-positive fractions 1 to 4, the same fractions that contained SapC (Fig. 2A, Top, Middle). By using this sedimentation approach, we also detected CD1b in liposome-positive fractions following incubation with SapC (Fig. 2A, Bottom). These findings are consistent with CD1b–SapC interactions that were previously reported (8). Importantly, CD1c-Fc sedimentation to the same liposome-positive sucrose fractions was specifically dependent on liposome-associated SapC molecules, because CD1c-Fc was not detected in liposome-positive fractions in the absence of SapC (Fig. 2B, Top). These findings are consistent with the assembly of a SapC–CD1–liposome complex, in which SapC binds liposomes by embedding into the liposomal bilayer and is then able to interact with CD1c-Fc on the liposome surface via protein-protein contacts. To test this concept, SapC, CD1c-Fc, and liposomes were incubated at pH 5 and then transferred to a sucrose gradient buffered to neutral pH to separate liposome-bound protein components from those present in the soluble phase. Following sucrose gradient fractionation and SDS/PAGE separation, SapC and CD1c-Fc were not detected in liposome-positive fractions, but instead were detected in liposome-negative fractions (Fig. 2B, Middle), a finding that can be explained by deprotonation of acidic amino acid residues at the elevated pH of the sucrose gradient, thereby triggering a pH-dependent release of SapC from the liposome surface (Fig. 1A, Bottom). These results suggest that CD1c-Fc association to liposomes is dependent on liposome-inserted SapC molecules. By comparison, SapB did not cosediment with liposomes to low-density fractions under acidic conditions, and correspondingly, CD1c-Fc fusion protein was not detected in liposome-positive fractions (Fig. 2B, Bottom).
Fig. 2.
SapC interacts with CD1c-Fc in a liposome-proximal topology. (A and B) Saposins and/or CD1 fusion proteins were incubated with liposomes (2 mM) at pH 5 as indicated. Samples were transferred to a sucrose gradient buffered to pH 5 (or pH 7) and centrifuged at 200,000 × g. Gradient fractions were collected, separated by SDS/PAGE, and visualized by silver staining. Proteins visualized by sliver staining are labeled at right.
Liposome Bilayer Enhances CD1c-Fc–SapC Interactions.
To examine possible direct SapC–CD1c protein contacts and the role of the lipid bilayer in SapC–CD1c-Fc interactions, we used a chemical crosslinking approach. SapC was coupled to a sulfhydryl-reactive cross-linker, and, in a separate reaction, CD1c-Fc was coupled to a sulfhydryl-containing cross-linker. After incubation with liposomes at pH 5.0, protein samples were resolved by nonreducing SDS/PAGE and immunoblotted with anti-SapC antibodies. Given that the molecular weights of SapC and CD1c-Fc are considerably different (∼10 kDa and 150 kDa, respectively), it predicts that SapC molecules crosslinked with CD1c-Fc would be detected at approximately 150 kDa by immunoblotting with anti-SapC antibodies whereas SapC molecules not associated with CD1c-Fc would be detected at approximately 10 kDa. In the absence of liposomes, SapC was capable of interacting with CD1c-Fc as evidenced by SapC detection at approximately 150 kDa (Fig. 3A, lane 3), suggesting that a lipid bilayer is not required for SapC–CD1c-Fc interaction. However, under the same experimental conditions, liposomes markedly enhanced SapC–CD1c-Fc interactions, as indicated by an increase in SapC detection at approximately 150 kDa (Fig. 3A, lane 4). To control for nonspecific cross-linked products that could result from random molecular collisions, a structurally related antigen presentation molecule, MHC class I fusion protein (MHCI-Fc), was coupled to a sulfhydryl-containing cross-linker as described for CD1c-Fc and incubated with SapC and liposomes at pH 5.0. Immunoblotting with anti-SapC antibodies revealed an absence of SapC signal at approximately 150 kDa (Fig. 3A, lane 5), thereby supporting specific protein–protein contacts between SapC and CD1c-Fc. Interestingly, SapC–CD1c-Fc cross-linked products were detected at neutral pH, suggesting that SapC, in a negatively charged form, has the potential to interact with CD1c-Fc (Fig. 3A, lane 6).
Fig. 3.
Liposome bilayer enhances CD1c-Fc–SapC interactions. (A) SapC and Fc fusion proteins were coupled to sulfhydryl-reactive and sulfhydryl-containing cross-linkers, respectively, and incubated with or without liposomes at pH 5.0 and pH 7.0. Samples were separated by nonreducing PAGE and immunoblotted with anti-SapC antibodies. (B) Total CD1c-Fc and MHC-I-Fc fusion proteins were separated by SDS/PAGE and visualized by silver stain.
SapC Facilitates CD1c-Fc Lipid Loading in Membrane-Proximal Context.
