Abstract
The conserved RNA-binding protein Hfq and its associated small regulatory RNAs (sRNAs) are increasingly recognized as the players of a large network of posttranscriptional control of gene expression in Gram-negative bacteria. The role of Hfq in this network is to facilitate base pairing between sRNAs and their trans-encoded target mRNAs. Although the number of known sRNA–mRNA interactions has grown steadily, cellular factors that influence Hfq, the mediator of these interactions, have remained unknown. We report that RelA, a protein long known as the central regulator of the bacterial-stringent response, acts on Hfq and thereby affects the physiological activity of RyhB sRNA as a regulator of iron homeostasis. RyhB requires RelA in vivo to arrest growth during iron depletion and to down-regulate a subset of its target mRNAs (fdoG, nuoA, and sodA), whereas the sodB and sdhC targets are barely affected by RelA. In vitro studies with recombinant proteins show that RelA enhances multimerization of Hfq monomers and stimulates Hfq binding of RyhB and other sRNAs. Hfq from polysomes extracted from wild-type cells binds RyhB in vitro, whereas Hfq from polysomes of a relA mutant strain shows no binding. We propose that, by increasing the level of the hexameric form of Hfq, RelA enables binding of RNAs whose affinity for Hfq is low. Our results suggest that, under specific conditions and/or environments, Hfq concentrations are limiting for RNA binding, which thereby provides an opportunity for cellular proteins such as RelA to impact sRNA-mediated responses by modulating the activity of Hfq.
The ∼100-nucleotide RyhB small regulatory RNA (sRNA), which was discovered 10 y ago in genome-wide screens in Escherichia coli (1, 2), has become a paradigm of a bacterial RNA regulator with a widely conserved function in iron homeostasis control (3). The sRNA is specifically transcribed under iron-depleted conditions and acts by a variety of antisense mechanisms on a large set of target mRNAs, many of which encode iron-containing and iron-storage proteins and are down-regulated by RyhB (4, 5). The hexameric RNA-binding protein Hfq was found to be essential for RyhB-mediated control of its targets (6, 7). Hfq acts as an RNA chaperone to expedite annealing of RyhB to its target mRNAs. In addition, the interaction between RyhB and Hfq recruits the major endo-ribonuclease RNase E for cleavage of the targets (8). Reciprocally, the sRNA in its free form is protected by Hfq from endonucleolytic cleavage; thus, Hfq ensures sufficiently high levels of the regulator (5, 9).
RyhB is one of many sRNAs that require association with Hfq for their activity (10). The importance of Hfq for global RNA regulation became apparent through studies using RNA coimmunoprecipitation with Hfq, which showed that the protein interacts with several hundred different sRNA and mRNA species (11, 12). Therefore, hfq-deficient cells exhibit pleiotropic effects, including decreased growth rates, increased sensitivity to stress, and attenuated virulence (reviewed in refs. 13 and 14). The extent of Hfq's involvement and the impact that Hfq has on RNA regulation highlight unresolved issues that concern Hfq availability and its subcellular localization. The estimated cellular copy number of Hfq ranges from ∼400 to ∼10,000 hexamers per cell (reviewed in ref. 10). As for Hfq localization, early studies indicated that most of the protein was associated with ribosomes, whereas a small fraction appeared to be associated with the nucleoid (15). A recent study concludes the opposite, showing that Hfq partitions with the cytoplasmic fraction rather than with 30S and 50S subunits or 70S monosomes (16). Another unresolved issue concerns functional partners. Hfq was shown to interact with components of the RNA decay machinery, including poly(A) polymerase I, polynucleotide phosphorylase, RNase E (10), and, recently, with Rho, the anti-termination factor (17). However, except for this still-unclear association with these proteins, no other proteins have been shown to interact with Hfq and/or to impact RNA regulation by Hfq.
In this study, we show that the major regulator of the stringent response, the ribosome-associated RelA protein, impacts sRNA–mRNA regulation by acting on Hfq and propose a mechanism whereby RelA increases the binding rate of RNAs to Hfq.
Results
RyhB Expression Inhibits Cell Growth in a relA-Dependent Manner.
