Abstract
Several classes of naturally occurring antimicrobials exert their antibiotic activity by specifically targeting aminoacyl-tRNA synthetases, validating these enzymes as drug targets. The aspartyl tRNA synthetase “Trojan horse” inhibitor microcin C7 (McC7) consists of a nonhydrolyzable aspartyl-adenylate conjugated to a hexapeptide carrier that facilitates active import into bacterial cells through an oligopeptide transport system. Subsequent proteolytic processing releases the toxic compound inside the cell. Producing strains of McC7 must protect themselves against autotoxicity that may result from premature processing. The mccF gene confers resistance against endogenous and exogenous McC7 by hydrolyzing the amide bond that connects the peptide and nucleotide moieties of McC7. We present here crystal structures of MccF, in complex with various ligands. The MccF structure is similar to that of dipeptide ld-carboxypeptidase, but with an additional loop proximal to the active site that serves as the primary determinant for recognition of adenylated substrates. Wild-type MccF only hydrolyzes the naturally occurring aspartyl phosphoramidate McC7 and synthetic peptidyl sulfamoyl adenylates that contain anionic side chains. We show that substitutions of two active site MccF residues result in a specificity switch toward aromatic aminoacyl–adenylate substrates. These results suggest how MccF-like enzymes may be used to avert various toxic aminoacyl–adenylates that accumulate during antibiotic biosynthesis or in normal metabolism of the cell.
Keywords: tRNA synthetase inhibitor, self-immunity, reaction mechanism, protein engineering
The influx of genome sequence data has resulted in the identification of various targets in pathogenic species for antibiotic development. Among promising targets are the 20 aminoacyl-tRNA synthetases (aaRSs) that are essential for protein synthesis. Aminoacyl-tRNA synthetases catalyze the attachment of the correct amino acid to cognate tRNA and thus are fundamental to translation of the genetic code (1, 2). The acylation reaction first involves the formation of an enzyme bound aminoacyl–adenylate complex, followed by the nucleophilic attack of the 2′ or 3′ hydroxyl of the terminal ribose of the tRNA on the adenylate to form the aminoacyl-tRNA and adenosine monophosphate (1). Inhibition of either one of these two enzymatic steps disrupts tRNA charging, which would stall elongation of growing polypeptide chains, followed by limited bacterial growth and attenuation of virulence in vivo (3).
One subset of aaRS inhibitors consists of nonhydrolyzable analogs of the aminoacyl–adenylate intermediate that is generated during the aaRS reaction. However, synthetic aminoacyl–adenylates analogs are poor drug candidates because (i) they inhibit human aaRSs, and (ii) most lack in vivo activity due to impermeability into bacterial cells (4). Nature has overcome both of these limitations by employing a Trojan horse strategy, in which the drug molecule is conjugated with a specialized module that facilitates active transport into bacterial cells where antibiotic activity is exerted after intracellular processing. Examples of naturally occurring antibiotics that employ this Trojan horse strategy include the LeuRS inhibitor agrocin 84 (in which the toxic group is linked through a phosphoramidate bond to a d-glucofuranosyloxyphosphoryl moiety) (5), the SerRS inhibitor albomycin (a toxic group covalently linked to a hydroxymate siderophore) (6), and the AspRS inhibitor microcin C7 (Fig. 1A, 1) (McC7; consisting of a modified aspartyl-adenylate linked to a six-residue peptide carrier).
Fig. 1.