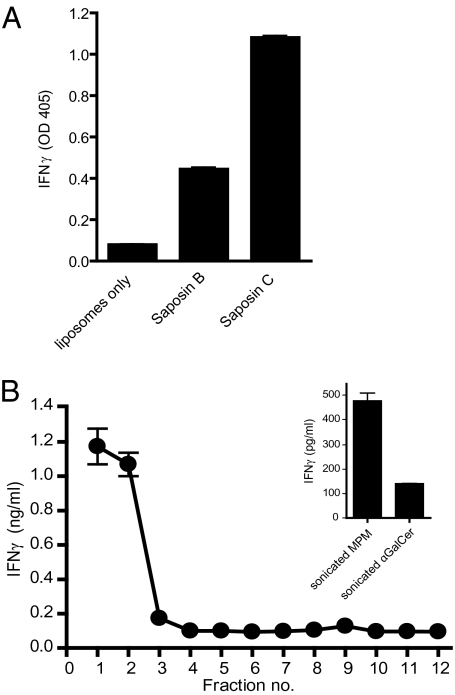
To examine functional loading of CD1 molecules, we developed a cell-free CD1 lipid loading reconstitution assay in which lipid antigens were provided as components of liposomes and recombinant saposins were tested for their lipid loading capacity in the presence of recombinant CD1 molecules expressed as IgG2a Fc-fusion proteins (13). Here, saposins and CD1c-Fc were incubated with liposomes supplemented with a mycobacterial lipid antigen presented by CD1c [0.1% mannosyl-β-1-phosphomycoketide (MPM)] (14, 15) at pH 5.0, and, after incubation, CD1c-Fc was immobilized on protein G-coated plate wells. MPM-reactive, CD1c-restricted T cells (CD8-1 T cells) were then added to plates, and T-cell activation was measured by IFN-γ release. We found SapC was a more efficient lipid-transfer mediator of MPM to CD1c than was SapB (Fig. 4A). In the absence of lipid-loading mediators, T-cell activation was not detected, suggesting that, without lipid-loading facilitators, CD1c-Fc molecules alone lack the capacity to extract MPM from liposomes. Given that, in this experimental system, SapC–CD1c-Fc interactions can occur in a liposome-proximal context as well as in the solvent-phase, we wished to determine the CD1 lipid loading capacity under these two circumstances. We examined the topological context of saposin-mediated CD1 lipid loading by combining the previously described sucrose gradient separation approach with a cell-free CD1 lipid loading reconstitution assay. Liposomes were supplemented with 0.1 mol % MPM and incubated with SapC. As before, SapC/liposome samples were transferred to an acidic sucrose gradient and liposome-associated SapC was separated from SapC in the solvent phase by ultracentrifugation. Following sucrose gradient fractionation, each gradient fraction was incubated with CD1c-Fc fusion protein separately and then CD1c-Fc was immobilized on protein G-coated plates. After fusion protein immobilization, MPM-reactive T cells were then added to the plates and CD1c lipid loading was measured by IFN-γ secretion. By using this approach, IFN-γ release was not detected in high-density sucrose fractions where SapC is present, but not bound to liposomes (Fig. 4B, fractions 8–12). In contrast, we found that nearly all the stimulatory activity for CD1c-restricted T cells was detected only in low-density, liposome-positive fractions (Fig. 4B, fractions 1–3), where CD1c-Fc fusion protein interacts with SapC bound to liposomes (as previously shown in Fig. 2A), suggesting that CD1c-Fc lipid loading only occurs in gradient fractions where SapC is associated with liposomes.
Fig. 4.
SapC facilitates CD1c-Fc lipid loading in a membrane-proximal context. (A) Lipid-transfer mediators facilitate CD1c-Fc loading. Liposomes (1 mM) with 0.1% mol MPM were incubated with saposin proteins (5.4 μM) as indicated and CD1c-Fc fusion protein in sodium citrate buffer, pH 5.0. Fusion protein samples were immobilized on protein G-coated plates. After washing with PBS solution, CD1c-restricted T cells (CD8-1 T cells) were added and incubated in complete media for 20 h. IFN-γ released by activated T cells was measured by ELISA. (B) Liposome-embedded SapC facilitates CD1c-Fc lipid loading. SapC was incubated with liposomes (1 mM total lipid) supplemented with 0.1% mol MPM in citrate buffer, pH 5.0. Liposome and SapC solutions were transferred to a sucrose gradient buffered to pH 5.0 and centrifuged at 200,000 × g. Gradient fractions were collected and dialyzed against citrate buffer, pH 5.0. CD1c-Fc fusion protein was added to each fraction and incubated. Samples were immobilized on protein G–coated plate. CD8-1 T cells (CD1c-restricted, MPM-reactive cells) were added. IFN-γ released by activated T cells was measured by ELISA. Inset: Sonicated MPM (4 μM) or αGalCer (100 ng/mL) were incubated with CD1c-Fc. After immobilization on protein G-coated plates, T cells were added to plates, and IFN-γ released by activated T cells was measured by ELISA.
SapB Forms Protein–Lipid Complexes with CD1 Lipid Antigens.
Although stable interactions between SapB and liposomes were not detected in our liposome sedimentation system (Fig. 1B), it is possible that SapB may instead interact with lipid bilayers by using a mechanism that employs transient membrane interactions to facilitate CD1 lipid loading. A crystal structure of SapB reveals two clasping SapB monomers accommodating a lipid in the nonpolar pocket formed by the clasping chains. This suggests that SapB may be capable of extirpating lipids from membrane bilayers to produce soluble SapB–lipid complexes (16). Therefore, we hypothesized that SapB might not require stable contacts with lipid bilayers but may instead form stable complexes with lipids after extracting them from membranes to facilitate CD1 lipid loading. Thus, we sought to identify SapB–lipid complexes in the absence of membranes to support a unique topology that differentiates the two saposin-function models as exemplified by SapB- vs. SapC-facilitated CD1 lipid loading.
