Abstract
Reversibly switchable fluorescent proteins (RSFPs) have attracted widespread interest for emerging techniques including repeated tracking of protein behavior and superresolution microscopy. Among the limited number of RSFPs available, only Dronpa is widely employed for most cell biology applications due to its monomeric and other favorable photochemical properties. Here we developed a series of monomeric green RSFPs with beneficial optical characteristics such as high photon output per switch, high photostability, a broad range of switching rate, and pH-dependence, which make them potentially useful for various applications. One member of this series, mGeos-M, exhibits the highest photon budget and localization precision potential among all green RSFPs. We propose mGeos-M as a candidate to replace Dronpa for applications such as dynamic tracking, dual-color superresolution imaging, and optical lock-in detection.
Keywords: (F)PALM/STORM, mEos2, mGeos, superresolution microscopy
Photoactivatable fluorescent proteins (PAFPs) are molecules that switch to a new fluorescent state in response to irradiation with light of a specific wavelength (1, 2). There are three classes of PAFPs: dark-to-bright photoactivators, irreversible photoconverters, and reversible highlighters (1). The third class of PAFPs, also termed RSFPs (reversibly switchable fluorescent proteins), can switch between the active and nonactive states multiple times, thus offering a distinct advantage over the first two classes. Only RSFPs can be used for repeated measurements of protein movements in live cells (3) and for the emerging technique of optical lock-in detection (OLID) (4). Moreover, RSFPs possess all the fundamental requirements to be useful for rewritable ultrahigh density optical data storage (5, 6). In addition, RSFPs demonstrate great potential for use in recently developed superresolution microscopy techniques, such as reversible saturable optical fluorescence transition (RESOLFT) microscopy with ultralow light intensities (7, 8) and photoactivated localization microscopy (PALM) (9) or stochastic optical reconstruction microscopy (STORM) (10), or fluorescence photoactivation localization microscopy (FPALM) (11) [collectively referred as (F)PALM/STORM].
In the applications mentioned above, different optical characteristics are emphasized for different purposes. For example, high photon output and photostability are desirable for higher precision in superresolution microscopy, whereas improvements in switching rate will result in a faster sampling rate in OLID (4). Unfortunately, there is only a limited number of RSFPs in the palette of PAFPs, and further innovations are required to optimize their optical properties for the desired applications. Dronpa is currently the RSFP of choice for most cell biology applications, mainly because it is monomeric and because it is the brightest among the PAFPs. However, the poor localization precision potential of Dronpa (26 nm) (12) limits its wide application in (F)PALM/STORM experiments, which could partly be due to its fast off-switching kinetics and hence less photon output per switching cycle. An attempt to find Dronpa variants with slower off-switching rate has led to the identification of PDM1-4 (13). Unfortunately, PDM1-4 was prone to form tetramers and did not show improvement in photon output. In addition, PDM1-4 has worsened photostability and less contrast between on and off-states (see Table S1). mEos2 is a monomeric, irreversible green-to-red photoconverter which demonstrates the highest localization precision potential in (F)PALM/STORM (14). In the present study, we identified a series of green mEos2 mutants with reversibly switching property. The developed green mEos2 mutants, hereafter collectively termed mGeos (stands for monomeric Green Eos), differ from each other in only the first position of the tripetide (XYG) of the chromophore and demonstrate varied optical characteristics such as photoswitching rates, photon outputs, photostabilities, and acid sensitivities. This diversity makes them potentially useful tools for various applications in diffraction-limited and superresolution imaging.
Results
Development of a New Series of Green RSFPs.
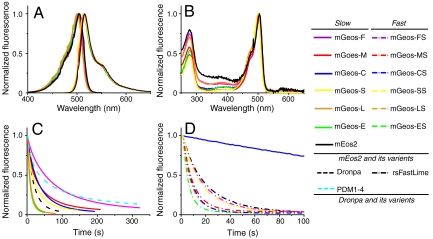
To eliminate the green-to-red conversion, we first tried to mutate mEos2 at two key sites (His62 and Glu212) (Fig. S1), which are important in photoactivated conversion. Glu212 has been shown to be a key residue for the provision of a proton that allows the formation of the red chromophore upon UV illumination (15). However, with the exception of the dim fluorescence of E212D, no Glu212 mutants demonstrated fluorescence, suggesting that Glu212 also plays a pivotal role in the formation of the green chromophore (16). It has been shown that the first position of the tripetide (HYG) of the chromophore is important for green-to-red conversion in EosFP (17). We thus performed series mutations in the same position in mEos2 (His62) and found that many of them yielded green species with no conversion ability. Interestingly, the His62 mutants of mEos2 can be switched on and off for many times and constitute a unique series of green RSFPs, which we named mGeos here. Similar to Dronpa, mGeos could be turned off by 488-nm laser light and be reactivated by 405-nm laser. mGeos demonstrate similar spectral properties with maximum excitation between 501 nm and 505 nm, and maximum emission between 512 nm and 519 nm, similar quantum yields (0.72–0.85) and extinction coefficients (52,000–77,000 M-1 cm-1) (Fig. 1 A and B and Table 1). Notably, some mGeos demonstrate extremely high brightness (which is the product of extinction coefficient and quantum yield). For example, the brightest mGeos, mGeos-C (with a cysteine mutation at residue His62), is 81%, 186%, and 133% as bright as Dronpa, EGFP, and mEos2, respectively.
