Abstract
Wild-type, full-length (40- and 42-residue) amyloid β-peptide (Aβ) fibrils have been shown by a variety of magnetic resonance techniques to contain cross-β structures in which the β-sheets have an in-register parallel supramolecular organization. In contrast, recent studies of fibrils formed in vitro by the Asp23-to-Asn mutant of 40-residue Aβ (D23N-Aβ1–40), which is associated with early onset neurodegeneration, indicate that D23N-Aβ1–40 fibrils can contain either parallel or antiparallel β-sheets. We report a protocol for producing structurally pure antiparallel D23N-Aβ1–40 fibril samples and a series of solid state nuclear magnetic resonance and electron microscopy measurements that lead to a specific model for the antiparallel D23N-Aβ1–40 fibril structure. This model reveals how both parallel and antiparallel cross-β structures can be constructed from similar peptide monomer conformations and stabilized by similar sets of interactions, primarily hydrophobic in nature. We find that antiparallel D23N-Aβ1–40 fibrils are thermodynamically metastable with respect to conversion to parallel structures, propagate less efficiently than parallel fibrils in seeded fibril growth, and therefore must nucleate more efficiently than parallel fibrils in order to be observable. Experiments in neuronal cell cultures indicate that both antiparallel and parallel D23N-Aβ1–40 fibrils are cytotoxic. Thus, our antiparallel D23N-Aβ1–40 fibril model represents a specific “toxic intermediate” in the aggregation process of a disease-associated Aβ mutant.
Keywords: Alzheimer’s disease, amyloid structure, solid state NMR
Alzheimer’s disease (AD) is thought to be a consequence of aggregation of the amyloid β-peptide (Aβ) into amyloid fibrils or related assemblies in brain tissue. Numerous structural studies of Aβ fibrils and oligomers have been reported (1–17), motivated by the dual goals of contributing to preventive and therapeutic approaches to AD and of elucidating the biophysical basis for amyloid formation. A defining structural characteristic of an amyloid fibril is the presence of a cross-β motif; i.e., a ribbon-like β-sheet running the length of the fibril, with β-strands approximately perpendicular to and interstrand hydrogen bonds approximately parallel to the long fibril axis. Studies of the 40-residue and 42-residue forms of Aβ (Aβ1–40 and Aβ1–42) have shown that these peptides can form multiple distinct fibril structures (11, 18, 19), but that the cross-β motifs within wild-type (WT) Aβ fibrils are invariably comprised of in-register parallel β-sheets (1, 5, 6, 8–10, 13, 14). Parallel β-sheets have also been found in Aβ1–40 oligomers (4) and in amyloid fibrils formed by amylin (20, 21), α-synuclein (22), β2-microglobulin (23, 24), prion proteins of yeast (25–27) and mammalian PrP (28, 29); in contrast, antiparallel β-sheets have been found in fibrils formed by Aβ fragments with 15 or fewer residues (30–32) and in amyloid-like crystals of certain Aβ fragments (33). These observations suggest that in-register parallel β-sheets might be a universal feature of amyloid structures in which the polypeptide chain contains more than one β-strand-forming segment.
Surprisingly, recent solid state nuclear magnetic resonance (SSNMR) measurements on polymorphic samples of fibrils formed by the Asp23-to-Asn, or Iowa, mutant of Aβ (D23N-Aβ1–40) have revealed anomalous β-sheet structures (15). The SSNMR data can be explained if a large fraction of the D23N-Aβ1–40 fibrils contain antiparallel β-sheets. This mutation leads to familial, early onset neurodegeneration involving extensive cerebral amyloid angiopathy (34). It is therefore possible that antiparallel β-sheet structures exert distinct pathogenic effects.
In this paper, we describe a procedure for producing structurally homogeneous D23N-Aβ1–40 fibrils that contain antiparallel β-sheets, based on differences in fibril extension rates between parallel and antiparallel structures. Structural homogeneity then permits a series of SSNMR and electron microscopy measurements that lead to a detailed structural model for the antiparallel cross-β motif within D23N-Aβ1–40 fibrils. The model explains why both parallel and antiparallel structures are possible in D23N-Aβ1–40 fibrils and suggests that antiparallel cross-β motifs may also exist in other cases. We present evidence that antiparallel β-sheet structures nucleate efficiently but are metastable with respect to conversion to parallel structures, as well as experiments showing that both antiparallel and parallel D23N-Aβ1–40 fibrils are toxic in neuronal cell cultures. The antiparallel D23N-Aβ1–40 structure may therefore be a considered a “toxic intermediate” along the path to thermodynamically stable amyloid fibrils (which we also find to be neurotoxic).
Results
Purification of Antiparallel β-sheet Fibrils by Seeding and Filtration.