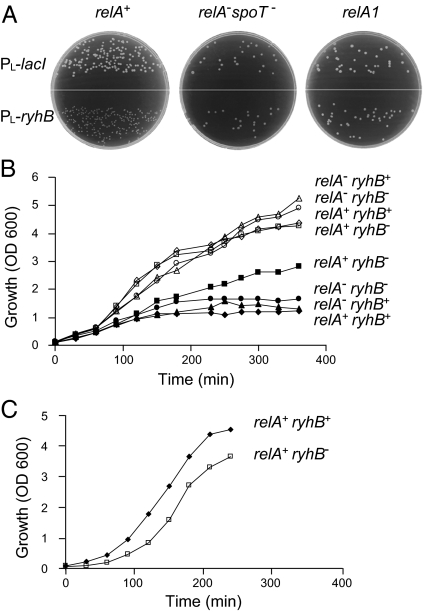
To screen for regulators that mediate the effects of sRNAs on cell physiology, plasmids expressing selected sRNAs were introduced into a collection of mutant strains of E. coli. We noted that constitutive expression of RyhB inhibits colony growth of bacteria carrying the wild-type relA allele, whereas RyhB expression in a strain deleted of both the relA and the spoT genes has no effect on growth; i.e., this latter strain formed normal-size colonies (Fig. 1A). RelA, the major regulator of the E. coli stringent response, is a ribosome-dependent (p)ppGpp synthetase that is activated in response to amino acid starvation, whereas SpoT, the second protein regulator of this response, exhibits dual functions as both a ppGpp synthetase and a hydrolase. Normal-size colonies were also detected with a strain harboring wild-type spoT and a relA1 mutant allele (Fig. 1A), which carries an IS2 insertion and permits residual ppGpp synthetase activity (18), indicating that the growth inhibition by RyhB is correlated with relA. Importantly, however, because the growth inhibition phenotype was detected in rich medium where there is no ppGpp synthesis, we predicted that the phenotype must be unrelated to ppGpp or RelA synthetase activity.
Fig. 1.
Expression of RyhB inhibits growth of relA+ cells. (A) High-level expression of RyhB leads to growth arrest of relA+ cells. Cultures of relA+, relA−spoT−, or relA1 carrying PL-ryhB or a control plasmid PL-lacI were grown on LB plates for 18 h overnight (ON) at 37 °C. (B) RyhB inhibits growth of relA+ cells under iron-limiting conditions. ON cultures of wild-type and mutants were diluted 1/100 in LB medium and allowed to grow aerated at 37 °C to OD600 of 0.1. Thereafter, half of the cultures were exposed to the iron chelator 2,2′-dipyridyl (200 μM). Growth of treated (solid symbols) and untreated cultures (open symbols) was measured at the indicated times (relA+ is also spoT+, whereas relA− is also spoT−). (C) The recovery of the arrested strain relA+ryhB+ is faster than that of relA+ryhB−. Cultures treated with 2,2′-dipyridyl as in B were grown ON. Thereafter, the cultures were diluted in fresh LB medium, and growth was monitored as indicated.
Conditions of limiting iron induce transcription of the chromosomally encoded ryhB gene. Under these conditions, cultures expressing RyhB and wild-type RelA show growth inhibition, whereas cells deficient for ryhB show normal growth (Fig. 1B). As also observed in Fig. 1A, the presence of RyhB has no effect in strains lacking relA and spoT. The RyhB-RelA–mediated growth arrest is advantageous once iron-supplemented rich medium is available again (Fig. 1C); when diluted further in rich medium, the recovery of the arrested wild-type cells is faster than that of ryhB-deficient cells, suggesting that RyhB-mediated adaptation to iron starvation is facilitated by RelA (Fig. 1C).
Regulation of a Subset of RyhB Target mRNAs Requires relA.