Fig. 1.
The spectra and photochemical kinetics of mGeos. (A) The excitation (measured at the emission maximum) and emission (measured at the excitation maximum) spectra of photoactivatable mGeos and mEos2. (B) The absorbance spectra of mGeos and mEos2 in their bright states. (C) Single switching kinetics for His62 mutant mGeos, Dronpa, and PDM1-4. (D) Single switching kinetics for His62 and F173S double mutant mGeos and rsFastLime. The blue solid line is mGeos-C, used as a control. The laser intensity in (C) is 15-fold higher than in (D).
Table 1.
Characteristics of mGeos, mEos2, and Dronpa
| Abs. (nm) | Emi. (nm) | QY | ϵ-max (M-1 cm-1) | pKa | Relaxation half-time (min) | |
| Dronpa* | 503 | 522 | 0.68 | 125,000 | 5 | 840 |
| mGeos-F | 504 | 515 | 0.85 | 53,135 | 5 | 205 |
| mGeos-M | 503 | 514 | 0.85 | 51,609 | 4.5-5 | 169 |
| mGeos-C | 505 | 516 | 0.81 | 76,967 | 6 | 223 |
| mGeos-S | 501 | 512 | 0.76 | 64,602 | 5-5.5 | 272 |
| mGeos-E | 501 | 513 | 0.75 | 69,630 | 6-6.5 | 848 |
| mGeos-L | 501 | 513 | 0.72 | 53,448 | 5-5.5 | 398 |
| mEos2 (Green)* | 506 | 519 | 0.84 | 56,000 | 5.6 | -- |
Characterization of the mGeos Family.
We performed photoswitching experiments in HeLa cells transfected with untagged mGeos under confocal microscopy. The results suggest that the majority of the mGeos demonstrate fluorescence enhancement of approximately 20-fold upon switching on from their off-states, which is equivalent to that of Dronpa but better than PDM1-4 (Table S1). Analysis of the photobleaching fraction per cycle showed that mGeos displayed varied photostabilities. For example, mGeos-I, -T and -V were much more photostable than Dronpa (Table S1), while the photostabilities of mGeos-M, -S, and -N were nearly the same as that of Dronpa but were better than PDM1-4, mGeos-C, and -F.
It is interesting that although these mGeos varied at only one residue in the first amino acid of the chromophore (XYG), they displayed different optical characteristics. First, they possessed varied photoswitching kinetics (Fig. 1 C and D, Fig. S2 and Table S1). For instance, the off-switching half-time of the slowest, mGeos-F, was 52.28 s, 1.45-fold longer than that of PDM1-4 and 3.78-fold longer than that of Dronpa. Several mGeos (mGeos-A, -E, -I, -L, -Q, -T, and -V) showed a much faster off-switching rate than did Dronpa (Table S1). To generate mutants with even faster switching rates, we added the F173S mutation (18) to mGeos and found several unique RSFPs with even faster off-switching rate. As an example, the off-switching half-time of the fastest mGeos, mGeos-ES (H62E and F173S mutant), was 33% of that of rsFastLime (19), the fastest off-switching RSFP known to date (Fig. 1D, Fig. S3). Moreover, we found that the different residues at the first site of the chromophore caused mGeos to possess different pKa values ranged from 4 to 7 in the on state (Table 1 and Fig. S4). mGeos-E demonstrated higher acid sensitivity (a pKa between 6–6.5) than did Dronpa and mEos2, which suggests that it could be used as an intracellular pH sensor. In contrast, mGeos-M showed a remarkable pH stability (a pKa between 4.5–5), which could be very useful when used as a fusion partner in acidic environments, such as the lumens of secretory vesicles and lysosomes. To further analyze photoswitching behavior of mGeos-X and Dronpa, purified biotin-conjugated mGeos-X and Dronpa were diluted and immobilized to streptavidin-coated cover glasses and the molecules that are turned on by 491 nm were counted. Using a previously described method (20–23), we observed more on/off switching of mGeos-E than those of other mGeos when excited by 491 nm alone (Fig. S5A). It is possible that mGeos-E can be weakly switched on by 491 nm because its dark-state exhibited higher absorbance at 491 nm than mGeos-M (Fig. S5B). We verified that spontaneous recovery of mGeos-E from dark to fluorescent state (Fig. S5 C–D) is very slow as compared with Dronpa-M159T (19) and mApple (24). Taken together, these properties suggest the potential application of mGeos-E in PALM with independently running acquisition (PALMIRA) (25).
mGeos Afford Excellent Fusion Partners Like mEos2.