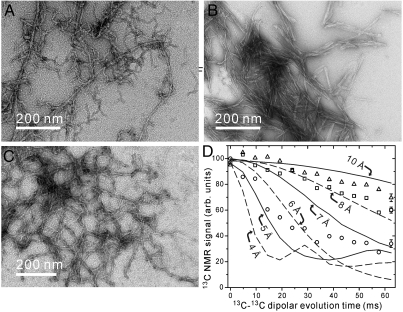
Peptide synthesis and fibril formation are described in Supporting Information. Fig. 1A shows a transmission electron microscope (TEM) image of fibrils formed from initially monomeric D23N-Aβ1–40 at 6 °C in incubation buffer (10 mM sodium phosphate, pH 7.4, 0.01% NaN3). This “parent fibril” sample is clearly heterogeneous, consistent with earlier results for de novo D23N-Aβ1–40 fibril preparations that indicated mixtures of parallel and antiparallel β-sheet structures (15, 16). Three approaches to purification of an antiparallel β-sheet structure were attempted. First, parent fibrils were subjected to repeated sonication and incubation, allowing the heterogeneous mixture to evolve towards the thermodynamically preferred structure at 6 °C (see thioflavin T fluorescence data in Fig. S1A). As shown in Fig. 1B, this treatment resulted in a more morphologically homogeneous state (henceforth called TE fibrils, for thermodynamic equilibrium). As shown in Fig. 1D, measurements of intermolecular 13C-13C dipole-dipole couplings showed a reduction in intermolecular distances among A21 methyl carbons in TE fibrils towards the 4.8 Å value expected in an in-register parallel β-sheet. Second, eight generations of seeded growth were performed at 6 °C in incubation buffer, starting with parent fibrils and following our previously described seeding protocol with 3 hr incubation periods in each generation (16). Repeated seeded growth also resulted in morphologically homogeneous fibrils, but again with parallel β-sheet structures (Fig. S1 B and C).
Fig. 1.
(A–C) TEM images of negatively stained D23N-Aβ1–40 fibrils. Images are shown for parent fibrils (A), fibrils in their thermodynamic equilibrium (TE) state after 500 h incubation with intermittent sonication (B), and fibrils prepared by two generations of the seeding/filtration (SFg2) protocol (C). The TE and SFg2 protocols select different fibril morphologies from the polymorphic parent sample. (D) Measurements of intermolecular dipole-dipole couplings among 13C labels at A21 methyl carbons in parent (squares), TE (circles) and SFg2 (triangles) fibrils, obtained with the PITHIRDS-CT SSNMR technique. Error bars represent uncertainty due to root-mean-squared noise in the 13C NMR spectra. Dashed and solid curves are simulated data for linear chains of 13C nuclei with the indicated spacings. Average intermolecular 13C-13C distances decrease in TE fibrils and increase in SFg2 fibrils, relative to distances in the parent fibrils.
Finally, a “seeding/filtration” protocol was devised that takes advantage of differences in the fibril extension rates of parallel and antiparallel structures. In this protocol, a sonicated aliquot of fibrils (i.e., seeds) was added to a monomeric D23N-Aβ1–40 solution, fibrils were allowed to grow in incubation buffer for 3 h at 6 °C, the resulting mixture was passed twice through 0.22 μm filters, additional monomeric D23N-Aβ1–40 was added to the filtrate, and the fibrils were allowed to grow at 6 °C for 24 h. This protocol selectively suppresses fibril structures that extend rapidly during the 3 h incubation period and hence has the opposite effect of standard seeded growth protocols. The seeding/filtration protocol was applied twice, starting with parent fibrils, to generate the fibrils shown in Fig. 1C (henceforth called SFg2 fibrils, for seeding/filtration generation 2). SFg2 fibrils have greater curvature than TE fibrils and apparently derive from the more curved fibrils in the parent sample. As shown in Fig. 1D, intermolecular distances among A21 methyl carbons in SFg2 fibrils are longer than the average values in the parent sample, consistent with purely antiparallel β-sheet structures in SFg2 fibrils.
Structural Restraints from SSNMR.