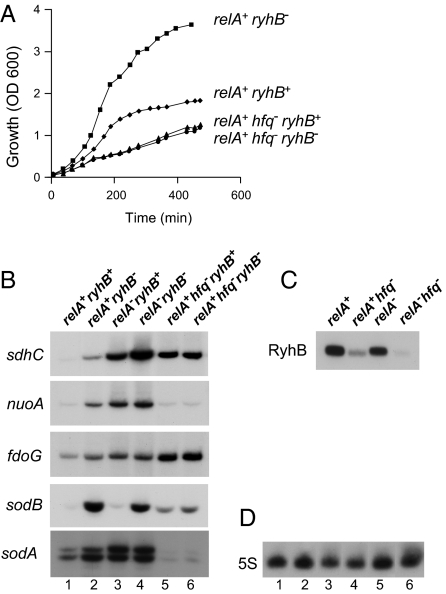
RyhB regulation of target mRNAs depends on the RNA-binding protein Hfq. In hfq-deficient cells, RyhB is unstable and therefore not active (Fig. 2C). We observed that relA-ryhB–dependent growth inhibition is lost in an hfq mutant (Fig. 2A), indicating that this phenotype is due to an effect of RyhB on its targets.
Fig. 2.
Regulation of a subset of RyhB targets requires relA. (A) The relA-ryhB–dependent growth arrest phenotype is no longer detected in the hfq mutant. ON cultures as indicated were diluted 1/100 in LB medium, grown to OD600 of 0.1, and exposed to the iron chelator 2,2′-dipyridyl (200 μM) (relA+ is also spoT+, whereas relA− is also spoT−). (B) Regulation of a subset of RyhB targets requires both Hfq and RelA. Primer extensions were carried out with RNA extracted from cultures grown in LB medium to early log phase and treated with 2,2′-dipyridyl for 1 h (relA+ is also spoT+, whereas relA− is also spoT−). (C and D) Northern analysis of RyhB sRNA (C) and 5S rRNA (D) in wild-type and mutants. The numbers 1–6 indicate strains as in B.
To learn whether RelA affects RyhB target regulation, we examined RNA levels of selected targets, under conditions of iron starvation, in wild-type and relA mutant strains. We selected previously predicted RyhB targets (19) as well as sodA, for which we found, by computational screens, that its ribosome-binding region is complementary to the core sequence of RyhB (Fig. S1). Furthermore, because we found that sodA regulation by RyhB is independent of fur (Fig. S1), it is conceivable that sodA is a direct target of RyhB. Analysis of the levels of RyhB target mRNAs (Fig. 2B) shows that, whereas RyhB regulation of nuoA, fdoG, and sodA is dependent on the presence of RelA, regulation of sdhC is only partly influenced by RelA, and sodB regulation is relA-independent. Importantly, RyhB RNA levels in a relA spoT mutant strain were comparable to those detected in the wild-type (Fig. 2C). Fig. 2D shows probing of 5S rRNA as a control for loading. These results indicate that regulation of a subset of RyhB targets requires both Hfq and RelA, whereas the regulation of other targets depends only on Hfq. The results also suggest that the observed growth inhibition is due to changes in expression of a subset of RyhB targets, changes that are mediated indirectly by RelA.
RelA Affects Binding of Hfq to RyhB in Vitro.
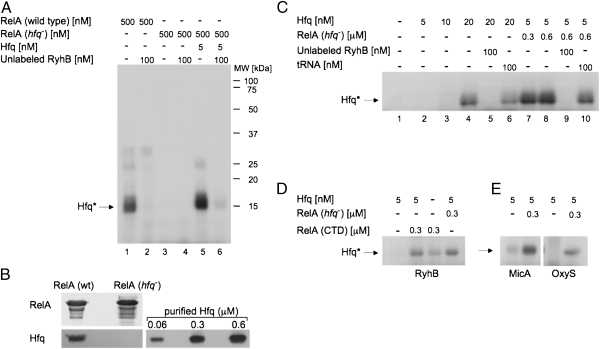
To learn about possible interactions between RelA, Hfq, and RyhB, we examined direct protein–RNA binding by cross-linking. In these experiments, reaction mixtures of labeled in vitro-synthesized RyhB and purified proteins were subjected to UV cross-linking. Thereafter, unbound, and thus unprotected, RNA residues were subjected to degradation by RNase A. Proteins covalently bound to labeled RNA residues were then detected in SDS gels. Intriguingly, we found that incubation of RyhB with purified RelA resulted in RyhB binding of a protein the size of Hfq. This observation led us to suspect that Hfq protein copurifies with RelA. Indeed, incubation of RyhB with RelA purified from an hfq-deficient mutant strain resulted in no binding of RyhB to that protein (Fig. 3A, lanes 1 and 3). Finally, Western analysis using Hfq-specific antibodies demonstrated the presence of Hfq protein in purified RelA (Fig. 3B). By measuring band intensity of Hfq in samples of purified RelA (Fig. 3B) we estimated that 240 μM of RelA harbor ∼0.24 μM of Hfq, and thus 500 nM of RelA carry 0.5 nM of Hfq. Accordingly, RyhB and RelA purified from hfq-deficient cells were incubated with 5 nM of Hfq. Fig. 3A (lane 5) shows binding of RyhB by Hfq, further demonstrating that the protein that binds RyhB RNA in RelA preparations is indeed Hfq.