We speculated that while the mutations at the external β-sheet sites would influence the oligomeric characteristics of mEos2, the His62 mutants at the internal chromophore site would not. Indeed, our gel filtration analysis of purified mGeos (mGeos-C, M, S, F, E, and L) showed oligomeric characteristics similar to that of mEos2 (Fig. S6), suggesting that they are suitable, just like their parent, for protein fusion and labeling in the study of living cells.
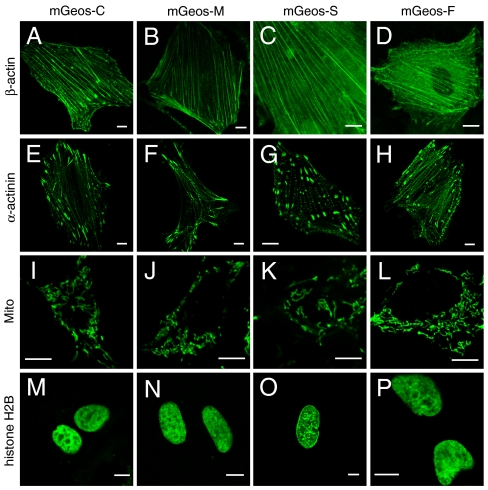
To further investigate mGeos’ performance in living cells, we observed mGeos fusion proteins transiently expressed in HeLa cells and HEK293 cells using confocal microscopy. Either C-terminal fused β-actin or N-terminal fused α-actinin localized, as expected, in the same manner as those labeled with mEos2 (14) or Dronpa (Fig. 2 and Fig. S7). The fusion of mGeos with a mitochondrial sorting peptide showed accurate localization in mitochondria (Fig. 2). We also observed that mGeos fused to H2B interfered with neither mitosis nor interphase (Fig. 2 and Fig. S7). These results suggest that mGeos can provide excellent fusion partners for biological imaging in living cells because they possess the same oligomeric characteristics as mEos2.
Fig. 2.
Confocal images of mammalian cells expressing mGeos-C (A, E, I, M), mGeos-M (B, F, J, N), mGeos-S (C, G, K, O) and mGeos-F (D, H, L, P) labeled proteins. (A)–(D) mGeos-β-actin (mouse) in HeLa cells; (E)–(H) α-actinin-mGeos (human) in HeLa cells; (I)–(L) mGeos-Mito (human, subunit VIII of human cytochrome C oxidase) in HEK293 cells; and (M)–(P) mGeos-histone H2B (human) in HeLa cells. Scale bars: 10 μm.
Slow Photoswitching mGeos are Suitable for (F)PALM/STORM Imaging.
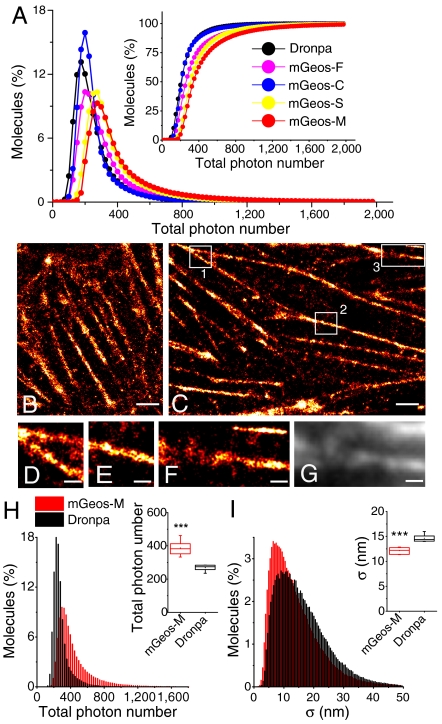
PAFPs used for superresolution imaging should have large photon output and a good balance between photobleaching and photoswitching (1, 2). We selected mGeos with slow off-switching kinetics (mGeos-N, -K, -F, -C, -S, and -M) for (F)PALM/STORM experiments in the hope for identifying better probes with higher number of emitted photons per switching cycle. mGeos fusions with β-actin were expressed in HeLa cells, and the total photon number per switching cycle and the localization precision potential were detected and analyzed as previously described (9). We found that mGeos-M had the highest number of photons collected per switching cycle and hence the highest localization precision potential of all the green RSFPs (Fig. 3A, Table S2). We propose that the long off-switching half-time of mGeos-M, which is 152% that of Dronpa, and its strong photostability may contribute to a higher photon output (Table S1). mGeos-S and -F are also excellent probes for superresolution imaging, with higher photon outputs (Fig. 3A, Table S2). To test the best localization precision potential we can achieve with mGeos-M, we optimized the gain, excitation laser power, pixel size and frame frequency. We obtained a median photon output of 387 and a localization precision potential of 12 nm for mGeos-M (Fig. 3 H–I), which is better than that of Dronpa (269 and 15 nm, respectively, collected under identical conditions) with statistical significance. As a result, finer actin structures could be observed when labeled with mGeos-M (Fig. 3C) than those labeled with Dronpa (Fig. 3B).
