Abstract
Complex I (NADH-ubiquinone oxidoreductase) in the respiratory chain of mitochondria and several bacteria functions as a redox-driven proton pump that contributes to the generation of the protonmotive force across the inner mitochondrial or bacterial membrane and thus to the aerobic synthesis of ATP. The stoichiometry of proton translocation is thought to be 4 H+ per NADH oxidized (2 e-). Here we show that a H+/2 e- ratio of 3 appears more likely on the basis of the recently determined H+/ATP ratio of the mitochondrial F1Fo-ATP synthase of animal mitochondria and of a set of carefully determined ATP/2 e- ratios for different segments of the mitochondrial respiratory chain. This lower H+/2 e- ratio of 3 is independently supported by thermodynamic analyses of experiments with both mitochondria and submitochondrial particles. A reduced H+/2 e- stoichiometry of 3 has important mechanistic implications for this proton pump. In a rough mechanistic model, we suggest a concerted proton translocation mechanism in the three homologous and tightly packed antiporter-like subunits L, M, and N of the proton-translocating membrane domain of complex I.
Keywords: cell respiration, proton pumping, conformational changes, phosphorylation potential
Complex I is the entry point of the respiratory chain in mitochondria and many bacteria and structurally by far the most complicated of the three respiratory chain complexes with protonmotive activity, viz. I, III, and IV. Complex I is an L-shaped integral membrane protein. Its hydrophilic arm, which protrudes out of the membrane on the negatively charged N side, contains the NADH-oxidizing FMN site, a set of iron sulfur centers, and a binding site for ubiquinone (1–3). The membrane domain includes seven subunits that in mitochondria are all encoded by mtDNA and that are present also in bacteria (see Fig. 1). Three of these subunits*, L, M, and N, are homologous to one another as well as to certain bacterial proton/cation antiporters (2, 3, 6). They share structural features, such as two disrupted transmembrane α-helices (4) and conserved lysines within the membrane domain, which appear to be important in the proton-pumping mechanism (7–9).
Fig. 1.
Complex I in the respiratory chain. The structure of the L-shaped molecule (4) has been placed in the membrane. The positions of the seven membrane subunits L, M, N, K, J, A, and H are roughly indicated. Each of the subunits L, M, and N is proposed to translocate one proton per turnover (blue arrows); translocation of a fourth proton might be catalyzed by the subunits in the heel of the boot-like structure (see text). The position of iron-sulfur cluster N2 is indicated by a yellow circle. The rough positions of two proposed ubiquinone molecules, QA and QB, are indicated. QA transmits electrons from N2 to QB. The latter is reduced to ubiquinol (QH2) with uptake of two “substrate protons” (black arrow). QH2 enters the membrane in exchange for an oxidized ubiquinone. The program VMD was used for visualization of the structure (5).
When considering possible mechanisms of redox-linked proton translocation by complex I, some kind of conformational coupling has appeared obvious due to the long distance between the redox-active cofactors in the hydrophilic domain and the likely proton-translocating subunits in the membrane domain. The crystal structures (4, 10) revealed a most uncommon long amphipathic α-helix that is part of the distal subunit L. It passes along almost the entire N surface of the membrane domain and is followed by a transmembrane helix on the proximal side of subunit N, thus “wrapping up” the three antiporter-like subunits. This helix has been suggested to communicate redox-dependent conformational changes in the hydrophilic domain to the membrane subunits (4, 10).
Here we first reexamine the stoichiometry of redox-coupled proton translocation in complex I and then explore the mechanistic implications of a pumping efficiency revised down from four to three protons per two electrons (i.e., during one turnover). This down-revision is based on energetic arguments, which set an upper limit on the number of protons that can be translocated per cycle without violation of the first law of thermodynamics and on the recently established H+/ATP ratio in ATP synthesis derived from structural information on the c-ring of ATP synthase (11). We then explore the functional implications of the lower ratio of 3 H+/2 e- and outline a rough mechanistic model of proton pumping by complex I.
Results and Discussion
The Mechanistic Proton Translocation Stoichiometry.
The stoichiometry of proton translocation by complex I has for a long time been known to exceed the 2 H+/2 e- of Mitchell’s original redox loop principle (12). The groups of Lehninger (13) and Azzone (14) reported ratios approaching 4 H+/2 e- by measuring proton release and charge translocation in mitochondria. Wikström (15) compared intramitochondrial alkalinization and charge translocation during succinate oxidation by ferricyanide (complex III alone) with that during oxidation of β-hydroxybutyrate by ferricyanide (complex I plus complex III) in rat-liver mitochondria and reported 2.7–2.8 times higher values in the latter case. Because the bc1 complex is known to effectively translocate 2 H+/2 e-†, the corresponding number for complex I would be 3.4–3.6, from which a stoichiometry of 4 H+/2 e- was inferred. Later, Galkin et al. (16) measured proton uptake coupled to NADH oxidation by ubiquinone in bovine heart submitochondrial particles and reported a stoichiometry of 4 ± 0.05. Somewhat lower values were reported for mitochondria from the yeast Yerrowia lipolytica, and for Yerrowia complex I reconstituted into liposomes (17) but nevertheless interpreted as 4 H+/2 e-.