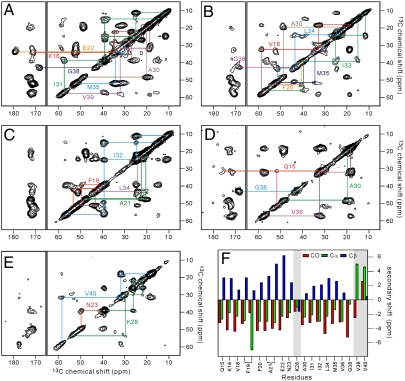
SFg2 fibril samples were prepared with uniform 15N,13C-labeling of certain residues, chosen to permit SSNMR measurements that probe various aspects of the molecular structure. Labeled residues were: K16, E22, A30, I31, M35, G38, and V39 (sample A); V18, F20, A30, I32, L34, M35, and G38 (sample B); F19, A21, L34, and I32 (sample C); Q15, A30, V36, and G38 (sample D); and N23, K28, and V40 (sample E). Fig. 2 shows two-dimensional (2D) 13C-13C NMR spectra of the five SFg2 samples, obtained with 2.4 ms finite-pulse radio-frequency-driven recoupling (fpRFDR) mixing periods (35, 36) so that strong crosspeaks connect chemical shifts of directly bonded, 13C-labeled sites. Except for F19 and A21, all residues show a single set of chemical shifts, consistent among samples A-E, indicating that all D23N-Aβ1–40 molecules have similar conformations and structural environments and that sample preparation is reproducible. F19 and A21 show two sets of chemical shifts with comparable intensities (Fig. 2C), possibly indicating comparable populations of two F19 side-chain conformations. The 13C linewidths of 1.6–2.3 ppm full-width-at-half-maximum are larger than in spectra of WT-Aβ1–40 and D23N-Aβ1–40 fibrils that contain in-register parallel β-sheets (13, 14, 16), reflecting a greater degree of internal disorder in SFg2 fibrils that may be associated with their greater curvature. Fig. 2F shows secondary shifts determined by comparison of 13C chemical shifts in Figs. 2 A–E with corresponding random coil shifts (37). Positive secondary shifts for Cβ and negative secondary shifts for CO and Cα sites are observed in residues 15–23 and 30–36, indicating β-strand conformations in these segments. Non-β-strand secondary shifts are observed at K28, V39, and V40. 13C chemical shifts and backbone ϕ and ψ dihedral angles predicted from these shifts by the TALOS+ program (38) are summarized in Table S1.
Fig. 2.
(A–E) 2D 13C-13C correlation spectra of SFg2 fibril samples A–E, with color-coded chemical shift assignment paths. (F) Secondary 13C chemical shifts obtained from the 2D 13C-13C spectra. Shaded areas highlight residues with non-β-strand secondary shifts.
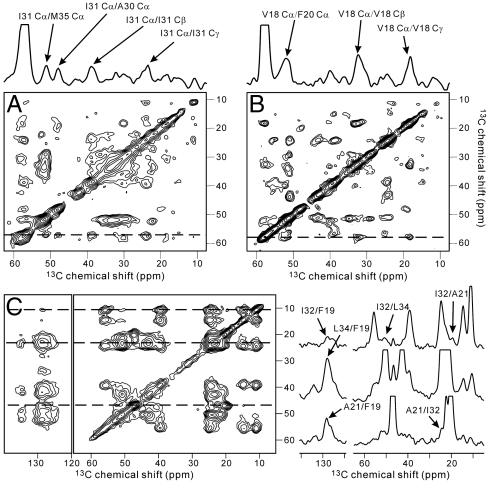
Restraints on the interstrand alignment within the antiparallel β-sheets were obtained from 2D proton-mediated 13C-13C exchange (2D CHHC) spectra. In previous studies of amyloid structures (31, 39), strong nonsequential Cα/Cα crosspeaks in 2D CHHC spectra have been shown to arise from the short interstrand Hα/Hα distances (ideally 2.2 Å) for residue pairs that align directly opposite one another in antiparallel β-sheets, allowing unambiguous determination of the registry of interstrand hydrogen bonding. Nonsequential Cα/Cα crosspeaks are not observed in 2D CHHC spectra of parallel β-sheets, regardless of registry (39), since no short interstrand Hα/Hα distances occur. Figs. 3 A and B show 2D CHHC spectra of samples A and B, in which I31 Cα/M35 Cα and V18 Cα/F20 Cα crosspeaks appear, comparable in intensity to intraresidue Cα/Cβ crosspeaks. The 2D CHHC spectrum of sample C (Fig. S2A) shows I32 Cα/L34 Cα crosspeaks. The ratio of the I31 Cα/M35 Cα crosspeak volume to the A30 Cα/I31 Cα crosspeak volume in Fig. 3A is 1.3 ± 0.2. I31 Cα/M35 Cα crosspeaks (but not A30 Cα/I31 Cα crosspeaks) are suppressed by isotopic dilution (Fig. S2 C and D), supporting the attribution of nonsequential crosspeaks to interstrand hydrogen bonding. These results indicate that both β-strand segments identified from secondary shifts form antiparallel β-sheets, with 17 + k↔21 - k registry in the sheet formed by residues 15–23 and 31 + k↔35 - k registry in the sheet formed by residues 30–36 (with M + k↔N - k denoting interstrand hydrogen bonding between residues M + k and N - k for integer k). The absence of A30 Cα/V36 Cα crosspeaks in the 2D CHHC spectrum of sample D (Fig. S2B) indicates that residues 31 and 35 define the edges of the C-terminal β-sheet.