Fig. 3.
Comparison between mGeos and Dronpa in superresolution microscopy. (A) Distribution of total photon number per burst in HeLa cells expressing β-actin fused with mGeos-M, mGeos-C, mGeos-F, mGeos-S, and Dronpa. (B)–(C) (F)PALM/STORM images of Dronpa-β-actin (B) and mGeos-M-β-actin (C) in a fixed HeLa cell. (D)–(F) Magnified images of the box 1–3 in (C), respectively. (G) TIRF image of area in (F). Scale: 2 μm in (B)–(C); 500 nm in (D)–(F). (H) Distribution of total photon number in HeLa cells expressing β-actin fused with mGeos-M (mean, 458; median, 387; n = 7) and Dronpa (mean, 299; median, 269; n = 6). (I) Distribution of position error in HeLa cells expressing β-actin fused with mGeos-M (mean, 14 nm; median, 12 nm; n = 7 cells) and Dronpa (mean, 17 nm; median, 15 nm; n = 6 cells). The sum of single molecule bursts analyzed for mGeos-M and Dronpa is 320,903 and 262,838, respectively. ***, P < 0.001 (Student’s t test).
Performance of Dual-Color (F)PALM/STORM Using mGeos-M and PAmCherry1.
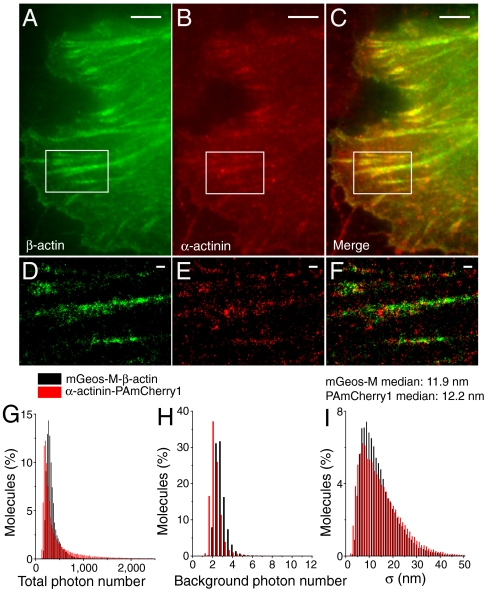
Determination of the relative positions of proteins within cells using multicolor superresolution microscopy is essential for understanding their biological function. For maximizing the ability to simultaneously observe different proteins as many as possible, it is desirable to have a palette of PAFPs that are separable in spectrum. Better green RSFPs are in need for multicolor (F)PALM/STORM because the currently used ones, PA-GFP (12) or Dronpa (12), produced limited photon outputs. To demonstrate the suitability of mGeos for dual-color (F)PALM/STORM, we labeled β-actin and α-actinin with mGeos-M and PAmCherry1, respectively, and coexpressed them in HeLa cells. Under TIRF imaging, β-actin and α-actinin seem to colocalize on stress fibers (Fig. 4 A–C). However, dual-color (F)PALM/STORM imaging revealed that punctate α-actinin periodically distributed in or between β-actin fibers (Fig. 4 D–F), as previously described (26, 27). Quantitative analysis revealed that mGeos-M and PAmCherry1 gave similar localization precision potential (approximately 10 nm) under our dual-channel imaging setup (Fig. 4 G–I). For dual-color imaging, it is critical to know to what extent each label correctly identifies its associated target proteins. This is assessed by means of dual-channel crosstalk experiments in Fig. S8. Our results demonstrated minimal leak through of mGeos into the red channel and virtually no overlap between mGeos-M and PAmCherry1 when they were attached to two differently localized proteins.
Fig. 4.