Whereas the stoichiometry of 4 H+/2 e- has thus been widely accepted, some skepticism might be warranted for the reason that the proton-translocating membrane arm of complex I contains three proton/cation antiporter-like subunits (see above), so that a mechanistic stoichiometry of three protons per oxidized NADH (or per turnover) may seem more plausible. On the other hand, the recent crystal structure of the membrane domain shows features outside the three antiporter-like subunits that were interpreted as a possible additional transmembrane proton transfer pathway (18). In the following, we take a new and independent look at this problem.
Thermodynamic Constraints.
The redox potential span between the electron donor (NADH) and the acceptor (ubiquinone) provides the driving force for proton translocation by complex I. On the basis of literature data for the measured redox poises of NAD and ubiquinone during state 4 oxidation of β-hydroxybutyrate by rat-liver mitochondria (19), Hinkle et al. (20) calculated a redox span of ΔEh ∼ 359 mV.
Because the driving redox reaction is the only energy source for the proton translocation coupled to it, energy conservation imposes a strict upper bound on the number n of protons translocated per electron in steady state,
| [1] |
where ΔμH+ is the proton electrochemical gradient. ΔEh - nΔμH+ is the potential lost to dissipation in complex I energy transduction, such that equality in Eq. 1 is achieved if the reaction proceeds without any dissipation. The stoichiometric ratio n under typical operating conditions must thus be strictly smaller than the bound in Eq. 1; how much smaller depends on the efficiency of coupling and the extent of leaks.
It is well established that the ΔμH+ of well-coupled rat-liver mitochondria respiring on β-hydroxybutyrate in state 4 is of the order of approximately 220–230 mV (20, 21). Inserting this value and the experimentally observed ΔEh of 359 mV in Eq. 1 yields a maximum H+/e- stoichiometry of n < 1.56–1.63 (i.e., a maximum H+/2 e- ratio of approximately 3.1–3.3). It is noteworthy that the widely accepted H+/2 e- ratio of 4 would require a driving force larger than approximately 440–460 mV if the measured range of ΔμH+ were considered accurate, implying a > 25% error in determining ΔEh. Alternatively, ΔμH+ must be lower than 180 mV in state 4 mitochondria if the determination of ΔEh is accepted.
In the above considerations the mechanistic proton pump stoichiometry (n) was assumed to be constant. Measurements of the H+/2 e- ratio are usually done under conditions where the ΔμH+ is very small (i.e., when there is virtually no force opposing proton translocation). If n can vary depending on the conditions, it is therefore possible that it is decreased at high ΔμH+—i.e., in the conditions of mitochondria respiring in state 4. It is noteworthy that under phosphorylating, state 3, conditions ΔμH+ falls to values near 170 mV (20, 21). In such conditions ΔEh is shifted to approximately 364 mV during respiration on β-hydroxybutyrate (calculated from data in ref. 19), in which case the maximum value of n is 2.14, and a maximum H+/2 e- ratio of 4 is possible. For this reason it will be important to obtain an independent measure of the H+/2 e- ratio under conditions of active ATP synthesis [i.e., in state 3 mitochondria (see below)].
De Jonge and Westerhoff (22) measured ΔEh and ΔμH+ in submitochondrial particles in near-equilibrium conditions of ATP-induced reversed electron transfer from succinate (via complex II) to NAD+, as catalyzed by complex I. Here again, Eq. 1 sets a boundary in steady state, and the authors found the mean value of n to be 1.6, which they interpreted as a H+/2 e- stoichiometry of 3 for complex I. In their work also the free energy of ATP synthesis (the phosphorylation potential, ΔGp) was determined, viz.
| [2] |
which is the driving force of the reversed electron transfer in these experiments. ΔGp,o is the free energy of ATP synthesis at standard conditions. Here, the equivalent of Eq. 1, viz.
| [3] |
is valid near equilibrium between ATP hydrolysis and protonmotive force, in which case m is the H+/ATP stoichiometry of the F1Fo ATP synthase. The mean and standard deviation of the three values of m reported in this work are 2.73 ± 0.32 (22). As the authors pointed out, the m obtained in this way is a maximum value, which at the time made them suggest that the H+/ATP stoichiometry is 2. In retrospect, their observation is quite remarkable because the true H+/ATP stoichiometry is 2.67, as we shall see below.