To examine the lipid antigen-binding capacity of SapB and SapC in the absence of membranes, we developed a nondenaturing electrophoresis approach (native-PAGE) to separate lipid-complexed saposin proteins from lipid-free saposin species. Here, we incubated SapB and SapC with the ganglioside GD1a at pH 5.0, and, after incubation, separated the saposin/GD1a samples by native PAGE. In the absence of the ganglioside GD1a, SapB was visualized as two rapidly migrating species that likely represent monomer and dimer conformations of SapB as previously described (Fig. 5A, lane 1) (16–18). Strikingly, at the highest concentration of GD1a tested, SapB migrated instead as a single band that had a slower electrophoretic mobility (Fig. 5A, lane 2, solid arrowhead). As the concentration of the ganglioside GD1a is reduced (Fig. 5A, lanes 2–5), there is a proportional decrease in the detection of SapB–lipid complex dimer (Fig. 5A, solid arrowhead), with a concomitant increase in the detection of both the SapB monomer and dimer (Fig. 5A, open and dashed arrowheads, respectively). In contrast to SapB, the electrophoretic mobility of SapC was not affected by the presence of the ganglioside GD1a at any concentration (Fig. 5B). These results suggest that SapB forms stable soluble complexes with lipids like GD1a whereas SapC does not.
Fig. 5.
SapB forms protein–lipid complexes with CD1 lipid antigens. (A) SapB and (B) SapC were incubated with decreasing concentrations of the ganglioside GD1a in sodium citrate (pH 5.0) buffer (lanes 2–5). SapB species (5 μg per sample) were separated by native-PAGE, and total protein was visualized by Coomassie staining. Lanes 6 in A and B show a sample of 84 μM GD1a in the absence of SapB or SapC, respectively. SapB and SapC monomers (open triangles), SapB and SapC dimers (dotted triangles), and SapB dimer–lipid complexes (closed triangles) are indicated at left. (C) SapB and SapC (5 μg) were incubated with αGalCer-biotin (1 μM) for 18 h at pH 5. Samples were neutralized with Tris, pH 9.0, and treated with 1% OsO4 for 10 min and separated by native-PAGE. In-gel detection of αGalCer-biotin was performed by incubating polyacrylamide gel with streptavidin-HRP (Lower). Total protein was visualized by Coomassie stain (Upper). BSA-biotin (0.01 μg) was used as a positive control.
Given that SapB was recently reported to facilitate αGalCer loading onto human CD1d molecules more efficiently than other saposins (9), we wished to determine if SapB forms protein–lipid complexes with CD1d-restricted antigens. We used a synthetic αGalCer lipid analogue that featured a biotin tag on the C2 position of the α-linked galactosyl headgroup (αGalCer-biotin) as a model CD1 presented antigen for our lipid-binding studies. We incubated the two saposin isotypes with αGalCer-biotin at pH 5.0 and separated saposins by native-PAGE. Following electrophoresis, αGalCer-biotin was detected by an in-gel streptavidin-HRP chemiluminescent method and total saposin content was visualized by Coomassie staining. In the absence of αGalCer-biotin, SapB and SapC appeared as two low molecular weight bands, which likely represent the monomer and dimer conformations of SapB and SapC (Fig. 5C, Upper) that have been previously reported (17). In the presence of αGalCer-biotin, the mobility of SapB and SapC was not significantly affected (Fig. 5C, Upper, second lane). However, in-gel detection of αGalCer-biotin revealed a significant signal that superimposed only with the SapB dimer band visualized by Coomassie stain (Fig. 5C, solid arrowhead). In contrast, αGalCer-biotin was not detected to associate with the monomeric or dimeric forms of SapC. These findings are consistent with the SapB X-ray crystal structure that shows that SapB binds to lipid cargo preferentially as a dimer. In addition, these findings indicate that under the same experimental conditions, SapB forms stable SapB-αGalCer-biotin complexes whereas SapC does not.
SapB Binds Short-Chain, but Not Long-Chain, CD1 Lipid Antigens.
Because CD1-restricted lipid antigens feature acyl chains ranging from C8 to approximately C90 in length, we tested whether the SapB hydrophobic pocket formed has the capacity to accommodate short- and long-chain lipid antigens. Here, 6x-histidine–tagged SapB was incubated alone or with a short- or long-chain form of glucose monomycolate (GMM), designated according to its overall mean chain length, C32 GMM or C80 GMM, under acidic conditions. After incubation, SapB–lipid complexes were isolated from free lipids by immobilized metal-affinity chromatography (IMAC) by using columns packed with nickel-chelating cellulose matrix. After extensive washing, SapB–lipid complexes were eluted from the columns with 0.2 M imidazole elution buffer. Eluates were spotted directly on TLC silica plates and salt was removed from spotted samples by running plates in deionized water. Silica plates were then sprayed with α-naphthol and charred to visualize glycolipids. In the absence of SapB, free-C32 GMM was not detected by α-naphthol staining in cellulose-nickel column eluates, indicating that, in the absence of SapB, free lipids lack the capacity to pass through the cellulose-nickel matrix under aqueous conditions (Fig. 6A). In contrast to free-lipids, control lipid GD1b and test lipid C32 GMM were detected by α-naphthol staining in column eluates only when incubated with SapB before IMAC separation (Fig. 6B, spots 1 and 2), indicating that GD1b and C32 GMM elution from cellulose-nickel columns is dependent on SapB and therein supporting the formation of GD1b- and C32 GMM-SapB complexes. The absence of glycolipid detection by α-naphthol staining of column eluates revealed that C80 GMM did not bind to SapB before IMAC separation (Fig. 6B, spot 3). To rule out bacterial glycolipids that may bind SapB during protein purification, SapB was incubated in the absence of the ganglioside GD1b or GMMs. Under these conditions, SapB eluates did not test positive for α-naphthol staining (Fig. 6B, spot 4). Together, these results suggest that, although the hydrophobic pocket formed by two clasping SapB monomers is capable of binding lipids with short to moderate acyl chains such as those exhibited by the ganglioside GD1b and C32 GMM, SapB monomers are unable to accommodate lipids with long acyl chains such as C80 GMM.