Fig. 3.
RelA stimulates binding of Hfq to RyhB in vitro. (A) RyhB RNA binds Hfq in RelA preparations. UV cross-linking of in vitro-synthesized 32P-labeled RyhB incubated with RelA protein purified from wild-type or hfq− cells. Where indicated, Hfq and/or unlabeled RyhB were added to the incubation mixtures. Proteins covalently bound to residues of labeled RNA were detected in SDS/PAGE (after boiling). Hfq protein bound to residues of labeled RyhB (Hfq*). (B) Detection of Hfq protein in samples of purified RelA. Twenty micrograms of RelA protein purified from wild-type or hfq mutant and samples of purified Hfq were separated by 15% SDS/PAGE after boiling in loading buffer. The upper part of the gel was stained with Coomassie blue dye, and the lower part of the same gel was analyzed using α-Hfq (Western blotting). (C) RelA stimulates RyhB binding by otherwise ineffective amounts of Hfq. UV cross-linking of labeled RyhB incubated with different concentrations of Hfq in the presence or absence of RelA purified from hfq− cells. Where indicated, unlabeled RyhB or yeast tRNA was added. (D) The C-terminal domain (CTD) of RelA is sufficient in stimulating RyhB binding by Hfq. UV cross-linking of labeled RyhB incubated with Hfq in the absence or the presence of RelA-CTD or RelA purified from hfq− cells. (E) RelA stimulates Hfq binding of unrelated RNAs as well. UV cross-linking labeled MicA and OxyS RNAs incubated with Hfq in the presence or the absence of RelA purified from hfq− cells. The proteins were detected as in A.
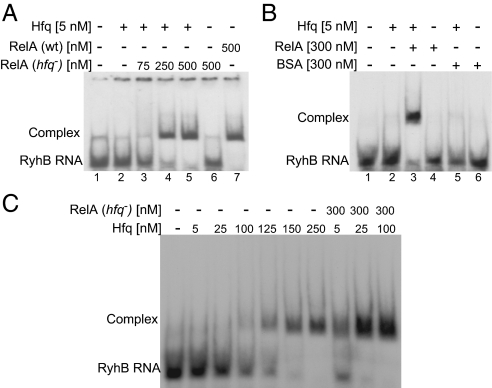
The observation that, in the presence of RelA, minute amounts of Hfq (0.5–5 nM) were sufficient to bind RyhB RNA prompted us to examine whether RelA stimulates RyhB binding via Hfq. Cross-linking experiments to detect binding of low amounts of Hfq to RyhB demonstrate that 5 and 10 nM of Hfq are insufficient to detect Hfq binding of RyhB (Fig. 3C, lanes 2 and 3). However, incubation of RyhB RNA with 5 nM Hfq in the presence of RelA purified from hfq− results in RyhB binding by Hfq (Fig. 3C, lanes 7 and 8). Addition of unlabeled RyhB significantly decreases the binding of the labeled RybB RNA, whereas addition of tRNA has little if any effect, indicating that Hfq binding to RyhB is specific (Fig. 3 A and C). Together, these results indicate that RelA augments RNA binding by Hfq. Likewise, gel mobility shift assays further demonstrate that RelA purified from wild-type cells leads to binding of RyhB, whereas RelA of hfq− shows no binding at all (Fig. 4A, lanes 6 and 7). Most importantly, these assays demonstrate that low amounts of Hfq are insufficient to bind RyhB, unless incubated in the presence of RelA (Fig. 4A, lanes 2–5). Quantitative analysis to estimate the apparent dissociation constant (Kapp) that represents the Hfq concentration at which 50% of labeled RyhB RNA is bound shows that Hfq binds RyhB with Kapp of ∼120 nM. In the presence of RelA, this value decreases dramatically, and RyhB binds Hfq with a much higher affinity (2.5 nM; Fig. 4C). BSA added as a control instead of RelA did not increase RyhB binding, indicating that RelA is a specific stimulator of Hfq activity (Fig. 4B, lanes 3 and 5). Moreover, UV cross-linking analysis of unrelated sRNAs such as OxyS and MicA shows a similar RelA-mediated stimulation of Hfq binding to RNAs as seen with RyhB (Fig. 3E). Thus, RelA renders Hfq a better binder of sRNAs.