Nanostructural distribution analyses of β-actin and α-actinin using two-color (F)PALM/STORM in HeLa cell. (A, B) TIRF images of mGeos-M-β-actin (Green) and PAmCherry1-α-actinin (Red). (C) Merged dual-color TIRF image, diffraction-limited. (D, E) (F)PALM/STORM images of mGeos-M-β-actin (Green) and PAmCherry1-α-actinin (Red), magnified from box in (A) and (B). (F) Merged image of mGeos-M-β-actin (Green) and PAmCherry1-α-actinin (Red). Scale bars: 2 μm in (A)–(C); 500 nm in (D)–(E). (G)–(I) Distribution analysis of total photon number per molecule, background photon number and position precision of mGeos-M-β-actin and α-actinin-PAmCherry1 coexpressed in HeLa cells.
Discussion
In summary, our study revealed a unique strategy for generating RSFPs of different optical characteristics by mutating the first position of chromophore residues, but not the sites in the vicinity of the chromophore. The distinguished properties of mGeos developed here expand our current tool kit of PAFPs and allow for broad applications in dynamic live cell imaging and superresolution imaging. For example, the excellent photostability of mGeos-I, -T, and -V enables long-term pulse-chase imaging. The faster on/off switching rate and higher photon output make mGeos-E and -L better tools than rsFastLime for repeated tracking and for the generation of high-contrast images of special structures and macromolecules using the OLID method. Furthermore, mGeos-M preserves the same oligomeric character and the approximately 10 nm localization precision potential of its parent mEos2 (14). As a dark-to-green PAFP with outstanding photochemical properties, mGeos-M is also suitable for multicolor superresolution microscopy when paired with other PAFPs of different colors. Thus, mGeos-M should be an excellent candidate to replace Dronpa for applications such as dynamic tracking, dual-color superresolution imaging, and optical lock-in detection. During the submission of this manuscript, a unique reversibly switchable enhanced green fluorescent protein (rsEGFP) was identified. The ability of rsEGFP to be reversibly photoswitched for many times enables superresolution imaging that is similar to stimulated emission depletion (STED) microscopy but operates at 106 times lower levels of light (8). Interestingly, some of the mGeos (mGeos-ES and -LS) have fast off-switching rates and multiple reversibility like rsEGFP, suggesting that these probes can find potential application in this unique, low-light-level RESOLFT microscopy.
Because mGeos have different photoswitching kinetics but only one different residue, located at the first position of XYG, they should provide a good start for a comparative study of the mechanism of reversible photoactivation in FPs using nuclear magnetic resonance or X-ray analysis. Light-driven switching of RSFPs generally involves an isomerization of the chromophore, frequently coupled with a change of its protonation state (18, 28, 29). Similar to Dronpa and its derivatives, mGeos show two absorption peaks: a major peak at approximately 502 nm and a minor peak at 388 nm (Fig. 1 and Fig. S4), which can be mapped to the (fluorescent) deprotonated and the (nonfluorescent) protonated forms of the chromophore (28). The existence of the two forms suggests that light might induce a structural rearrangement with the different protonation states of the chromophore. Based on the structures of the mEos2 homologs EosFP (15) and IrisFP (29), X in XYG of mGeos is located at the far end of the chromophore away from Tyr-63, but near Glu-212 and Gln-38, which play pivotal roles in stabilizing the planar structure and π-system of the chromophore by hydrogen-bonds through a water molecule. We speculate that different X in XYG may interact with different amino acids near the chromophore, and indirectly influence the protonation equilibria of the Tyr-63 by changing electrostatic surface potentials and the large π-system. For example, the hydrophobic amino acids (V/I/A/L) may interact with Leu-210 and/or Met-40 near the chromophore through hydrophobic-hydrophobic interaction, which may favor the protonated form of the chromophore upon light illumination to give the fast mGeos variants (Fig. S2). It is not clear how X residues could influence the isomerization between a trans (off) and cis (on) states. It is likely that different sized X residues at the distal side of the chromophore may use a unique way to affect chromophore isomerization. Refined structure determination and comparison between mGeos are needed to reveal the exact mechanism of photoswitching.
Materials and Methods
Design and Generation of a Series of Bright Green Dronpa-like Fluorescent Proteins Based on mEos2.