Fig. 3.
(A, B) 2D CHHC spectra of SFg2 fibril samples A and B, with 1D slices through 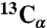 chemical shifts of I31 and V18 (dashed lines). Strong I31 Cα/M35 Cα and V18 Cα/F20 Cα crosspeaks indicate antiparallel β-sheets. (C) 2D RAD spectrum of SFg2 fibril sample C with 1D slices at the I32
chemical shifts of I31 and V18 (dashed lines). Strong I31 Cα/M35 Cα and V18 Cα/F20 Cα crosspeaks indicate antiparallel β-sheets. (C) 2D RAD spectrum of SFg2 fibril sample C with 1D slices at the I32 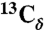 , L34
, L34 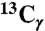 , and A21
, and A21 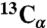 chemical shifts (dashed lines). Arrows indicate interresidue crosspeaks.
chemical shifts (dashed lines). Arrows indicate interresidue crosspeaks.
Contacts among amino acid side chains were identified from 2D 13C-13C exchange spectra obtained with 500 ms radio-frequency-assisted diffusion (RAD) mixing periods (40, 41). Nonsequential interresidue crosspeaks in such spectra generally indicate interatomic distances less than 8 Å (12). The 2D RAD spectrum of sample C (Fig. 3C) shows crosspeaks between aromatic signals of F19 and aliphatic signals of A21, I32, and L34, as well as crosspeaks among aliphatic sites of A21, I32, and L34. F19/I32 and F19/L34 crosspeaks indicate that D23N-Aβ1–40 adopts a “U-shaped” conformation that brings the N- and C-terminal β-sheets in contact with one another, as observed in WT-Aβ1–40 fibrils that contain in-register parallel β-sheets (12, 13). 2D RAD spectra of samples A, B, and E show crosspeaks between the E22 side-chain carboxyl signal and K16 aliphatic signals, between the G38 Cα signal and the A30 methyl signal, and between aliphatic signals of K28 and methyl signals of V40 (Fig. S3). K16/E22 crosspeaks are consistent with the 17 + k↔21 - k registry of the N-terminal β-sheet indicated by the 2D CHHC spectrum of sample B (Fig. 3B) and by earlier measurements of 15N-13C dipole-dipole couplings between L17 and A21 in D23N-Aβ1–40 fibrils (15). A30/G38 and K28/V40 crosspeaks indicate that the C-terminal residues of a given molecule are proximal to the residues between the two β-strands (i.e., the “bend” segment) of neighboring molecules.
The 17 + k↔21 - k registry of the N-terminal β-sheet suggests the possibility of favorable electrostatic interactions between oppositely charged K16 amino and E22 carboxylate side-chain groups. However, measurements of 15N-13C dipole-dipole couplings between K16 Nξ and E22 Cδ, using the frequency-selective rotational echo double resonance (REDOR) technique (42), yielded a negative result (Fig. S4A). Absence of direct K16-E22 salt bridges is attributable to exposure of the K16 and E22 side chains to solvent outside the fibril core. Similarly, direct pairing of oppositely charged K28 amino and C-terminal V40 carboxylate groups (15) was not observed (Fig. S4B).
Additional intermolecular structural restraints come from PITHIRDS-CT measurements that indicate 5.5 ± 0.5 Å intermolecular distances among G33 and F19 carbonyl carbons but relatively long intermolecular distances among A21 and A30 methyl carbons (Fig. S4D), and REDOR measurements that indicate 4.7 ± 0.3 Å distances between A30 methyl carbons and V36 amide nitrogens and between A21 methyl carbons and L17 amide nitrogens (Fig. S4E).
Electron Microscopy Indicates a Single Cross-β Unit.
Electron diffraction measurements on unstained SFg2 fibrils show the strong 4.8 Å feature that arises from the spacing between β-strands in a cross-β motif (Fig. S5 A and B). A histogram of mass-per-length (MPL) values of individual fibrils, obtained with the tilted-beam TEM (TB-TEM) technique (43), peaks at 10 ± 2 kDa/nm (Fig. S5 C and D). Since a single layer of D23N-Aβ1–40 molecules (4.3 kDa mass) in a cross-β motif would have MPL = 9.0 kDa/nm, the MPL histogram implies that individual SFg2 fibrils contain a single cross-β unit, in turn supporting our interpretation that interresidue contacts identified from SSNMR data are within one cross-β unit. We attribute the large-MPL tail of the histogram (Fig. S5D) to bundles of fibrils, which are not readily distinguished from single fibrils in the TB-TEM images themselves. In contrast, TB-TEM and other MPL measurements on WT-Aβ1–40 fibrils yield MPL histograms peaked at 18 kDa/nm and 27 kDa/nm (43), consistent with structures comprised of two and three cross-β units (11–13). The lower MPL of SFg2 fibrils may contribute to their greater curvature.