Fig. 4.
RelA stimulates RNA binding by Hfq. (A) RelA stimulates complex formation of RyhB with otherwise ineffective amounts of Hfq. Gel mobility shift of labeled RyhB incubated with Hfq and/or RelA purified from wild-type or hfq− cells. Bound and unbound labeled RyhB is indicated. (B) RyhB binding by Hfq in the presence of BSA or RelA (purified from hfq−). (C) The apparent dissociation constant of Hfq-RyhB is ∼120 nM. The value of Kapp decreases dramatically in the presence of RelA, and RyhB binds Hfq with a much higher affinity (2.5 nM).
The RelA protein harbors two functional domains: the N-terminal domain that contains the synthetase activity of RelA and the C-terminal domain (CTD) that is involved in oligomerization of RelA (20). To learn whether the domain carrying the synthetase activity is necessary to stimulate RNA binding by Hfq, we examined binding of RNA by Hfq in the presence of the CTD of RelA. We found the CTD of RelA to be sufficient in promoting binding of RyhB by Hfq (Fig. 3D). Together, these results indicate that the synthetase domain is not involved in Hfq stimulation, whereas the CTD of RelA is sufficient to affect Hfq. It is interesting to note that, as opposed to the full-length RelA that copurifies with some Hfq protein, the CTD of RelA, although purified from wild-type cells, shows very little binding of RNA by Hfq (Fig. 3D). Possibly, the association of the truncated protein with ribosomes is less efficient than that of the full-length RelA (see below).
RelA Affects Binding of Hfq to RyhB in Vivo.
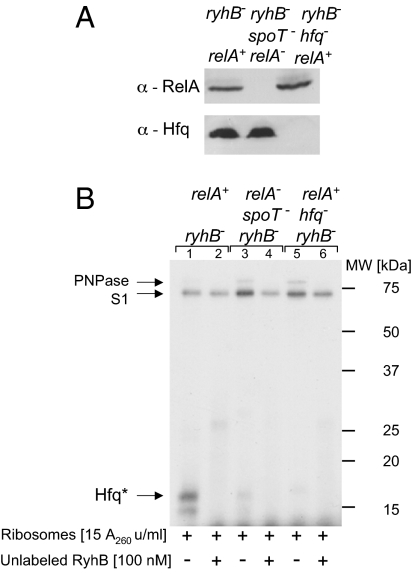
Because both proteins, RelA and Hfq, are thought to be associated with the translational machinery, we set out to analyze the effect of RelA on RyhB binding by Hfq in polysomal fractions extracted from wild-type and strains deficient in relA/spoT or hfq. Western blots of these ribosomes using antibodies raised against RelA and Hfq demonstrate that relA−spoT− ribosomes carry intact Hfq but no RelA and that hfq− ribosomes carry RelA and no Hfq, whereas wild-type ribosomes carry both proteins (Fig. 5A). UV cross-linking of labeled RyhB RNA incubated with wild-type ribosomes reveals binding of RyhB to Hfq as well as to two other proteins (Fig. 5B), which we identified by mass spectrometry analysis as ribosomal protein S1 and PNPase. We further confirmed the identity of PNPase using ribosomes extracted from a pnp mutant strain (Fig. S2). Inclusion of unlabeled RyhB in the reaction carrying wild-type ribosomes reduces Hfq binding of the labeled RyhB (Fig. 5B, lanes 1 and 2). Intriguingly, when RyhB is incubated with ribosomes extracted from cells deficient in relA/spoT, the sRNA does not bind Hfq although the protein is clearly present (Fig. 5B, lanes 3–4, and Fig. 5A). The dramatic decrease in RyhB binding by Hfq in ribosomes lacking RelA confirms our in vitro data showing that RelA promotes binding by Hfq. Similar results were obtained with ribosomes purified from cultures grown under conditions of iron starvation, suggesting that the rate of Hfq binding in relA+ is not affected by the iron starvation stress (Fig. S2). Adding RelA to the ribosomal fraction after its extraction failed to restore binding by Hfq (Fig. S3).