Fig. 6.
SapB binds and loads short, but not long, CD1 lipid antigens. (A) GD1b (8 μg) was passed through cellulose nickel columns and treated with 0.2 M imidazole buffer. Buffer alone (Left), column eluate (Center), and organic wash eluate (Right; 2:1 chloroform:methanol wash), were spotted directly on a TLC silica plate. Salt was removed by running plate with deionized water and were subsequently sprayed with α-naphthol to detect glycolipids (B) 6x-Histidine–tagged SapB (5 μg) was incubated on its own or with the ganglioside GD1b (4 μg), C32 GMM (8 μg), or C80 GMM (8 μg) at pH 5 for 16 h. Following incubation, samples were passed through columns packed with nickel chelating cellulose. After extensive washing, columns were treated with 0.2 M imidazole, and eluates were spotted directly on a TLC silica plate. Salt was removed by running the plate with deionized water. Plates were allowed to dry in a vacuum. Dried plates were sprayed with α-naphthol to detect glycolipids. Plates were then charred to visualize signal. (C) Sonicated C32 GMM (1 μg) or C80 GMM (1 μg) was incubated with saposin proteins (5.4 μM) as indicated and CD1b-Fc fusion protein in sodium citrate buffer, pH 5.0. Fusion protein samples were immobilized on protein G-coated plates. After washing with PBS solution, CD1b-restricted T cells (LDN5 T cells) were added and incubated in complete media for 20 h. IFN-γ released by activated T cells was measured by ELISA.
Next, to test whether SapB-complexed lipids are loaded onto CD1 molecules, we used a cell-free CD1 lipid loading reconstitution assay similar to that described in Fig. 4A. Here SapB was incubated with sonicated C32 GMM or C80 GMM in the presence of CD1b-Fc at pH 5.0. After incubation, CD1b-Fc was immobilized on protein G-coated plate wells, and, after extensive washing with PBS solution, GMM-reactive, CD1b-restricted T cells (LDN5 T cells) were added to plates, and T-cell activation was measured by IFN-γ release. In the absence of SapB, weak T-cell activation was detected in samples incubated with C32 GMM (Fig. 6C, lane 2), whereas C80 GMM did not stimulate T-cell activation vs. background (Fig. 6C, lane 4). These findings suggest that, in the absence of lipid-loading mediators, such as SapB, C32 GMM is loaded onto CD1b-Fc molecules more efficiently than C80 GMM under the acidic conditions tested. In the presence of SapB, C32 GMM-loaded molecules stimulated greater T-cell activation, as measured by IFN-γ release (Fig. 6C, lane 3), versus pH-dependent loading that was observed in the absence of SapB (Fig. 6C, lane 2). In marked contrast, SapB did not enhance C80 GMM lipid loading. Taken together, these findings suggest that SapB has the capacity to facilitate lipid loading of short-chain GMMs onto CD1b-Fc molecules, whereas it is unable to facilitate loading of long-chain GMMs.
Recent studies have implicated saposin function in CD1 lipid antigen loading (6, 8–10). These pioneering efforts have brought to light the importance of saposin proteins in CD1 immunobiology, which serve a critical function comparable to accessory proteins that facilitate peptide transport and loading onto MHC class I and II proteins. Whereas the key antigen processing step for MHC is cleavage of large proteins to small peptides, most native CD1 antigens are synthesized with alkyl chains that approximate the volume of the CD1 cavity. Considering that most CD1 antigens are derived from cellular membranes, a key cellular processing event for the CD1 system is extracting lipids from their normal state within bilayers or multimolecular aggregates and to transfer lipid monomers for CD1 loading. In this study, we describe two different saposin mechanisms for lipid mobilization that reveal how lipids with markedly different structures can achieve CD1 loading.