We produced unique reversibly photoactivatable proteins based on mEos2. This molecule was selected because its green state is brighter than that of EGFP and it is highly homologous to Dronpa, a reversibly photoactivatable green fluorescent protein. Based on a structure analysis of wild-type EosFP (15), two positions (Glu212 and His62) were selected for random mutations. Site-specific random mutations of mEos2 were performed based on pRSETa-mEos2 (Addgene plasmid 20341) using the polymerase incomplete primer extension (PIPE) method (30, 31). Nineteen kinds of E212X and 19 kinds of H62X plasmids were sequenced, purified and transfected into Escherichia coli strain BL21 (DE3). These colonies were analyzed using a widefield upright fluorescence microscope (Stereo Discovery V8, Carl Zeiss) and the blue light was generated by X-cite 120PC (mercury lamp) with proper filter set. Then mutants were analyzed by FV1000 microscopy (Olympus) to confirm if those mutants lost the conversion from green to red (as compared to mEos2 as a control) using 488 nm and 561 nm laser to image and 405 nm laser to activation. To separate these probes from mEos2, the new reversibly photoactivatable proteins were denoted mGeos (mGeos-X denotes mEos2 H62X mutant). To obtain mutants with faster photoswitching kinetics, we also used the PIPE method to introduce the F173S mutation into mGeos (which we termed mGeos-XS, indicating mEos2 H62X and F173S double mutant). As controls, rsFastLime (Dronpa V157G) and PDM1-4 (Dronpa K145N) were generated from Dronpa (Amalgaam Inc.) (3) using site-specific mutations.
Plasmid Construction.
All the mGeos’ cDNAs were cloned into the BamHI/Not I sites of pEGFP-N1 (Clontech) and the NheI/BglII sites of pEGFP-C1 (Clontech) to replace the EGFP gene and generate pmGeos-N1 and pmGeos-C1. The full-length Homo sapiens H2B and α-actinin (Proteintech Group, Inc., China) cDNAs with NheI/XhoI sites were PCR-amplified and inserted into pmGeos-N1. β-actin (Proteintech Group, Inc., China) was cloned into a pmGeos-C1 vector. To generate pmGeos-Mito, cDNAs of mGeos containing AgeI/NotI sites were PCR-amplified and swapped with the Hyper gene in the pHyper-dMito vector (Evrogen). Synthetic DNA primers for cloning and site-specific random mutations were purchased from Invitrogen. All mutants were sequenced (The Beijing Genomics Institute, China) before further analysis. The Restriction Enzymes were purchased from New England BioLab Inc.
Protein Expression and Purification.
Proteins of mEos2, Dronpa, EGFP, and all mGeos were expressed in the E. coli strain BL21(DE3) and purified using an Ni-NTA His-Bind resin (Qiagen), followed by a gel filtration step using a Superdex 200 column (GE Healthcare). For further analysis, purified proteins were concentrated by ultrafiltration and diluted in phosphate buffered saline (PBS).
Measurement of Spectral Properties and pKa.
Proteins were first diluted in PBS (pH = 7.4) to limit UV absorption under 0.1 so that quantum yields could be accurately measured. Then, the absorption, excitation/emission spectra were immediately recorded using an Agilent 8453 UV/V spectrophotometer and an Edinburgh Instrument FLS920, respectively. For the emission spectra, mGeos were irradiated with 503-nm light. To determine the fluorescence excitation spectra, the fluorescence was recorded at 513 nm. The fluorescence quantum yields and the molar extinction coefficients at the respective absorption maxima were determined relative to the reported value for mEos2 (14) (quantum yield: 0.84, molar extinction coefficient at 506 nm: 56,000 M-1 cm-1). For pKa measurement, purified mGeos dissolved in PBS (pH = 7.4) were diluted 1∶20 in either glycine-hydrochloric acid buffers (for pH ≤ 5) or sodium phosphate buffers (for pH≥6) with pH values ranged from 3–11, and the absorption spectra were immediately recorded. The pKa value was taken as the pH value where the absorption reached 50% of the maximum. To measure the off-state absorption of mGeos-E and -M, purified proteins were first illuminated by 491 nm laser to nonbright state and then measured as described above.
Relaxation Half-time Measurement.
The protocol was used as described before (19, 32). Purified proteins of mGeos-F, C, S, M, E, and L were first photoswitched to their off-states using a 40 mW 491 nm laser. Then, the relaxation curves from off-state into thermal equilibrium state were obtained in a Varioskan Flash spectral scanning multimode reader (Thermo Scientific) with 96-well optical bottom plates (Nunc) for about 14 h at 25 °C. A weak blue light (490 ± 6 nm) was used to read the data at 520 nm every 4 min during those period. And then, the determination of relaxation half-time for mGeos-F, C, S, M, E, and L were based on single exponential function fitting of the curves. The relaxation half-time of Dronpa was taken from ref. (19).
Cell Culture, Transfection, and Fixation.