Finally, we note that Bogachev et al. (23), working on whole Escherichia coli cells, reported an H+/e- ratio of “not less than 1.5” for complex I (i.e., ≥3 H+/2 e-).
Making use of the Measured ATP/2 e- Ratios.
In this section, we combine detailed structural information on FoF1-ATP synthase with literature values on mitochondrial P/O ratios to infer the H+/2 e- stoichiometry of complex I in state 3 conditions. Recently, Watt et al. (11) made the important discovery that the membrane Fo domain of the FoF1-ATP synthase has eight c subunits in all animal mitochondria, from which follows that eight protons are transferred across the membrane domain for each full rotation of the γ subunit. Because the catalytic F1 domain is an α3β3 trimer, a full rotation of the γ subunit results in the synthesis of 3 ATP molecules. This finding therefore settles the intrinsic H+/ATP stoichiometry at 8/3 = 2.67 in animal mitochondria. This ratio is also the relevant stoichiometry in submitochondrial particles, where the catalytic F1 domain faces the external medium. In mitochondria the situation is different, because transport of ADP3- into the mitochondrial matrix in exchange for ATP4- is associated with translocation of one electrical charge equivalent, and transport of the inorganic phosphate monoanion (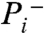 ) into the matrix is linked to cotransport of a proton (electroneutrally). Hence, the “cost” of making an extramitochondrial ATP from extramitochondrial ADP and Pi requires the translocation of 2.67 + 1 = 3.67 H+ ions.
) into the matrix is linked to cotransport of a proton (electroneutrally). Hence, the “cost” of making an extramitochondrial ATP from extramitochondrial ADP and Pi requires the translocation of 2.67 + 1 = 3.67 H+ ions.
The settlement of the (extramitochondrial) H+/ATP ratio to 3.67 in animal mitochondria opens up a new and independent test of the H+/2 e- ratios of proton translocation in the respiratory chain, because we can now make use of the extensive literature data on ATP/2 e- (P/O) ratios in mitochondria. This is because
| [4] |
i.e., the measured ATP/2 e- ratio for a certain span of the respiratory chain encompasses independent information on the proton translocation stoichiometry of that redox span. When determined in this way, the resulting H+/2 e- ratio is a minimum number relative to the true stoichiometry, because some proton leakage is likely between the generator and consumer of protonmotive force. Here, we assume that one full rotation of the γ subunit of the FoF1-ATP synthase results in the synthesis of exactly 3 ATP molecules, not less. Eq. 4 then yields the minimum H+/2 e- ratio under conditions of active, phosphorylating state 3 respiration as called for above.
Hinkle (24) has summarized virtually all the ATP/2 e- ratios reported in the literature and provided a valuable discussion of the different sources of error in these measurements. Here, we make use of the ATP/2 e- measurements published earlier by Hinkle et al. (20), again using rat-liver mitochondria for consistency. Table 1 summarizes this data, which was reported for the entire respiratory chain (β-hydroxybutyrate oxidation by O2 (i.e., complexes I + III + IV), for succinate oxidation by O2 (complexes III + IV), for succinate oxidation by ferricyanide (complex III), and for the oxidation of TMPD (+ ascorbate) by O2 (complex IV), in each case under conditions in which contribution of other parts of the chain was duly blocked by specific inhibitors.
Table 1.
Conversion of measured ATP/2 e- ratios to proton translocation stoichiometries in respiratory chain of rat-liver mitochondria
| Complex | ATP/2 e- | H+/2 e- (efficiency,%) |
| I + III + IV | 2.27 ± 0.08 | 8.32 (92%) |
| III + IV | 1.48 ± 0.04 | 5.43 (91%) |
| III | 0.49 ± 0.02 | 1.80 (90%) |
| IV | 0.98 ± 0.09 | 3.59 (90%) |
| I (I + III + IV minus III + IV) | 0.79 | 2.90 (97%) |
| I (I + III + IV minus III minus IV) | 0.80 | 2.90 (97%) |
The ATP/2 e- ratios are from Hinkle et al. (20) for rat-liver mitochondria oxidizing β-OH-butyrate (I + III + IV) or succinate (III + IV) by O2, succinate by ferricyanide (III), or TMPD (+ascorbate) by O2 (IV). The H+/2 e- ratios are derived from the ATP/2 e- ratio using Eq. 4 and a value of 3.67 for the H+/ATP ratio. Per cent efficiency is reported for the assumption that the effective mechanistic H+/2 e- stoichiometries† are 3, 2, and 4, respectively, for complexes I, III, and IV (i.e., 9 for I + III + IV and 6 for III + IV). The two ATP/2 e- ratios for complex I are derived from the measured ratios, as indicated.