Double-Layered Antiparallel β-sheet Model.
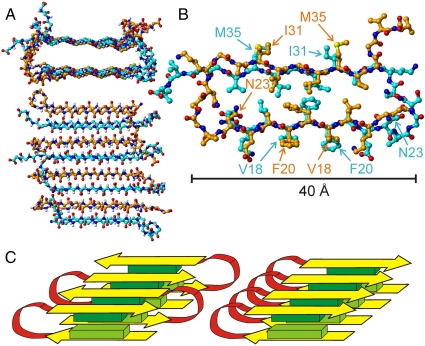
Atomic coordinates consistent with the experimental data were generated with Xplor-NIH (44). Details of the structural modeling procedure are given in Supporting Information. Residues 1–14 were omitted, as no restraints were obtained for these residues and these residues are likely to be partially or fully disordered (5, 7, 8, 11–13). Eight copies of the peptide were included. Experimentally based restraints (summarized in Table S2) included: (i) dihedral angle restraints for residues 16–23, 30–36, and 39; (ii) approximate distance restraints connecting F19 and I32, F19 and L34, A21 and I32, I32 and L34, A30 and G38, K28 and V40, and K16 and E22; (iii) hydrogen bonding restraints for the V18-F20, I31-M35, and I32-L34 pairs identified in 2D CHHC spectra; (iv) quantitative intermolecular F19-F19, G33-G33, A30-V36, and A21-L17 distance restraints derived from the PITHIRDS-CT and REDOR data. In addition, backbone N-H bond vectors of residues 17–21 and 31–35 were restrained to align approximately with the long axis of the fibril in order to enforce a cross-β motif. For the ten final structures (superimposed in Fig. S6), the root-mean-squared deviation among coordinates in residues 16–34 of the central pair of molecules in the cross-β octamer was 0.86 Å for backbone atoms and 1.50 Å for non-hydrogen atoms.
The resulting model has sufficient precision to reveal the principal aspects of secondary, tertiary, and quaternary structure and to suggest the principal interactions that stabilize the antiparallel cross-β motif within D23N-Aβ1–40 fibrils. As shown in Fig. 4, side chains of L17, F19, A21, A30, I32, L34, and V36 create a purely hydrophobic central core, running the entire length of the fibril. Together with the backbone hydrogen bonds of residues 17–21 and 31–35, hydrophobic interactions in the core are apparently the most important stabilizing interactions. Charged side chains of K16, E22, and K28, as well as the C-terminal carboxylate group of V40, are exposed to solvent outside of the core. As discussed above, we did not find evidence for close electrostatic interactions among these charges (Fig. S4 A and B). Residues 37–40 are disordered in our antiparallel D23N-Aβ1–40 fibril model. In contrast, these residues participate in the parallel β-sheet structure and in contacts between cross-β units in WT-Aβ1–40 fibrils (11–13, 17), although possibly with reduced order (7, 19). Disorder in residues 37–40 in antiparallel D23N-Aβ1–40 fibrils may be a consequence of the fact that these fibrils are comprised of only one cross-β unit. The relatively small number of ordered residues in antiparallel D23N-Aβ1–40 fibrils (about 50% of the amino acid sequence, versus 75% in parallel WT-Aβ1–40 fibrils) may contribute to the thermodynamic preference for parallel structures, as discussed further below.
Fig. 4.
Structure of residues 15–40 in antiparallel D23N-Aβ1–40 (SFg2) fibrils. (A) Backbone atoms of the full eight-molecule system used for structure development, viewed parallel (Top) and perpendicular (Bottom) to the fibril axis. Carbon atoms are colored orange or cyan in molecules with alternating orientations within the antiparallel cross-β motif. This is one example of 10 similar structures determined from the experimental restraints (PDB file 2LNQ). See Fig. S6 for superpositions of the 10 structures. (B) Central pair of molecules, showing all non-hydrogen atoms and viewed parallel to the fibril axis. (C) Schematic representation of the double-layered antiparallel cross-β motif (Left), showing β-strands in yellow, the intervening loop in red, and groups of hydrophobic side chains as light green and dark green blocks. A schematic representation of the double-layered parallel cross-β motif identified in earlier studies of WT-Aβ1–40 fibrils (Right) is shown for comparison.