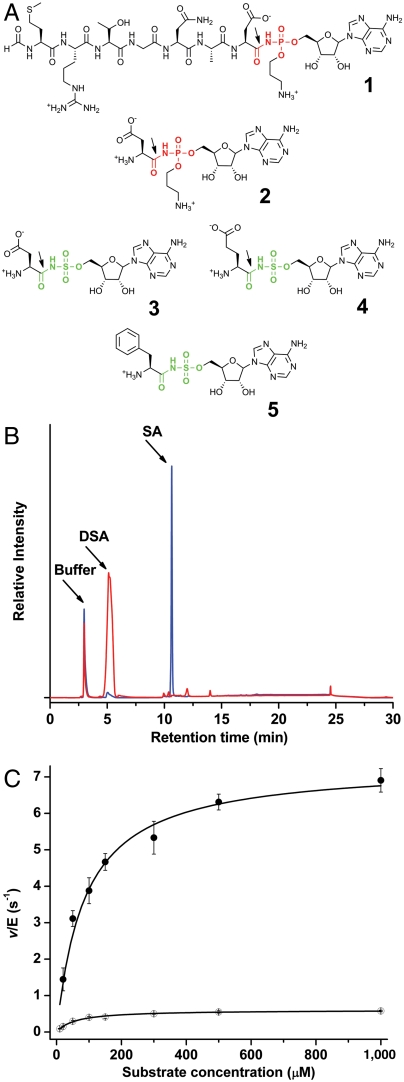
Chemical structures of microcin C7 and analogs and demonstration of MccF hydrolysis activity. (A) Chemical structures of microcin C7 (1) and its processed form (2), along with synthetic sulfamoyl adenylates of aspartate (3), glutamate (4), and phenylalanine (5). The site of hydrolysis by wild-type MccF is marked by an arrow. (B) Hydrolysis of DSA by wild-type MccF enzyme demonstrated by HPLC separation the substrate and reaction product, sulfamoyl adenosine. (C) Michaelis–Menten kinetic curves for the hydrolysis of DSA (●) and ESA (○) by wild-type MccF enzyme.
Following active transport into target cells through the YejABEF transporter, the McC7 heptapeptide precursor is processed by nonspecific cellular aminopeptidases to release the toxic compound, processed McC7 (7) (Fig. 1A, 2). Producing organisms must protect themselves against the premature, unwanted processing that can result in toxicity. In the case of agrocin 84 and albomycin, the antibiotic biosynthesis clusters encode additional tRNA synthetases (LeuRS in the case of agrocin 84, SerRS in the case of albomycin), which are resistant to inhibitors. The McC7 biosynthetic cluster does not encode a tRNA synthetase but instead, employs specialized enzymes that act on processed and/or intact McC7 in the producing cells. One such autoimmunity determinant is encoded by the mccF gene, whose product proteolytically hydrolyzes the amide bond connecting the terminal aspartate and modified AMP in McC7 (8). MccF was shown to inactivate both intact and processed forms of McC7. The enzyme functions only on substrates that bear an acidic aspartyl (aspartyl sulfamoyl adenylate, DSA) (Fig. 1A, 3) or glutamyl sulfamoyl adenylate (ESA) (Fig. 1A, 4) moiety conjugated to adenosine, with diminished activity also observed for seryl conjugated adenosine (8).
In order to elucidate determinants of substrate recognition, we have determined the high-resolution crystal structure of wild-type MccF and of a catalytically inactive variant in complex with natural and synthetic substrates. Analysis of these structures reveals how the repertoire of substrates accepted by serine carboxypeptidases can be expanded to include nucleotide-containing peptide antibiotics through π-stacking interactions. The crystal structures, along with kinetic characterization of structure-guided mutants, provide a firm structural framework for understanding the substrate recognition, and catalysis by MccF, which are further borne out by our engineering experiments that yield an enzyme with a drastically altered substrate scope.
Results
In Vitro Kinetics of MccF-Catalyzed Reaction.
We had previously demonstrated that overexpression of the mccF gene results in McC7 resistance, and that recombinant MccF cleaved the amide bond that connects the peptidyl and nucleotidyl moieties of McC7 (8). Similar effects were also observed with the synthetic DSA and ESA (Fig. 1B). We further investigated the MccF-catalyzed reaction by determining steady-state kinetic parameters.