Our findings demonstrate that saposins B and C enhance CD1 lipid loading to varying degrees. These data are consistent with previous in vitro studies that have shown a hierarchical CD1 lipid loading capacity among all saposin isoforms (9, 10). Given that SapB and SapC are the best understood saposins, our studies centered on delineating the mechanisms used by these saposins to facilitate CD1 lipid loading. Our results indicate that SapC, but not SapB, has the capacity to stably associate with liposome membranes, as evidenced by the detection of SapC in liposome-positive fractions following sucrose gradient sedimentation. These findings support a mechanism in which SapC inserts directly into the lipid bilayer, thereby disrupting the tightly packed lipids that comprise the target membrane. These SapC–liposome interactions are reminiscent of SapC–membrane interactions observed in the context of glycosphingolipid (GSL) degradation whereby SapC inserts into membrane bilayers to provide lysosomal hydrolases greater accessibility to GSL substrates embedded in model membranes (19–22). Because membrane-associated SapC facilitates GSL degradation by positioning hydrolytic enzymes, such as β-glucosidase, next to disorganized membrane environments where GSL substrates are accessible (23, 24), we tested whether SapC also interacts with CD1 molecules on the surface of model membranes. Our studies reveal that CD1c-Fc interacts with liposome-associated SapC molecules at pH 5.0, as shown by the detection of SapC and CD1c-Fc in liposome-positive fractions following sucrose gradient sedimentation. Release of SapC from liposomes at pH 7.0 resulted in the loss of CD1c-Fc detection in liposome-positive fractions, thereby supporting the notion that SapC interacts with CD1c-Fc on the liposome surface. Given these data, we predict that liposome-associated SapC disrupts the liposome bilayer and positions CD1c-Fc, through direct physical interaction, in a disrupted bilayer environment where lipid antigens may be more easily acquired (Fig. S2A). In this capacity, SapC may recruit CD1 molecules to perturbed regions of cellular membranes, where it may function as a “locater” that identifies the site of lipids that are accessible for CD1 loading amid the vast landscape of endocytic membranes.
Although our liposome sedimentation studies indicate that SapC and CD1c-Fc interact in a membrane-associated topological context, this experimental strategy was not suitable to examine SapC–CD1c contacts that may occur in the soluble phase. As a result, we used a two-step crosslinking strategy to detect protein–protein interactions in the soluble phase and on the liposome surface. In the presence of liposomes, the signal of the SapC–CD1c-Fc crosslinked product was noticeably stronger than in the absence of liposomes, suggesting that, although SapC and CD1c-Fc have the capacity to interact in the soluble phase, the membrane-associated conformation and topology of SapC enhances SapC–CD1c-Fc interactions. NMR studies have shown that, in the absence of lipid particles, SapC adopts a compact structure that buries hydrophobic residues in its protein core, whereas, in the presence of SDS micelles, it adopts an open V-shaped conformation (25). The dramatic differences between soluble-phase and membrane-associated SapC protein structure have the potential to affect protein–protein contacts between SapC and CD1c-Fc molecules and may explain the enhanced detection of the crosslinked SapC–CD1c-Fc product observed in our analysis.
Because our liposome sedimentation and crosslinking studies suggest that CD1c-Fc is recruited to the liposome surface by membrane-associated SapC molecules, we hypothesized that CD1c-Fc molecules could access liposome-embedded lipid antigens by co-opting a similar membrane-associated mechanism used by lysosomal hydrolases to mediate GSL degradation (19, 22, 23, 26). In support of this hypothesis, our results demonstrate that only liposome-associated SapC molecules had the capacity to load CD1c-Fc and evoke T-cell activation when the antigen is provided in a membrane-bound form (Fig. 4B). In marked contrast, T-cell stimulation was not detected in liposome-negative fractions. These results suggest that SapC-facilitated CD1 lipid loading may occur primarily on the surface of target membranes. In this context, we hypothesize that the CD1c lipid binding groove is positioned next to the sites of SapC membrane perturbation, thereby facilitating the transfer of membrane-embedded lipids to CD1c molecules (Fig. S2A). A related SapC-mediated CD1 lipid loading model was proposed by Winau et al. in which CD1b acts like a “scoop” that is held by SapC on the membrane surface (8). Here, CD1b is predicted to sample (or “scoop up”) lipid antigens from a “container” represented by the perturbed lipids surrounding membrane-inserted SapC molecules. In this context, we predict that the α1 and α2 domains of CD1c interact with membrane-embedded SapC molecules and with the lipids present in the disorganized membrane environment that border membrane-embedded SapC molecules. Taken together, our results suggest that CD1c-Fc loading occurs in a membrane-associated topology whereby SapC membrane perturbation is essential for access to membrane-embedded lipids and SapC–CD1c-Fc interactions position CD1c toward the accessible lipids bordering SapC on the liposome surface.
The findings for SapB contrast strikingly with those noted for SapC. SapB showed no stable association with liposomes in sedimentation studies compared with SapC. Given that our studies indicate that SapB enhances MPM lipid loading of CD1c-Fc molecules in a cell-free CD1 lipid loading reconstitution assay, and other studies have demonstrated that SapB is the dominant saposin that loads CD1d-Fc proteins (9), we reasoned that, unlike the membrane-proximal topology of SapC-facilitated CD1 loading, SapB may load CD1 molecules in a soluble-phase topology as a SapB–lipid antigen complex that leaves the membrane.