Cytotoxicity of D23N-Aβ1–40 Fibrils.
We investigated the toxicity of D23N-Aβ1–40 fibrils in cultures of primary rat embryonic hippocampal neurons, using direct measurements of neuronal survival following the addition of fibrils to the culture medium as previously described (11, 45). Fibrils with both antiparallel and parallel β-sheet structures exhibit significant toxicities at fibrillized peptide concentrations of 1 μM or greater (Fig. S7). Within the statistical limits of our measurements, the toxicities of antiparallel and parallel fibrils are indistinguishable from one another.
Discussion
Generality of the Double-Layered Antiparallel Cross-β Motif.
Antzutkin et al. (8) pointed out that alignment of the hydrophobic segments of Aβ1–40 in an in-register parallel (but not antiparallel) structure maximizes favorable hydrophobic interactions within a planar β-sheet, suggesting that cross-β motifs formed by any sequence with multiple hydrophobic segments would be comprised of in-register parallel β-sheets. For amyloid fibrils formed by Gln-and Asn-rich sequences, where “polar zipper” interactions are believed to be important stabilizing interactions (46, 47), Chan et al. (25) pointed out that polar zipper interactions would also be maximized in an in-register parallel β-sheet, regardless of the ordering of Gln and Asn residues. Only electrostatic repulsions obviously destabilize an in-register parallel structure, but these can be reduced or eliminated by placing charged groups outside the fibril core or by pairing negatively and positively charged groups.
Experimental results for Aβ1–40 (6, 8, 10, 11, 13, 14), Aβ1–42 (5, 6, 9), Aβ10–35 (1, 9), α-synuclein (22), amylin (20, 21), β2-microglobulin (23, 24), Ure2p (27), Sup35 (26), and PrP (28, 29) fibrils support the idea that in-register parallel β-sheets might be a universal structural feature of amyloid fibrils when they are formed by full-length peptides and proteins. For HET-s fibrils, a “quasi–in-register” parallel β-sheet structure was also found, with homologous protein segments forming a parallel cross-β motif (48). Although antiparallel β-sheets were found in Aβ16–22 (32, 39), Aβ34–42 (30), and Aβ11–25 (31) fibrils, these peptides contain only a single hydrophobic segment. All hydrophobic groups can then interact within a planar antiparallel β-sheet (32, 39), which is apparently favored by electrostatic interactions between oppositely charged groups at the N-and C-termini. Antiparallel β-sheets in certain cross-β microcrystals (33) can be explained similarly.
The existence of antiparallel β-sheets in D23N-Aβ1–40 fibrils necessitates a revision of our understanding of the factors that determine β-sheet organization, particularly in fibrils that are stabilized by hydrophobic interactions. The argument for parallel structures put forth by Antzutkin et al. (8) assumes that the cross-β motif is purely two-dimensional (i.e., lies in a single plane). If the cross-β motif develops in three dimensions, through alternation of β-strand segments with bends or loops within a multilayered cross-β unit, then both parallel and antiparallel structures can produce favorable alignments of hydrophobic segments within each β-sheet layer, as well as favorable hydrophobic contacts between layers. Fig. 4C illustrates this point. The double-layered antiparallel cross-β motif that we have identified in D23N-Aβ1–40 fibrils may therefore exist in amyloid fibrils that are formed by other peptides with similar distributions of hydrophobic residues. In principle, antiparallel cross-β motifs with more than two layers could be formed by sequences that contain more than two hydrophobic β-strands.
The 17 + k↔21 - k registry of the N-terminal β-sheet in SFg2 fibrils allows all L17, F19, and A21 side chains to be in the central core and all K16 and E22 side chains to be solvent-exposed. Similarly, the 31 + k↔35 - k registry of the C-terminal β-sheet allows all A30, I32, L34, and V36 side chains to be in the central core. In contrast, M + k↔N - k registries with odd values of N-M would produce lower-symmetry structures (i.e., two molecules in the asymmetric unit, rather than one), for example with F19 side chains alternately in and out of the central core. The experimentally determined registries for SFg2 fibrils also maximize the intermolecular alignment of hydrophobic L17-A21 segments within the N-terminal β-sheet and hydrophobic A30-V36 segments within the C-terminal β-sheet. Thus, the structure determined from our SSNMR data appears to maximize contacts among hydrophobic groups, both within each β-sheet and between the β-sheets.