Table 1 also lists the corresponding H+/2 e- ratios based on Eq. 4, and the value in parentheses gives the stoichiometric efficiency assuming that the effective H+/2 e- ratio† is 4, 2, and 3, respectively, for complexes IV, III, and I. Note that the H+/2 e- value for I + III + IV approaches 9 with an efficiency similar to those of the other combinations that do not include complex I. If the mechanistic H+/2 e- ratio were 4 for complex I, the ratio would be 10 for I + III + IV and the efficiency only 83% (i.e., very different from the other combinations that do not include complex I).
Note also that subtracting the ATP/2 e- ratio for the III + IV combination (1.48) from that of the I + III + IV combination (2.27) yields an ATP/2 e- ratio of 0.79 for complex I. In good agreement with this, subtracting the individual ATP/2 e- values of complexes III (0.49) and IV (0.98) from the value for I + III + IV (2.27) yields 0.80 for complex I (Table 1). An ATP/2 e- ratio of approximately 0.8 corresponds to a minimum H+/2 e- ratio of 2.9, which is 97% of 3.0 (but 73% of 4.0). A 97% coupling efficiency for complex I, when considered together with the individual coupling efficiencies of complexes III and IV, indeed agrees very well with the efficiency of 92 % for the combined function of I + III + IV (Table 1). This analysis is also in agreement with the previous general notion that complex I is a particularly efficient redox-linked proton pump (i.e., with minimal energy dissipation).
From this data it is clear that in the phosphorylating state 3 of mitochondria, the H+/2 e- ratio of complex I still seems to be 3, even though the thermodynamics would now allow a stoichiometry of 4 due to the lowered ΔEh/ΔμH+ ratio relative to state 4.
Uptake and Release of Protons Linked to the Redox Chemistry.
The chemistry of the reaction catalyzed by complex I comprises oxidation of the two-electron donor NADH bound to a site near the cofactor FMN in the polar domain of the complex on the N side of the membrane (1–3). Beyond FMN there is a chain of purely electron-carrying iron-sulfur centers, which ends up at the Fe/S cluster N2 nearest to the membrane. This arrangement implies that the proton release associated with oxidation of NADH will occur on the N side (i.e., the mitochondrial matrix or the bacterial cytoplasm; see Fig. 1). We note here that a second proton will be released on the N side upon rereduction of NAD+ to NADH, as catalyzed by various dehydrogenases. The electrons will finally reduce ubiquinone (Q), and the two protons required for this must be taken from the N side (Fig. 1), because protonmotive force would be dissipated if they were taken from the P side. Thus, the proton release and uptake due to the redox chemistry will not contribute directly to the creation of protonmotive force by complex I. Yet, the proton uptake that is linked to the ubiquinone reduction chemistry may nevertheless be an important component of the proton-pumping mechanism (cf below).
Ubiquinone.
In the complex I structure a fairly well-established ubiquinone binding-site lies in a cavity near cluster N2, but this site lies some 25 Å away from the N-side surface of the membrane (Fig. 1) (1–3). Apparently, it is Q in this site that can form a fast-relaxing semiquinone radical in conditions of high protonmotive force, because this radical has been shown to lie at an approximately 12 Å distance from cluster N2 based on the measured magnetic interaction between the two (25). Complex I isolated from both E. coli (26) and bovine heart mitochondria (27) contain a single ubiquinone molecule per FMN, which is presumably bound to this site. The redox potential of the Q/Q- couple for the bound Q in complex I has been estimated to be more negative than -300 mV from rapid freeze experiments (26) and theoretical calculations (28). In agreement with this, the Em,7 of the Q/Q- couple is -240 mV (versus NHE) in 80% ethanol (29), and the half-reduction potential (E1/2) is -361 mV in dimethylformamide (30). Overall, this would mean that there is virtually no drop in redox potential between the NAD+/NADH and the bound Q/Q- couples, from which follows that the entire energy drop occurs beyond transfer of the first electron to the bound ubiquinone.
We envisage two major possibilities. In the first, the single Q bound near cluster N2 becomes half-reduced and fully reduced, after which it diffuses into the membrane domain in exchange for an oxidized Q from the membrane. In this scenario, it would be the half-reduced anionic semiquinone that would initiate the events of proton translocation.
Alternatively, the situation might be analogous to bacterial reaction centers (and photosystem II) with two quinone binding sites (A and B; ref. 31) of which one (B) exchanges readily with the quinone pool in the membrane (32; see Fig. 1). There is indeed some evidence for ubiquinone binding to those four membrane subunits that reside in the “heel” of the boot-like structure of complex I (33–37). It is possible, therefore, that the more tightly bound ubiquinone (QA) in the cavity beneath cluster N2 accepts the electrons from N2, and delivers them further to another more loosely bound quinone (QB) nearer to the membrane proper, which readily exchanges with the membrane pool (Fig. 1). In this case, QA may shuttle between the QA and  states, whilst double reduction and protonation may occur only at QB (Figs. 1 and 2; see below).
states, whilst double reduction and protonation may occur only at QB (Figs. 1 and 2; see below).