In order to determine the kinetic parameters for ESA hydrolysis reaction, we utilized a continuous coupled assay that monitored the formation of glutamate upon hydrolysis of ESA. Kinetic measurements for the hydrolysis of DSA were performed by direct detection of reaction product (sulfamoyl adenosine) using analytical HPLC. Steady-state kinetic parameters for the hydrolysis of processed McC7 could not be determined using either of these methods. However, as the mode of binding of processed McC7 is essentially identical to that of DSA (see below), we assume that the kinetic parameters for processed McC7 hydrolysis would be very close to those for DSA. The catalytic efficiency (kcat/Km) for the hydrolysis of DSA was 9.37E4 M-1 s-1, and is approximately eightfold greater than that for ESA (1.13E4 M-1 s-1) (Fig. 1C and Table 1). Mutation of the active site catalytic serine residue to alanine rendered the enzyme completely devoid of measurable catalytic activity. Notably, for the wild-type enzyme, no hydrolysis could be observed with sulfamoyl adenylates that contained aromatic amino acids, such as phenylalanyl sulfamoyl adenylate (FSA) (Fig. 1A, 5).
Table 1.
Steady-state kinetic parameters for DSA and ESA hydrolysis by MccF wild-type and mutant enzymes
| Km, μM | kcat, s-1 | kcat/Km, M-1 s-1 | Relative kcat/Km* | |
| DSA | ||||
| Wild type | 74.31 | 6.96 | 9.37E4 | 1.000 |
| N220A | 186.32 | 2.22 | 1.19E4 | 0.126 |
| K247A | 133.92 | 3.27 | 2.44E4 | 0.260 |
| S118A | ND | ND | ||
| ESA | ||||
| Wild type | 51.64 | 0.58 | 1.13E4 | 1.000 |
| W186F | 218.77 | 0.39 | 1.78E3 | 0.157 |
| W186A | ND | ND | ||
| R246A | 154.27 | 0.56 | 3.64E3 | 0.322 |
| N220A | 105.89 | 0.62 | 5.85E3 | 0.518 |
| K247A | 238.92 | 0.08 | 3.51E2 | 0.031 |
| N220A/K247A | ND | ND | ||
ND, no detectable activity precluded kinetic parameter determination.
*The kcat/Km of mutant enzyme relative to wild-type enzyme.
MccF is a Member of the Serine ld-Carboxypeptidase Superfamily.
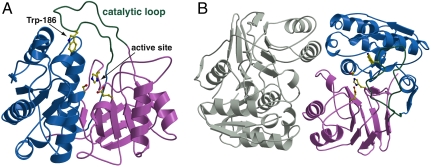
The structure of wild-type MccF has been determined to 1.2-Å resolution by single wavelength anomalous dispersion data collected from crystals of selenomethionine-labeled protein (relevant data collection statistics are given in Table S1). The structure of MccF can be divided into two distinct domains: an amino terminal region (residues Pro7–Thr164) consisting of four parallel β-sheets flanked by α-helices, and a carboxy-terminal region (Lys209–Ser341) composed of a seven-strand β-sandwich with three α-helices situated along one face of the strands. The two domains are linked through an unusually long loop (Arg165–Gly208), which we term the “catalytic loop” (Fig. 2A). Numerous contacts with the side- and main-chain atoms of both domains fix the position of the catalytic loop. Consistent with the behavior of MccF in solution as a dimer, the crystallographic asymmetric unit consists of an MccF homodimer (Fig. 2B) and a total surface area of over 3,681 Å2 is occluded upon dimerization. The overall topology of MccF is similar to ld-carboxypeptidases that cleave the linkage between meso-diaminopimelic acid and d-alanine during peptidoglycan recycling (Protein Data Bank ID code 1ZRS; Z score 14.2; rmsd of 1.9 Å over 274 aligned Cα atoms) (9). However, the catalytic loop is absent from ld-carboxypeptidases and is therefore unique to MccF (Fig. S1).
Fig. 2.
Overall three-dimensional crystal structure of MccF. (A) Structure of MccF monomer showing the three domains: amino terminal domain (blue), carboxy-terminal domain (pink), and catalytic loop (green). (B) Structure of the MccF homodimer showing the relative positions of the three domains with one monomer colored as above and the second monomer colored gray.