In support of this hypothesis, our findings indicate that, in the presence of the ganglioside GD1a, SapB was detected as a single protein species that showed a slower electrophoretic mobility in native PAGE than SapB species in the absence of the ganglioside GD1a. The GD1a-induced change in SapB mobility suggests that, similarly to the SapB-phospholipid structure that was reported by Ahn et al. (16), SapB forms soluble complexes with the ganglioside GD1a, whereby SapB binds to GD1a as a homodimer, and thus the additional size and/or conformation change imparted by the GD1a lipid accounts for the slower electrophoretic mobility that was observed for SapB in the presence of the ganglioside GD1a. In marked contrast, SapC mobility was not affected by the presence of GD1a in any of the GD1a lipid concentrations tested, suggesting that, unlike SapB, SapC lacks the capacity to form soluble SapC–lipid complexes under the experimental conditions tested.
We then tested the potential of SapB to form soluble protein–lipid complexes with CD1d-restricted lipid antigens. In-gel detection of SapB samples shows that αGalCer-biotin preferentially binds to SapB dimers, as indicated by the αGalCer-biotin signal that superimposes only with the Coomassie-stained SapB dimer band (Fig. 5C). Notably, under the same experimental conditions, αGalCer-biotin did not impact the electrophoretic mobility or associated with SapC by in-gel detection. Together, these data suggest that SapB employs a soluble protein–lipid complex topology, rather than a membrane-associated topology, to facilitate CD1 lipid loading (Fig. S2B).
The crystal structure of the SapB-phospholipid complex reveals that the SapB dimer's hydrophobic pocket has a volume of approximately 900 Å3 (16), which is smaller than all known CD1 grooves, suggesting that only lipids of moderate carbon acyl-chain length may fit. Given that CD1b molecules can bind and present mycolic acids that can reach C90 in length (27–29) and thus fill or exceed the 2,300-Å3 volume of the CD1b lipid-binding hydrophobic channels, we hypothesized that microbial lipid antigens of such length might not be able to form protein–lipid complexes with SapB. Our findings indicate that SapB forms protein–lipid complexes with GD1a, GD1b, C32 GMM, and αGalCer-biotin, but is not capable of complexing with C80 GMM. These observations support the notion that the length of lipid antigens is an important determinant of SapB binding and, thus, SapB-mediated CD1 lipid loading. Based on our SapB lipid binding studies and GMM loading studies, we demonstrate that long-chain lipid antigens, such as mycobacteria-derived mycolates, are not spontaneously loaded onto CD1b molecules, but instead may depend on SapC-mediated membrane-proximal topology to load onto CD1b molecules. In fact, in cellular studies, only recombinant SapC was able to reconstitute CD1b antigen presentation in prosaposin-deficient, CD1b-transfected antigen presenting cells pulsed with long-chain mycolates (8).
In addition to acyl-chain variability, acyl-chain unsaturations are a common feature of cellular and microbial lipids (7). These chemical groups have a major effect on the shape of lipids, which is commonly underappreciated. Acyl chain unsaturations (alkene groups) generate a kink and limit flexibility in the acyl chain structure (30). Thus, alkene groups or other acyl-chain modifications may cause lipids to adopt a shape that is not compatible with the dimensions of the SapB hydrophobic pocket or may prevent acyl chains from adoptively fitting into the hydrophobic pocket by restricting its chain. Our studies do not directly address SapB lipid specificity, but, given the limited volume of the SapB dimer structure, we speculate that lipid volume and conformation are important determinants of SapB–lipid complex formation. These limitations should not apply to the membrane-perturbation properties of SapC, which may allow it to facilitate the loading of lipid antigens not able to associate stably with SapB and so represent a mechanism of lipid transfer that is more universal and not limited by length, saturation, or other specificity constraints. These contrasting mechanisms may be necessary to facilitate CD1 loading of endogenous and microbial lipid antigens that, collectively, feature broad structural and biophysical diversity and are not likely to be loaded onto CD1 molecules by a single loading mechanism. Moreover, given that SapA and SapD also potentiate GSL degradation, it is possible that the members of the saposin family have evolved differential mechanisms to promote both GSL degradation and CD1 loading of diverse lipid structures. Future studies are needed to determine the mechanisms that are used by SapA and SapD to facilitate CD1 lipid loading. The CD1 lipid loading mechanisms proposed in this study are similar to the “solubilizer” and “liftase” mechanisms proposed for SapB and SapC in GSL degradation, respectively (22, 31). Our results suggest that differential saposin-mediated lipid loading mechanisms are necessary to address the diversity of lipids that can be sampled by CD1 molecules.
Materials and Methods
CD1c-, CD1d-, and MHCI-Fc Fusion Protein Expression and Purification.
Recombinant CD1b, CD1c, CD1d, and MHCI were expressed as IgG2a fusion protein in a CHO cell protein expression system as previously described (13). Conditioned media from each CHO cell transfectant was passed through a protein A–Sepharose column by gravity flow. After washing the column with PBS solution, fusion proteins were eluted with 100 mM sodium acetate, pH 4.3, and collected in tubes preloaded with 1 M Tris, pH 9.0, to rapidly neutralize the elution buffer pH and thereby reduce acid-mediated protein hydrolysis.
Liposome Preparation.
Small unilamellar vesicles were prepared by mixing phosphatidylcholine, cholesterol, and bis(monoacylglycero)phosphate at the concentrations indicated. After mixing, organic solvents were evaporated under a nitrogen stream, and the resultant lipid film was hydrated by using a 50 mM sodium citrate (pH 5.0) solution. Lipid vesicles were frozen and thawed six times to ensure equilibrium between the bulk solvent and vesicle-trapped aqueous volume. Lipid samples were passed through two stacked polycarbonate membranes (80-nm pore diameter) 21 times by using an extruder apparatus (Avanti Polar Lipids).