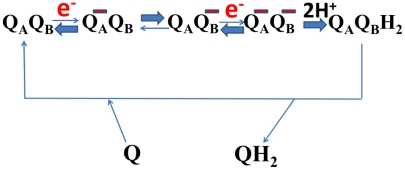
Fig. 2.
Proposed oxidoreduction function of two ubiquinone molecules in complex I. QA is the ubiquinone proposed to be bound near iron-sulfur cluster N2, whereas QB is another ubiquinone bound more weakly near the membrane domain (see Fig. 1). QA is insulated from protonation and its one-electron reduction is thermodynamically unfavorable (see text). Uptake of the second electron to yield the doubly negatively charged pair is again unfavorable. The doubly charged ubiquinone pair is proposed to drive conformational changes in the LMN subunit triad (see Fig. 3 A and B), which are reversed on protonation of QB. Reduced QB then exchanges with the membrane pool for oxidized Q.
Mechanistic Principles.
Too little is known as yet in order to formulate a detailed mechanism of proton-pumping by complex I, but if the H+/2 e- stoichiometry of proton translocation is indeed 3, the pumping mechanism can hardly be composed of two identical power strokes coupled to two one-electron redox events involving the same ubiquinone molecule (i.e., Q + e- → Q- and Q- + e- + 2H+ → QH2) (38). Assuming such a two-stroke mechanism, a fractional stoichiometric efficiency with 1.5 H+ pumped per stroke is necessarily a result of an average over an ensemble of enzymes, half of which pump two protons per stroke and the other half only one. Such a variation in the stoichiometry of individual enzymes must be the result of slippage in the pump mechanism and should thus be suppressed in an efficient energy transducer. Moreover, by virtue of being at the stoichiometric midpoint between approximately 50% each of 2 H+/e- and 1 H+/e- power strokes, such variations would make the pump stoichiometry sensitive to small perturbations in transmembrane potential and the pH on both sides of the membrane. In contrast, for a robust pump whose stoichiometry is insensitive to small changes in conditions we expect a nearly integer stoichiometry on average.
The 3 H+/2 e- stoichiometry thus suggests that proton translocation as catalyzed by the LMN triad is coupled to a single redox transition. The three homologous antiporter-like subunits L, M, and N may undergo a set of conformational changes, each taking up one proton from the N side of the membrane and releasing it on the P side, as proposed by Sazanov et al. (4, 18). We suggest that the three membrane subunits cooperate tightly with one another in synchronous reaction steps of proton uptake, reorientation of proton binding sites, and proton release (Fig. 3 A and B).
Fig. 3.
Two possible scenarios for proton pumping by complex I. The corners of the cube represent eight different states distinguished by (i) the cooperative sidedness of the proton-transferring groups of subunits L, M, and N (x axis; P or N connectivity), (ii) the presence or absence of negative charge at the catalytic ubiquinone(s) (y axis; red arrows indicating electron uptake), and (iii) the protonation state of the key groups in subunits L, M, and N (z axis; uptake and release of H+). The four P and N states have protonic connectivity to the P and N side of the membrane, respectively. In case A, the orientation with proton access from the N side has the lowest energy in the absence of electrons or protons at the respective active sites (state 1), and the protonatable groups in the L, M, and N subunits have a pKa well above the pH of the aqueous N phase. In case B, the lowest energy state (state 1) has the L, M, and N subunits protonically accessible to the P side, and electron transfer to the quinone(s) is required for the switch to the N orientation. Note that the states marked with the red minus sign correspond to those where electron transfer to ubiquinone yields net negative charge that drives conformational transitions in the L, M, and N subunits (i.e., state 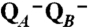 in Fig. 2).
in Fig. 2).  refers to the chemical protons taken up into reduced QB to form the quinol, QH2, whereby the negative charge is neutralized. We stress that cycles A and B here are only two examples out of several possible ones.
refers to the chemical protons taken up into reduced QB to form the quinol, QH2, whereby the negative charge is neutralized. We stress that cycles A and B here are only two examples out of several possible ones.
A key question regarding such a mechanism concerns the means by which the driving force from the redox reaction is transmitted to the set of LMN subunits in the membrane arm. Euro et al. (9) suggested that the nature of the transmission might be a cooperative two-phase electrostatic process that may be initiated by the negative charge of the ubisemiquinone anion (or the quinol anion, Q2- or QH-) bound at the ubiquinone binding site and transmitted to the membrane arm, causing a cooperative conformational change in the L, M, and N subunits. The second phase could be initiated by quenching of the negative charge at the quinone by its protonation. We think it is doubtful that the long α-helix of subunit L is involved as a mechanical piston in transmitting the conformational changes (4, 18), because extensive softening of the structure of this helix by site-directed mutagenesis had generally little effect on activity (39). As a possible alternative, the long α-helix might have a stabilizing effect on the LMN heterotrimer, clamping it together during its conformational transitions (39).