Fig. 5.
RelA affects Hfq binding to RyhB in polysomal fractions. (A) Detection of RelA and Hfq in polysomal fractions extracted from the indicated strains using α-Hfq or α-RelA (using Western blotting). (B) RyhB binding by Hfq takes place in ribosomes of relA+ only. UV cross-linking of labeled RyhB incubated with ribosomes extracted from the indicated strains at 22 °C for 15 min is shown. The proteins were analyzed by SDS/PAGE as in Fig. 3A. Arrows indicate Hfq, S1, and PNPase proteins. Hfq protein bound to residues of labeled RyhB (Hfq*).
RelA Promotes Oligomerization of Hfq.
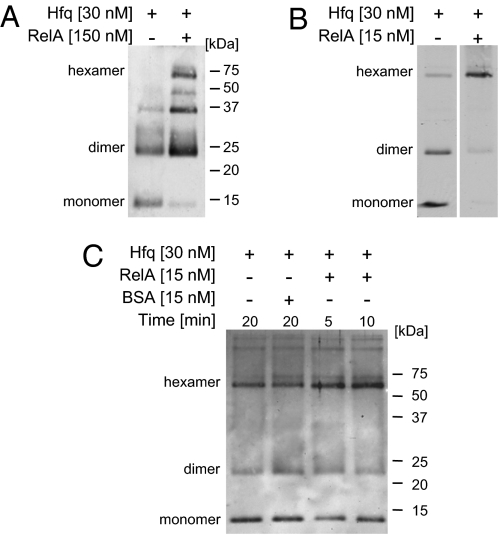
Hfq functions as a homo-hexameric ring in RNA binding. To learn about the effect of RelA on Hfq, we compared the oligomerization status of Hfq protein in the presence and in the absence of RelA. The proteins, incubated and cross-linked using glutaraldehyde, were analyzed by SDS/PAGE. Incubation of Hfq alone (30 nM at 23 °C) reveals the presence of a monomer and two multimeric forms of ∼24 and 35 kDa (Fig. 6A). However, incubation of Hfq in the presence of high concentrations of RelA (150 nM) results in the appearance of Hfq hexamers and in an increase in the levels of other multimeric forms, whereas the levels of the monomer decrease dramatically. Moreover, incubation of Hfq (30 nM on ice) with a much lower concentration of RelA (15 nM), to change the ratio between RelA and Hfq in favor of Hfq, results in a dramatic decrease in the levels of both forms, Hfq monomers and dimers, whereas the levels of the hexamers increase significantly (Fig. 6B). In contrast, incubation of Hfq with BSA has no effect on the pattern of Hfq multimers (Fig. 6C). Likewise, incubation of an increased concentration of Hfq (100 nM) without RelA barely impacted the pattern of multimerization (Fig. S4). Together, these results indicate that RelA is a required functional partner of Hfq, stimulating its multimerization. We propose that, by stimulating the formation of the Hfq active structure, RelA affects the binding of RNA by Hfq.
Fig. 6.