Antiparallel β-sheets with odd values of N-M occur in fibrils formed by residues 11–25 of Aβ (Aβ11–25), where the registry is 17 + k↔20 - k at pH 7.4 and 17 + k↔22 - k at pH 2.4 (31). SSNMR spectra of Aβ11–25 fibrils show sharp lines and single sets of chemical shifts, indicating high symmetry (i.e., one molecule in the asymmetric unit). In this case, stacking of many antiparallel β-sheets perpendicular to the long fibril axis, as suggested by Lu, et al. (49), can account for the high symmetry, as well as the ribbon-like appearance of Aβ11–25 fibrils (31). High-symmetry structures with odd values of N-M are not possible when each peptide molecule contributes to two β-sheet layers, as in Fig. 4.
Competition Between Parallel and Antiparallel Cross-β Structures.
Although antiparallel D23N-Aβ1–40 fibrils are long-lived under typical conditions, they are thermodynamically metastable: a mixture of antiparallel and parallel structures evolves toward purely parallel structures by gradual dissolution of the antiparallel fibrils and growth of the parallel fibrils (Fig. 1D and Fig. S1A). The extension rate of antiparallel fibrils is also less than that of parallel fibrils, causing parallel structures to dominate after several generations of seeded growth (Fig. S1 B and C) but allowing the antiparallel SFg2 structure to be purified by our seeding/filtration protocol. Yet antiparallel fibrils can be the major component of the mixture of structures that appears when D23N-Aβ1–40 monomers self-assemble spontaneously, without seeding. This observation can be understood only if the rate of spontaneous nucleation of antiparallel structures is greater than the rate of spontaneous nucleation of parallel structures. A high nucleation rate (but low extension rate) for antiparallel structures leads to a high abundance of relatively short antiparallel fibrils, as observed experimentally (Fig. 1A). Eventually, the antiparallel fibrils would shrink and disappear as D23N-Aβ1–40 molecules transfer to the thermodynamically more stable parallel fibrils, but this process is very slow (unless all fibrils are kept short by intermittent sonication as in Fig. S1A).
Why are parallel D23N-Aβ1–40 fibril structures more stable than antiparallel structures? As discussed above, parallel structures involve a larger number of ordered residues, longer β-strand segments, and interactions between cross-β units that are not present in antiparallel structures (assuming that parallel D23N-Aβ1–40 fibril structures closely resemble parallel WT-Aβ1–40 structures). Other factors may possibly include a more optimal packing of hydrophobic side chains or the presence of polar zipper interactions in rows of Q15, N23, or N27 side chains in parallel structures.
Why are antiparallel WT-Aβ1–40 fibrils not observed? Substitution of charged D23 side chains for the uncharged N23 side chains in Fig. 4B may introduce destabilizing electrostatic interactions. In certain parallel WT-Aβ1–40 fibrils, electrostatic destabilization is avoided by pairing of D23 and K28 side chains, but the D23-K28 interactions in parallel fibrils have been shown to be intermolecular, not intramolecular (5, 12). Intermolecular D23-K28 interactions (involving pairs of molecules separated by at least 9.6 Å along the fibril axis) may be incompatible with an antiparallel structure similar to that in Fig. 4B. Alternatively, antiparallel WT-Aβ1–40 fibrils may be stable or metastable, but may nucleate more slowly than parallel WT-Aβ1–40 fibrils (perhaps due to the conformational preferences of WT-Aβ1–40 monomers or oligomers that precede nucleation). A more definitive understanding of the kinetic and thermodynamic balance between antiparallel and parallel structures in Aβ fibrils may result from future experimental and theoretical studies.
Materials and Methods
SSNMR data were acquired at 9.39 T (100.4 MHz 13C NMR frequency) and room temperature, using a Varian InfinityPlus spectrometer and a Varian 3.2 mm magic-angle-spinning (MAS) probe. Samples for SSNMR were pelleted for 2 h at 435,000 × g, lyophilized, loaded into the MAS rotor, and rehydrated with deionized H2O (1.0 μL/mg, except in PITHIRDS-CT measurements). Samples contained 3–4 mg of D23N-Aβ1–40. Spectra of rehydrated samples did not change during data acquisition or after storage for 6 mo. PITHIRDS-CT data were acquired at 20.00 kHz MAS with pulsed spin-lock detection as previously described (13, 50). 2D 13C-13C spectra with fpRFDR mixing were acquired at 20.00 kHz MAS with 15.0 μs 13C π pulses in the mixing period, 110 kHz proton decoupling, and two-pulse phase modulation (51) in t1 and t2. CHHC spectra were acquired at 20.00 kHz MAS as previously described (31, 39), with 140 μs 13C-1H and 1H-13C cross-polarization periods and a 200 μs 1H-1H spin diffusion period between t1 and t2. 2D RAD spectra were acquired at 10.00 kHz MAS. Frequency-selective REDOR data were acquired at 5.00 kHz MAS as previously described (12, 15). Measurement times were roughly 24 h, 16 h, 60 h, 48 h, and 48 h in PITHIRDS-CT, 2D fpRFDR, CHHC, 2D RAD, and frequency-selective REDOR experiments, respectively. 13C chemical shifts are relative to tetramethylsilane.