Based on the recent structure (18), the LMN subunits are very tightly coupled. It is therefore conceivable that mechanical tension in response to the buildup of uncompensated charge at the Q-binding site(s) is released by a concerted conformational change in all three subunits. With regard to how ubiquinone is involved more precisely in the mechanism, we currently favor the QA/QB hypothesis discussed above, which could lead to the reaction sequence described in Fig. 2. Negative charge is built up by electron transfer to form the 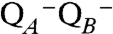 state, which triggers a concerted conformational transition in the LMN triad that changes the sidedness of proton accessibility and ultimately results in the uptake and release of the pumped protons. This conformational change also alters the structure around the Q site(s) to permit the uptake of the two substrate protons from the N side of the membrane, which then neutralize the charge. By this, the electrostatic imbalance driving the conformational change is reversed, and reduced quinol in the B site exchanges for an oxidized quinone from the membrane pool. Fig. 3
A and B uses a cubic reaction diagram to show two possible reaction sequences of such a redox-linked proton pump.
state, which triggers a concerted conformational transition in the LMN triad that changes the sidedness of proton accessibility and ultimately results in the uptake and release of the pumped protons. This conformational change also alters the structure around the Q site(s) to permit the uptake of the two substrate protons from the N side of the membrane, which then neutralize the charge. By this, the electrostatic imbalance driving the conformational change is reversed, and reduced quinol in the B site exchanges for an oxidized quinone from the membrane pool. Fig. 3
A and B uses a cubic reaction diagram to show two possible reaction sequences of such a redox-linked proton pump.
Finally, we note that Sazanov et al. (18) indicated possible proton translocation channel structures not only in the L, M, and N subunits but also at the interface of subunits N, K, J, and A (i.e., in the “heel” of the boot structure of complex I) with the idea that these latter structures might be responsible for translocation of “the fourth proton.” Whereas our analysis above provides strong evidence for pumping of three rather than four protons per turnover under phosphorylating, state 3, conditions of mitochondria, it cannot exclude a 4 H+/2 e- stoichiometry under conditions of very low protonmotive force. Such conditions are indeed typical in the direct measurements of the pump stoichiometry where the observed ratio has significantly exceeded 3 (13–17) A possible scenario is, therefore, that the N, K, J, and A subunits in the heel serve as a variable transmission: At low ΔEh/ΔμH+ ratios it does not function; at high ratios, it facilitates translocation of a fourth proton per cycle (Fig. 1). Considering that the electron transfer function of complex I is responsible for most of the electron input into the respiratory chain in many organisms, it might indeed be advantageous to regulate the degree of coupling between electron flux and proton translocation.
Electron flux in the respiratory chain is largely limited by the flux catalyzed by complex I. An overly tight coupling between quinone reduction and proton pumping to maximize the H+/e- stoichiometry might thus endanger the overall electron flux in the entire respiratory chain. Such an obstruction would be expected to favor formation of reactive oxygen species, mainly superoxide (40). Limiting the H+/2 e- ratio to 3 during active oxidative phosphorylation in mitochondria, even though the thermodynamic constraints would allow a ratio of 4 (see above), might also be important to optimize mitochondrial power output (i.e., the energy transduced per unit time) at the expense of stoichiometric efficiency (41).
Conclusions
In order to put a sharper bound on the stoichiometry of the proton pump in complex I, we exploited the fact that the function of this molecular machine is tightly coupled to the operation of the other enzymes in the respiratory chain. Remarkably, recent structural information about the rotary motor ATP synthase driven by the protons pumped in complex I contains critical information on the complex I stoichiometry. This holistic view, in which coupled energy-transducing elements are analyzed together, not separately, should prove useful also for other processes involving multiple molecular machines.
A reexamination of a wide range of literature data suggests that complex I operates with a stoichiometry of proton translocation of 3 H+/2 e- (i.e., lower than the commonly accepted value of 4 H+/2 e-). This reduced stoichiometry is consistent with the recently determined H+/ATP ratio of the mitochondrial F1Fo-ATP synthase of animal mitochondria and of the measured ATP/2 e- ratios for different segments of the mitochondrial respiratory chain. Mechanistically, a reduced stoichiometry of 3 protons per cycle could be achieved in a set of concerted proton translocation steps involving all three of the homologous antiporter-like subunits L, M, and N, possibly driven by the buildup of uncompensated charge in the Q-binding sites. While we currently do not have any more specific evidence in support of such a model, we hope that this discussion will stimulate further studies of complex I as a functionally important molecular machine responsible for primary energy transduction in cell respiration.