Cell-Free CD1 Lipid Loading Reconstitution Assay.
MPM (embedded in unilamellar liposomes), C32-GMM, or C80 GMM (as sonicated particles) were incubated with saposins and CD1 fusion protein at pH 5.0. After incubation, samples were neutralized with 1 M Tris 9.0, and samples were transferred to protein G-coated plates. CD1 fusion proteins were allowed to bind to plates for 120 min. Plates were then washed three times with PBS solution, and CD8-1 or LDN5 T cells were added to the plates at 100,000 cells per well. T cells were cultured for 18 h, and supernatants were harvested and tested for IFN-γ release by ELISA.
Sucrose Gradient Sedimentation.
A three-step sucrose gradient was used to separate liposomes from proteins present in solution. Density gradient consisted of 50%, 25%, and 10% sucrose steps. Liposome/protein samples were centrifuged at 200,000 × g for 3 h at 4 °C. Fractions were collected starting from the top of the centrifugation tube (low-density sucrose fractions). Samples were separated by SDS/PAGE and visualized by silver staining. Where indicated, liposomes were detected in low-density sucrose gradient fraction after centrifugation by supplementing liposomes with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) and measured the OD405 of each fraction collected after centrifugation.
CD1c-Fc Thiolation and SapC Derivatization with 1-Ethyl-3-[3-Dimethylaminopropyl]Carbodiimide Hydrochloride/3-[2-Pyridyldithio]Propionyl Hydrazide (PDPH) and Sulfosuccinimidyl 6-(3′-[2-pyridyldithio]-propionamido)hexanoate Cross-Linkers.
CD1c-Fc was treated with 0.17 μM sulfosuccinimidyl 6-(3′-[2-pyridyldithio]-propionamido)hexanoate for 90 min at room temperature. Samples were passed through Microspin desalting columns (Pierce) to remove unreacted sulfosuccinimidyl 6-(3′-[2-pyridyldithio]-propionamido)hexanoate reagent (Pierce). Where indicated, samples were treated with 10 mM DTT to yield sulfhydryl groups and passed through a desalting column to remove reducing agent. SapC was incubated with 10 mM 3-[2-pyridyldithio]propionyl hydrazide and 1 mM 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride for 90 min, and samples were passed through Microspin desalting columns to remove unreacted reagents.
SapC-CD1c-Fc Protein Crosslinking Assay.
PDPH-derivatized SapC and thiolated fusion proteins (CD1c-Fc or MHCI-Fc) were incubated with liposomes under acidic conditions for 120 min. Samples were separated by nonreducing SDS/PAGE and were subsequently transferred to PVDF membrane by using a semidry transfer apparatus. PVDF membrane was blotted with anti-SapC antibodies (Santa Cruz Biotechnology) for 1 h and detected with an anti–rabbit-HRP secondary antibody.
GD1a-Binding Assay.
The ganglioside GD1a was dried in glass test tubes by placing samples under a stream of nitrogen. Lipids were hydrated with 50 mM sodium citrate, pH 5.0, for 30 min and then sonicated for 1 to 2 min in a water bath sonicator. Saposins (5 μg per sample) were added to each sonicated lipid sample, mixed well, and incubated at 37 °C overnight. Samples were separated by native-PAGE and visualized by Coomassie staining.
α-GalCer-Biotin Chemiluminescent Detection.
αGalCer-biotin was incubated with saposin molecules overnight as described in the previous section. Before separation by native-PAGE, samples were neutralized with 1M Tris, pH 9.0, and treated with 1% OsO4 for 10 min. After native-PAGE separation, gel was fixed with 15% TCA for 60 min and then washed with PBS solution to remove fixative. A streptavidin–HRP conjugate was used to detect saposin–αGalCer-biotin complexes.
Isolation of SapB–Lipid Complexes and Lipid Detection.
The ganglioside GD1b, synthetic C32 GMM, or purified C80 GMM from Mycobacterium smegmatis were dried in glass test tubes by placing samples under a stream of nitrogen. Lipids were hydrated with 50 mM sodium citrate, pH 5.0, 0.25% CHAPS for 30 min, and then sonicated for 1 to 2 min in a water-bath sonicator. Saposins (5 μg per sample) were added to each sonicated lipid sample, mixed well, and incubated at 37 °C overnight. Samples were then passed twice through cellulose-nickel columns. After extensive washing with PBS solution, samples were eluted with 0.2 M imidazole. Eluates were spotted directly on silica gel 60 F254 TLC plates (EMD), and imidazole was removed by running the plate in deionized water. TLC plates were then dried and sprayed with α-naphthol/sulfuric acid and charred to visualize glycolipids. The long-chain GMM lipid referred to as C80 GMM in this report was purified from M. smegmatis and has an alkyl chain length range of C60 to C90. We refer to the long-chain GMM as C80 GMM to be consistent with similar designations of purified long-chain GMMs in the literature (11, 32).