RelA enhances Hfq multimerization. (A) [RelA]>[Hfq]. Hfq was incubated with or without RelA for 15 min at 23 °C, and then the proteins were cross-linked using 0.4% freshly diluted glutaraldehyde for 5 min. Cross-linking was stopped with 200 mM fresh glycine, and the proteins were boiled in loading buffer and analyzed on 15% SDS/PAGE. Western blotting using α-Hfq. (B) [RelA]<[Hfq]. Hfq was preincubated for 15 min in binding buffer on ice. Thereafter, RelA was added where indicated, and the mixtures were incubated for another 20 min on ice. The proteins were cross-linked for 1 min, treated, and analyzed as described in A. (C) BSA has no effect on Hfq multimerization. The proteins Hfq, RelA, and BSA were incubated on ice for the indicated times. The proteins were cross-linked for 1 min, treated, and analyzed as described in A.
Discussion
In this study, we show that expression of RyhB inhibits bacterial growth in a relA-dependent manner. The observation that a subset of RyhB targets requires RelA suggests that the growth inhibition phenotype results from an effect of RelA on RyhB's control of its target genes. As previously observed with a number of systems that use growth inhibition to prevail over stress conditions, so too the RyhB-RelA–dependent growth inhibition is of benefit once rich medium is available again; when diluted further in rich medium, the recovery of the arrested wild-type cells is faster than that of ryhB-deficient cells, suggesting that RelA plays an important role in RyhB-mediated adaptation to iron starvation. Similar results were obtained with RyhB and the island-encoded ortholog IsrE of Salmonella (21). Under conditions of iron depletion, the growth of wild-type Salmonella was inhibited, whereas in a strain lacking both ryhB and isrE, no growth inhibition was observed (21).
The effect of RelA on RyhB control of its targets is indirect and mediated by Hfq. We propose that RelA, by enhancing multimerization of Hfq oligomers, increases the level of the active form of Hfq, thereby enabling binding of RNAs, whose intrinsic affinities for this protein are low. Consistent with this argument, we found that the relA-dependent RyhB target mRNAs sodA, nuoA, and fdoG exhibit a twofold lower affinity for Hfq than the RelA-independent target sodB (Fig. S5). These observations support the emerging notion (22–25) that Hfq levels are not excessively high and that under some conditions Hfq becomes scarce. We measured Hfq in our relA+ and relA− cells and found the amounts to be ∼5,000 and ∼4,000 hexamers per cell, respectively. Thus, the estimated concentration of Hfq in those cells is approximately 8 and 6.5 μM, respectively (Fig. S6). Although the concentration of Hfq seems high, this value is far from indicative of how much of the Hfq protein is available for binding because this protein engages with the largest group of RNA regulators in bacteria, the small base-pairing RNAs. The concept that highly expressed sRNAs might titrate Hfq was originally proposed for OxyS, an sRNA that was observed to down-regulate RpoS levels by decreasing the levels of available Hfq (23). Later studies showed that overexpression of the Hfq-dependent ArcZ sRNA dramatically changed the profile of Hfq-bound RNAs in vivo; the total number of Hfq-bound mRNAs was significantly reduced upon increased expression of ArcZ (22). Likewise, it was shown that transcription of a single sRNA and/or a single target mRNA can result in the sequestration of Hfq and thus in Hfq depletion (25). Most recently, Moon and Gottesman (24) showed that Hfq is limiting and that sRNAs can compete with each other for Hfq to different extents, changing the outcome of gene regulation. We show here that RelA enables binding of RNAs by otherwise ineffective amounts of Hfq. Moreover, we show that Hfq associated with polysomes extracted from wild-type and the relA mutant exhibits differential binding. This strongly indicates that binding by Hfq is affected not only by the total concentration of Hfq in the cytoplasm, but also by the abundance of unoccupied Hfq in specific environments. For example, the association of Hfq with the translational machinery or with the RNA destabilization machinery is just as critical. These results point to another level of regulation in which Hfq scarcity plays a role in modulating gene expression by sRNAs.