Acknowledgments.
G.H. is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. M.W. is supported by grants from the Sigrid Jusélius Foundation, Biocentrum Helsinki, and the Academy of Finland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
*We use the subunit nomenclature of the E. coli complex I.
†For cytochrome bc1 (Complex III) and cytochrome c oxidase (Complex IV) these stoichiometries do not correspond to the number of protons released on the P side of the membrane (which are four and two per 2e-, respectively) but to the thermodynamically effective stoichiometry, which is the relevant entity considering the driving force for ATP synthesis. Thus, for each two electrons transferred, Complex IV translocates four electrical charges across the membrane and takes up four protons from the N side of the membrane, two of which are released on the P side. This is thermodynamically equivalent to translocation of four protons, because proton release on the P side will not significantly contribute to the concentration term of the protonmotive force (ΔpH). The inverse is true for Complex III. Here, for every two electrons, two protons are taken up from the N side, and four are released on the P side, which is thermodynamically equivalent to translocation of two protons.
References
- 1.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 2.Tocilescu MA, Zickermann V, Zwicker K, Brandt U. Quinone binding and reduction by respiratory complex I. Biochim Biophys Acta. 2010;1797:1883–1890. doi: 10.1016/j.bbabio.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Hirst J. Towards the molecular mechanism of respiratory complex I. Biochem J. 2010;425:327–339. doi: 10.1042/BJ20091382. [DOI] [PubMed] [Google Scholar]
- 4.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–447. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 5.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 6.Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 7.Amarneh B, Vik S. Mutagenesisof subunit N of the Escherichia coli complex I identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry. 2003;42:4800–4808. doi: 10.1021/bi0340346. [DOI] [PubMed] [Google Scholar]
- 8.Torres-Bacete J, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T. Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1. Conserved charged residues essential for energy-coupled activities. J Biol Chem. 2007;282:36914–36922. doi: 10.1074/jbc.M707855200. [DOI] [PubMed] [Google Scholar]
- 9.Euro L, Belevich G, Verkhovsky MI, Wikström M, Verkhovskaya ML. Conserved lysine residues of the membrane subunit nuo M are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (Complex I) Biochim Biophys Acta. 2008;1777:1166–1172. doi: 10.1016/j.bbabio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Hunte C, Zickermann V, Brandt U. Functional modules and the structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 11.Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation, Glynn Research Ltd, Bodmin, Cornwall, 1966. Biochim Biophys Acta. 2011;1807:1507–1538. doi: 10.1016/j.bbabio.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Reynafarje B, Lehninger AL. The K+/site and H+/site stoichiometry of mitochondrial electron transport. J Biol Chem. 1978;253:6331–6334. [PubMed] [Google Scholar]
- 14.Pozzan T, Miconi V, Di Virgilio F, Azzone GF. H+/site, charge/site, and ATP/site ratios at coupling sites I and II in mitochondrial e- transport. J Biol Chem. 1979;254:10200–10205. [PubMed] [Google Scholar]
- 15.Wikström M. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 1984;169:300–304. doi: 10.1016/0014-5793(84)80338-5. [DOI] [PubMed] [Google Scholar]
- 16.Galkin AS, Grivennikova VG, Vinogradov AD. H+/2 e- stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 1999;451:157–161. doi: 10.1016/s0014-5793(99)00575-x. [DOI] [PubMed] [Google Scholar]
- 17.Galkin A, Dröse S, Brandt U. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim Biophys Acta. 2006;1757:1575–1581. doi: 10.1016/j.bbabio.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 19.Muraoka S, Slater EC. The redox states of respiratory chain components in rat-liver mitochondria II. The “crossover” on the transition from state 3 to state 4. Biochim Biophys Acta. 1969;180:227–236. doi: 10.1016/0005-2728(69)90109-1. [DOI] [PubMed] [Google Scholar]
- 20.Hinkle PC, Kumar MA, Resetar A, Harris DL. Mechanistic stoichiometry of mitochondrial oxidative phosphorylation. Biochemistry. 1991;30:3576–3582. doi: 10.1021/bi00228a031. [DOI] [PubMed] [Google Scholar]