Electron microscopy was performed with an FEI Morgagni microscope at 80 kV. Negatively stained images and MPL data for unstained samples were obtained as previously described (16, 43), using a side-mounted Advanced Microscopy Techniques (AMT) Advantage HR CCD camera. Electron diffraction patterns of unstained D23N-Aβ1–40 fibrils were recorded with a bottom-mounted AMT XR-550 B sCMOS camera at a nominal 350 mm camera length. Diffraction angles were calibrated with thallium chloride crystals (Fig. S5B).
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute on Aging, within the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2LNQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111305109/-/DCSupplemental.
References
- 1.Benzinger TLS, et al. Propagating structure of Alzheimer’s β-amyloid((10–35)) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed M, et al. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat Struct Mol Biol. 2010;17:561–U56. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chimon S, et al. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 5.Lührs T, et al. 3D structure of Alzheimer’s amyloid-β1–42 fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torok M, et al. Structural and dynamic features of Alzheimer’s Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 7.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide solvent protection analysis demonstrates that amyloid-β1–40 and amyloid-β1–42 form different fibrillar structures under identical conditions. Biochem J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antzutkin ON, et al. Multiple quantum solid state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci USA. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s β-amyloid fibrils from electron microscopy and solid state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 10.Balbach JJ, et al. Supramolecular structure in full-length Alzheimer’s β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 12.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Seeded growth of β-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tycko R, Sciarretta KL, Orgel J, Meredith SC. Evidence for novel β-sheet structures in Iowa mutant β-amyloid fibrils. Biochemistry. 2009;48:6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiang W, Yau WM, Tycko R. Structural evolution of Iowa mutant β-amyloid fibrils from polymorphic to homogeneous states under repeated seeded growth. J Am Chem Soc. 2011;133:4018–4029. doi: 10.1021/ja109679q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury CS, et al. Studies on the in vitro assembly of Aβ1–40: Implications for the search for Aβ fibril formation inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 19.Kodali R, Williams AD, Chemuru S, Wetzel R. Aβ1–40 forms five distinct amyloid structures whose β-sheet contents and fibril stabilities are correlated. J Mol Biol. 2010;401:503–517. doi: 10.1016/j.jmb.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 21.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Margittai M, Chen J, Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 23.Ladner CL, et al. Stacked sets of parallel, in-register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J Biol Chem. 2010;285:17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debelouchina GT, Platt GW, Bayro MJ, Radford SE, Griffin RG. Intermolecular alignment in β2-microglobulin amyloid fibrils. J Am Chem Soc. 2010;132:17077–17079. doi: 10.1021/ja107987f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JCC, Oyler NA, Yau WM, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryndushkin DS, Wickner RB, Tycko R. The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-β structure: Evidence from solid state NMR. J Mol Biol. 2011;409:263–277. doi: 10.1016/j.jmb.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc Natl Acad Sci USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tycko R, Savtchenko R, Ostapchenko VG, Makarava N, Baskakov IV. The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: Evidence from solid state nuclear magnetic resonance. Biochemistry. 2010;49:9488–9497. doi: 10.1021/bi1013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lansbury PT, et al. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 31.Petkova AT, et al. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J Mol Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 32.Balbach JJ, et al. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer’s β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 33.Sawaya MR, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 34.Grabowski TJ, Cho HS, Vonsattel JPG, Rebeck GW, Greenberg SM. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann Neurol. 2001;49:697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 35.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 36.Bennett AE, et al. Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys. 1998;108:9463–9479. [Google Scholar]
- 37.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C, and 15N random coil NMR chemical shifts of the common amino-acids. 1. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tycko R, Ishii Y. Constraints on supramolecular structure in amyloid fibrils from two-dimensional solid state NMR spectroscopy with uniform isotopic labeling. J Am Chem Soc. 2003;125:6606–6607. doi: 10.1021/ja0342042. [DOI] [PubMed] [Google Scholar]
- 40.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 41.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 42.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate 13C-15N distance measurements in uniformly 13C,15N-labeled peptides. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 43.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci USA. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwieters CD, Kuszewski JJ, Clore GM. Using XPLOR-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- 45.Mattson MP, et al. β-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Melckebeke H, et al. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 49.Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG. Exploiting amyloid fibril lamination for nanotube self-assembly. J Am Chem Soc. 2003;125:6391–6393. doi: 10.1021/ja0341642. [DOI] [PubMed] [Google Scholar]
- 50.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126 doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 51.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.