Supplementary Material
Acknowledgments
We are grateful to A.N. Odyniec, M. Brigl, G.F.M. Watts, and T.Y. Cheng for suggestions and excellent technical assistance. This work was supported by National Institutes of Health (NIH) Grants AI028973 and AI063428 (to M.B.B.), DK36729 and NS36681 (to G.A.G.), and AR048632 and AI049313 (to D.B.M. and A.K.); a Howard Hughes Medical Institute Gilliam Fellowship (to L.L.); the Burroughs Wellcome Fund (D.B.M. and A.K.); a Personal Research Chair from Mr. James Bardrick (to V.B., N.V., and G.S.B.); a Royal Society Wolfson Research Merit Award (to V.B., N.V., and G.S.B.); the Medical Research Council (V.B., N.V., and G.S.B.); Wellcome Trust Grant 084923/B/08/Z (to V.B., N.V., and G.S.B.); and a Netherlands Organization for Scientific Research Grant (to A.J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200764109/-/DCSupplemental.
References
- 1.Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol. 2009;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 2.Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. doi: 10.1146/annurev.cellbio.24.110707.175347. [DOI] [PubMed] [Google Scholar]
- 3.Landsverk OJ, Bakke O, Gregers TF. MHC II and the endocytic pathway: regulation by invariant chain. Scand J Immunol. 2009;70:184–193. doi: 10.1111/j.1365-3083.2009.02301.x. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan BA, Nagarajan NA, Kronenberg M. CD1 and MHC II find different means to the same end. Trends Immunol. 2005;26:282–288. doi: 10.1016/j.it.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Sprong H, van der Sluijs P, van Meer G. How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 6.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 7.Moody DB. T Cell Activation by CD1 and Lipid Antigens. New York: Springer; 2007. [Google Scholar]
- 8.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 9.Yuan W, et al. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc Natl Acad Sci USA. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 12.de Alba E, Weiler S, Tjandra N. Solution structure of human saposin C: pH-dependent interaction with phospholipid vesicles. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]
- 13.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 14.van Summeren RP, Moody DB, Feringa BL, Minnaard AJ. Total synthesis of enantiopure beta-D-mannosyl phosphomycoketides from Mycobacterium tuberculosis. J Am Chem Soc. 2006;128:4546–4547. doi: 10.1021/ja060499i. [DOI] [PubMed] [Google Scholar]
- 15.Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 16.Ahn VE, Faull KF, Whitelegge JP, Fluharty AL, Privé GG. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc Natl Acad Sci USA. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John M, Wendeler M, Heller M, Sandhoff K, Kessler H. Characterization of human saposins by NMR spectroscopy. Biochemistry. 2006;45:5206–5216. doi: 10.1021/bi051944+. [DOI] [PubMed] [Google Scholar]
- 18.Vogel A, Schwarzmann G, Sandhoff K. Glycosphingolipid specificity of the human sulfatide activator protein. Eur J Biochem. 1991;200:591–597. doi: 10.1111/j.1432-1033.1991.tb16222.x. [DOI] [PubMed] [Google Scholar]
- 19.Alattia JR, Shaw JE, Yip CM, Privé GG. Molecular imaging of membrane interfaces reveals mode of beta-glucosidase activation by saposin C. Proc Natl Acad Sci USA. 2007;104:17394–17399. doi: 10.1073/pnas.0704998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 21.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Wilkening G, Linke T, Sandhoff K. Lysosomal degradation on vesicular membrane surfaces. Enhanced glucosylceramide degradation by lysosomal anionic lipids and activators. J Biol Chem. 1998;273:30271–30278. doi: 10.1074/jbc.273.46.30271. [DOI] [PubMed] [Google Scholar]
- 23.Salvioli R, Tatti M, Ciaffoni F, Vaccaro AM. Further studies on the reconstitution of glucosylceramidase activity by Sap C and anionic phospholipids. FEBS Lett. 2000;472:17–21. doi: 10.1016/s0014-5793(00)01417-4. [DOI] [PubMed] [Google Scholar]
- 24.Vaccaro AM, Tatti M, Salvioli R, Ciaffoni F, Gallozzi E. Correlation between the activity of glucosylceramidase and its binding to glucosylceramide-containing liposomes. Role of acidic phospholipids and fatty acids. Biochim Biophys Acta. 1990;1033:73–79. doi: 10.1016/0304-4165(90)90196-4. [DOI] [PubMed] [Google Scholar]
- 25.Hawkins CA, de Alba E, Tjandra N. Solution structure of human saposin C in a detergent environment. J Mol Biol. 2005;346:1381–1392. doi: 10.1016/j.jmb.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Qi X, Grabowski GA. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem. 2003;278:31918–31923. doi: 10.1074/jbc.M302752200. [DOI] [PubMed] [Google Scholar]
- 27.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 28.Beckman EM, et al. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 29.Grant EP, et al. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauch J, et al. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508–47515. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaccaro AM, et al. Function of saposin C in the reconstitution of glucosylceramidase by phosphatidylserine liposomes. FEBS Lett. 1993;336:159–162. doi: 10.1016/0014-5793(93)81631-9. [DOI] [PubMed] [Google Scholar]
- 32.Enomoto Y, et al. Temperature-dependent biosynthesis of glucose monomycolate and its recognition by CD1-restricted T cells. Biochem Biophys Res Commun. 2005;337:452–456. doi: 10.1016/j.bbrc.2005.09.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.