How RelA stimulates the formation of the Hfq-active structure is yet to be resolved. The RelA protein of E. coli is a ribosome-dependent (p)ppGpp synthetase that is activated in response to amino acid starvation. We show that the RelA-CTD protein lacking the synthetase activity is sufficient to promote binding of RNAs by Hfq. Moreover, the in vivo assays were carried out in rich medium, and in vitro binding and multimerization assays were carried out in the absence of ATP; thus, neither ppGpp nor RelA synthetase activity are involved in Hfq multimerization. Furthermore, the ratio between RelA and unbound Hfq that is available for RNA binding is unknown. Therefore, whether RelA stimulates multimerization of Hfq in a stoichiometric way or catalytically is not clear. Studies of the mechanism of action of RelA have shown that binding of RelA to the ribosome is predominantly influenced by mRNA and that its release correlates with ppGpp synthesis (26). On the basis of that data, the authors proposed that RelA hops between blocked ribosomes, providing an explanation for how low intracellular concentrations of RelA (1/200 ribosomes) can accomplish regulation. In this context, it is interesting to note the report by Peregrín-Alvarez et al. (27) in which integration of experimental and computational interaction data on E. coli proteins identified functional interactions between Hfq and spoT; between relA and spoT; and between spoT and some components of the ribosome (27). Still, the mechanistic details of how the low abundance RelA protein affects multimerization of Hfq require elucidation.
The finding that relA-deficient polysomal fractions carrying the Hfq protein fail to enable binding of RNA by Hfq is intriguing. Ribosome-associated molecular chaperones are thought to be the first line of defense against protein aggregation as translating polypeptides emerge from the ribosome (28). By analogy, it is possible that the RelA-Hfq system is an antecedent mechanism aimed at controlling translation of newly synthesized incoming mRNAs.
Materials and Methods
UV Cross-linking Assays.
Binding reactions of 20 μL contained 1 nM labeled RyhB, Hfq, and/or RelA at the indicated concentrations and binding buffer C (SI Materials and Methods). The reactions were incubated at 22 °C for 15 min and then transferred to a chilled (4 °C) metal block and irradiated with 254 nm light (20,000 μJ/cm2) for 5 min, using the UV Stratalinker model 1800/2400 instrument (Stratagene). Samples were then transferred to tubes and treated with 100 mg/mL RNase A for 30 min at 37 °C. The proteins labeled through RNA residues were separated by 12 or 18% SDS/PAGE. UV cross-linking assays with polysomal fractions were carried out essentially as described above except that the binding mixtures contained 15 OD260 ribosome units/mL and binding buffer A (SI Materials and Methods). All protein samples were boiled before SDS/PAGE analysis.
Extraction of Polysomal Fraction.
Cell cultures were grown in 2× Yeast Tryptone medium to OD600 of 2.0, and low-salt washed ribosomes were extracted as described (29). The presence of RelA and Hfq in these fractions was examined using rabbit anti-RelA antibody (kindly provided by Gad Glaser, Hebrew University, Jerusalem, Israel) and by rabbit anti-Hfq antibody (generated against the CTD of Hfq by Genmed Synthesis).
Supplementary Material
Acknowledgments
We thank Gad Glaser for generously providing protocols, strains, RelA, RelA-CTD, and RelA antibodies; Gisela Storz for strains, Hfq antibodies, and Hfq; and Giani Deho for the KG202 strain. We appreciate the technical assistance of Simi Koby, Gilly Padalon-Brauch, Gal Nechooshtan, and Yaniv Baum. This work was supported by Grants 499/06 and 911/09 from the Israel Science Foundation, founded by the Israel Academy of Sciences and Humanities, and Grant 3-00000-6010 from The Ministry of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113113109/-/DCSupplemental.
References
- 1.Argaman L, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 2.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geissmann TA, Touati D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Bläsi U. Interaction of the RNA chaperone Hfq with mRNAs: Direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 8.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang A, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 12.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 15.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vecerek B, Beich-Frandsen M, Resch A, Bläsi U. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 2010;38:1284–1293. doi: 10.1093/nar/gkp1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabhi M, et al. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–2816. doi: 10.1038/emboj.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- 19.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 21.Padalon-Brauch G, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papenfort K, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang A, et al. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 27.Peregrín-Alvarez JM, Xiong X, Su C, Parkinson J. The modular organization of protein interactions in Escherichia coli. PLOS Comput Biol. 2009;5:e1000523. doi: 10.1371/journal.pcbi.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: The first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6:157–162. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 29.Krohn M, Wagner R. A procedure for the rapid preparation of guanosine tetraphosphate (ppGpp) from Escherichia coli ribosomes. Anal Biochem. 1995;225:188–190. doi: 10.1006/abio.1995.1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.