HEK293 cells and HeLa cells were cultured in DMEM complete medium (Gibco) supplemented with 10% fetal bovine serum and maintained at 37 °C and 5% CO2 in a humidified incubator. Cells were then transiently transfected using Lipofectamine™ 2000 (Invitrogen) in accordance with the manufacturer’s protocol. After transfection, the cells were grown in DMEM complete medium for 24 h. For (F)PALM/STORM imaging, cells were grown in IMEM (Gibco) or DMEM complete medium without phenol red. Cells were recultured on cover slips (Fisher Scientific) for another 24 h and then fixed with 4% (w/v) para-formaldehyde (for (F)PALM/STORM imaging, 3% (w/v) para-formaldehyde and 0.5% glutaraldehyde were used) in PBS for 20 to 40 min at room temperature, washed three to five times with PBS and stored in PBS until imaging.
Analysis of the Switching Kinetics of mGeos.
To analyze the switching kinetics of mGeos, we used an FV1000 Laser Scanning Confocal Microscope (Olympus) to acquire images. HeLa cells were transfected with pmGeos-N1 plasmids and fixed with 4% (w/v) para-formaldehyde in PBS (pH = 7.4) on cover slips. To activate and bleach mGeos we used lasers with wavelengths of 405 nm and 488 nm, respectively, and recorded the off-switching images. We repeated on/off switching until we observed considerable photobleaching. During the entire experiments, scanning speed, gain, amplification, and off-set value were held constant. Because the difference in photoswitching half-times between mGeos-X and mGeos-XS, we used different 488 nm laser powers (the laser intensity and density were 9 μW and 127 mW/cm2 for Fig. 1C and Fig. S2, 0.6 μW and 8 mW/cm2 for Figure 1D, 2.4 μW and 34 mW/cm2 for Figure S3, respectively). In all switching experiments, the 405 nm laser power was kept the same (intensity: 24 μW; density: 338 mW/cm2). The values of off-switching half-time for mGeos-X, Dronpa, PDM1-4, mGeos-XS, and rsFastLime in Table S1 were calculated based on curves in Fig. 1C and 1D. The off-switching fluorescence percentage and bleaching percentage in Table S1 was the mean value measured based on cycles in Figs. S2 and S3. The ROI value was marked and read out by Image J software (NIH) and analyzed using Igor Pro 5.0 (Wavemetrics).
Confocal Microscopy and Cell Imaging.
Confocal microscopy images of mGeos were obtained using an FV500 Laser Scanning Confocal Microscope (Olympus). All the figures were produced using a 60X 1.4 NA objective and a 488-nm laser. Images were quantified and analyzed using an Olympus FluoView Viewer (Japan).
(F)PALM/STORM Setup.
(F)PALM/STORM imaging of Dronpa and mGeos was performed as previously described (9, 33). We used an Olympus IX71 inverted microscope equipped with a 100X, 1.45 numerical aperture (NA) oil objective (Olympus PLAN APO). The fluorescence signals were acquired using an electron-multiplying charge-coupled device (EMCCD) camera (Andor iXon DV-897 BV) and high-quality filters (Chroma, 49002 ET-GFP) for mGeos-X and Dronpa imaging. The maximum power near the rear pupil of the objective was 8.74 mW for the 405 nm laser (LASOS), 4.13 mW for the 491 nm laser (Cobolt Dual Calypos™), and 2.54 mW for the 561 nm laser (Cobolt Jive). The intensity of the 405 nm laser was set so that it only activated a few molecules in each frame. For two-color imaging, we used filters from Semrock (Di01 - R488/561 - 25 × 36 and FF01 - 523/610 - 25) and added an Optosplit II (Cairn) with three filters from Semrock (FF03 - 525/50 - 25, FF01 - 617/73 - 25, and FF560 - Di01 - 25 × 36). In fixed-cell experiments, samples transfected with reversibly photoactivatable proteins were imaged using a 100X oil objective with a 1.6X amplification adaptor. The EMCCD camera gain was set to 300 throughout all (F)PALM/STORM imaging experiments, and a frame rate of 5 Hz was used.
Single Molecule Immobilization for On/Off Switching Analysis.
For single molecule localization measurements in vitro, fluorescence proteins were labeled as described previously (9). Purified mGeos were incubated with biotinamido hexanoic acid 3-sulfo-N-hydroxysuccinide ester (BAC-SulfoNHS) (BK101, Sigma) in a sodium phosphate buffer solution for approximately 30 min, at which point the biotin-labeled proteins were isolated by gel filtration column (Sephadex G-25M). To keep fluorescence proteins immobilized, cleaned cover glasses were treated with 5 μL of 1 mg mL-1 biotin-BSA and 5 μL of 1 mg mL-1 streptavidin sequentially for 5 min. The cover glasses were then washed twice, and 5 μL of 0.01 μM biotinylated mGeos were incubated on cover glasses. Finally, the cover glasses were placed in PBS buffer (pH = 7.4). Single molecules were imaged as described above. Using 491 nm laser alone, on/off switching (or blinking) events were calculated as the number of localizations within an interval localization spacing of 30 nm.