- 21.Nicholls DG. The influence of respiration and ATP hydrolysis on the proton-electrochemical gradient across the inner membrane of rat-liver mitochondria as determined by ion distribution. Eur J Biochem. 1974;50:305–315. doi: 10.1111/j.1432-1033.1974.tb03899.x. [DOI] [PubMed] [Google Scholar]
- 22.De Jonge PC, Westerhoff HV. The proton-per-electron stoichiometry of ‘site 1’ of oxidative phosphorylation at high protonmotive force is close to 1.5. Biochem J. 1982;204:515–523. doi: 10.1042/bj2040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogachev AV, Murtazina RA, Skulachev VP. H+/e- stoichiometry for NADH dehydrogenase I and dimethyl sulfoxide reductase in anaerobically grown Escherichia coli cells. J Bacteriol. 1996;178:6233–6237. doi: 10.1128/jb.178.21.6233-6237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinkle PC. P/O ratios of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2005;1706:1–11. doi: 10.1016/j.bbabio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Yano T, Dunham WR, Ohnishi T. Characterization of the ΔμH+-sensitive ubisemiquinone species (SQNf) and the interaction with cluster N2: New insight into the energy-coupled electron transfer in Complex I. Biochemistry. 2005;44:1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- 26.Verkhovskaya M.L, Belevich N, Euro L, Wikström M, Verkhovsky MI. Real-time electron transfer in respiratory complex I. Proc Natl Acad Sci USA. 2008;105:3763–3767. doi: 10.1073/pnas.0711249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinzawa-Itoh K, et al. Bovine heart NADH-ubiquinone oxidoreductase contains one molecule of ubiquinone with ten isoprene units as one of the cofactors. Biochemistry. 2010;49:487–492. doi: 10.1021/bi9016318. [DOI] [PubMed] [Google Scholar]
- 28.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Rich PR. Electron and proton transfers through quinones and cytochrome bc complexes. Biochim Biophys Acta. 1984;768:53–79. doi: 10.1016/0304-4173(84)90007-7. [DOI] [PubMed] [Google Scholar]
- 30.Prince RC, Dutton PL, Bruce JM. Electrochemistry of ubiquinones, menaquinones and plastoquinones in aprotic solvents. FEBS Lett. 1983;160:273–276. [Google Scholar]
- 31.Wraight CA. In: Biophysical and Structural Aspects of Bioenergetics, Intraprotein Proton Transfer—Concepts and Realities from the Bacterial Photosynthetic Reaction Center. Wikström M, editor. Cambridge, UK: RSC Publishing; 2005. pp. 273–313. [Google Scholar]
- 32.Ohnishi ST, Salerno JC, Ohnishi T. Possible roles of two quinone molecules in direct and indirect proton pumps of bovine heart NADH-quinone oxidoreductase (complex I) Biochim Biophys Acta. 2010;1797:1891–1893. doi: 10.1016/j.bbabio.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao MC, et al. Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J Biol Chem. 2004;279:32360–32366. doi: 10.1074/jbc.M403885200. [DOI] [PubMed] [Google Scholar]
- 34.Kao MC, et al. Characterization of the membrane domain subunit NuoJ (ND6) of the NADH quinone oxidoreductase from Escherichia coli by chromosomal DNA manipulation. Biochemistry. 2005;44:3562–3571. doi: 10.1021/bi0476477. [DOI] [PubMed] [Google Scholar]
- 35.Kao MC, Nakamaru-Ogiso E, Matsuno-Yagi A, Yagi T. Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry. 2005;44:9545–9554. doi: 10.1021/bi050708w. [DOI] [PubMed] [Google Scholar]
- 36.Kervinen M, Pätsi J, Finel M, Hassinen IE. A pair of membrane-embedded acidic residues in the NuoK subunit of Escherichia coli NDH-1, a counterpart of the ND4L subunit of the mitochondrial complex I, are required for high ubiquinone reductase activity. Biochemistry. 2004;43:773–781. doi: 10.1021/bi0355903. [DOI] [PubMed] [Google Scholar]
- 37.Schuler F, Casida JE. Functional coupling of PSST and ND1 subunits in NADH: Ubiquinone oxidoreductase established by photoaffinity labeling. Biochim Biophys Acta. 2001;1506:79–87. doi: 10.1016/s0005-2728(01)00183-9. [DOI] [PubMed] [Google Scholar]
- 38.Brandt U. A two-state stabilization-change mechanism for proton-pumping complex I. Biochim Biophys Acta. 2011;1807:1364–1369. doi: 10.1016/j.bbabio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Belevich G, Knuuti J, Verkhovsky MI, Wikström M, Verkhovskaya M. Probing the mechanistic role of the long α-helix in subunit L ofrespiratory Complex I from Escherichia coli by site-directed mutagenesis. Mol Microbiol. 2011;82:1086–1095. doi: 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kussmaul L, Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci USA. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stucki JW. The optimal efficiency and the economic degrees of coupling of oxidative phosphorylation. Eur J Biochem. 1980;109:269–283. doi: 10.1111/j.1432-1033.1980.tb04792.x. [DOI] [PubMed] [Google Scholar]