(F)PALM/STORM Images Analysis.
Data analysis and superresolution image reconstruction were performed as previously described (9, 34). Briefly, a wavelet transform algorithm (35) with a proper threshold was used for single molecule detection, and the localizations of the molecules were determined by finding local maxima with a mask of 5 × 5 pixel. If two molecules occurred at the same pixel (± 1) in successive frames, they were considered to arise from the same molecule, and they were summed across the common fit window (7 × 7 pixel). Then the raw images of molecules in the fit windows were background subtracted and the pixel values were converted to numbers of photons using the camera calibration factor provided by EMCCD manufacturer (Andor). After that, all molecules were fitted with a two dimensional Gaussian using MLE_GPU algorithm (36) to obtain the number of photons (N) and the standard deviation of the PSF (s). The background noise per pixel (b) was determined by taking the standard deviation of the intensity (in photons) of an illuminated area in a raw image where no single molecules are visible. Finally, the localization precision potential for each localized molecule was calculated using Eq. 17 in ref. (37). For Fig. 3A and Table S2, we used a low threshold (k = 3) in wavelet transformation in order to collect more molecules in burst events. For Fig. 3 B–I, we used a higher threshold (k = 4) for better resolution in superresolution images.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Loren L Looger and Addgene for providing mEos2 cDNA. This work was supported by grants from the Major State Basic Research Program of P. R. China (2010CB912303, 2010CB833701), the National Science Foundation of China (30870564, 90913022, 31130065, 311708118, 31127901), and the Chinese Academy of Sciences Project (KSCX2-1W-J-3, KSCX2-EW-Q-11).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113770109/-/DCSupplemental.
References
- 1.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 3.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 4.Marriott G, et al. Optical lock-in detection imaging microscopy for contrast-enhanced imaging in living cells. Proc Natl Acad Sci USA. 2008;105:17789–17794. doi: 10.1073/pnas.0808882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer M. Reversible molecular photoswitches: a key technology for nanoscience and fluorescence imaging. Proc Natl Acad Sci USA. 2005;102:9433–9434. doi: 10.1073/pnas.0504264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam V, et al. Data storage based on photochromic and photoconvertible fluorescent proteins. J Biotechnol. 2010;149:289–298. doi: 10.1016/j.jbiotec.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grotjohann T, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shroff H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno H, et al. Higher resolution in localization microscopy by slower switching of a photochromic protein. Photochem Photobiol Sci. 2010;9:239–248. doi: 10.1039/b9pp00124g. [DOI] [PubMed] [Google Scholar]
- 14.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc Natl Acad Sci USA. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. Base catalysis of chromophore formation in Arg96 and Glu222 variants of green fluorescent protein. J Biol Chem. 2005;280:26248–26255. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- 17.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adam V, et al. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc Natl Acad Sci USA. 2008;105:18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiel AC, et al. 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem J. 2007;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annibale P, Scarselli M, Kodiyan A, Radenovic A. Photoactivatable fluorescent protein mEos2 displays repeated photoactivation after a long-lived dark state in the red photoconverted form. J Phys Chem Lett. 2010;1:1506–1510. [Google Scholar]
- 21.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nat Methods. 2011;8:527–528. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- 22.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: Unraveling the effects of photoblinking. PLoS One. 2011;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egner A, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J, Worth NF, Rolfe BE, Campbell GR, Campbell JH. Heterogeneous distribution of isoactins in cultured vascular smooth muscle cells does not reflect segregation of contractile and cytoskeletal domains. J Histochem Cytochem. 2000;48:1441–1452. doi: 10.1177/002215540004801101. [DOI] [PubMed] [Google Scholar]
- 28.Andresen M, et al. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci USA. 2007;104:13005–13009. doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiedenmann J, et al. From EosFP to mIrisFP: Structure-based development of advanced photoactivatable marker proteins of the GFP-family. J Biophotonics. 2011;4:377–390. doi: 10.1002/jbio.201000122. [DOI] [PubMed] [Google Scholar]
- 30.Klock HE, Lesley SA. The polymerase incomplete primer extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 31.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 32.Andresen M, et al. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat Biotechnol. 2008;26:1035–1040. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- 33.Ji W, et al. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci USA. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould TJ, Verkhusha VV, Hess ST. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olivo-Marin JC. Extraction of spots in biological images using multiscale products. Pattern Recogn. 2002;35:1989–1996. [Google Scholar]
- 36.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nature Methods. 2010;7:373–U352. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.