A catalytic triad, composed of Ser118, His311, and Glu243, is situated at the interface between the two domains (Fig. 2A), consistent with the earlier report that mutation at Ser118 results in a loss of catalytic activity (8). The side chain of Glu243 is buried in a polar environment with one of the carboxylate oxygen atoms hydrogen bonded to the Nδ1 of His311 (2.9 Å) and the second carboxylate oxygen atom hydrogen bonded to the Oη of Tyr137 (2.5 Å) and to Nδ2 of Asn218 (3.0 Å). The Nϵ2 of His311 is appropriately placed at a distance of 2.8 Å from the Ser118 side-chain hydroxyl where it can abstract the proton to generate the catalytic alkoxide anion for nucleophilic attack at the scissile peptide bond (10). Rather unexpectedly, the Ser118 side chain exists in two different conformations and a hydrogen-bonding interaction with His311 is possible in only one of these conformations.
We also determined the 1.5-Å resolution crystal structure of the MccF Ser118Ala mutant and found it to be nearly identical to the wild-type enzyme structure (Table S1). Thus, the loss of catalytic activity for the mutant is due to the loss of the catalytic serine side-chain hydroxyl nucleophile. Subsequent discussions of the substrate cocrystal complexes of the Ser118Ala MccF are descriptive of the productive state for substrate binding to the wild-type enzyme.
The Catalytic Loop Contributes Directly to Substrate Engagement.
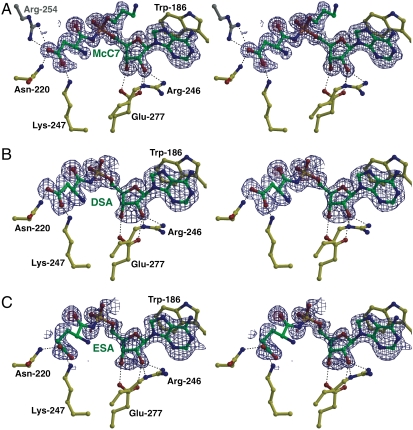
In order to delineate the molecular basis for substrate recognition, we determined the 1.3-Å resolution cocrystal structure of Ser118Ala MccF in complex with processed McC7. Difference Fourier analysis reveals unambiguous electron density corresponding to processed McC7 in the vicinity of the active site (Fig. 3A). Following the Schechter and Berger nomenclature (11) for serine proteases, we refer to the nucleotide binding site as the P1 site and the aspartyl side chain of processed McC7 occupies the P1′ site. The interactions at the P1 site are primarily modulated by π-stacking interactions of the adenine ring of McC7 against the indole side chain of Trp186, located within the catalytic loop. The 2′ and 3′ hydroxyl groups of the ribose sugar are respectively positioned 2.9- and 2.7-Å away from the side-chain carboxylate oxygen atoms of Glu277 and engage in hydrogen-bonding interactions (Fig. 3A). The 2′ hydroxyl is also positioned 2.7-Å away from guanidine side chain of Arg246, which in turn is also hydrogen bonded to Ser183 side-chain hydroxyl. At the P1′ site, one of the carboxylate oxygen atoms of processed McC7 aspartate is positioned 3.1-Å away from the Nζ of Lys247 and the other oxygen atom is 2.9-Å away from Nδ2 of Asn220. This carboxylate oxygen atom is also engaged in an interaction with the Oγ1 of Thr221 (3.2 Å) and the guanidino side chain of an alternate conformation of Arg254 from the other MccF monomer (N-O distances of 3.1 and 2.9 Å).
Fig. 3.
Stereoviews of MccF-S118A active site with substrates bound. (A) Stereoview showing the active site features of the MccF-S118A in complex with processed McC7. The MccF carbon atoms are shown in yellow ball-and-stick and the McC7 carbon atoms are colored in green. Superimposed is a difference Fourier electron density map (contoured at 2.7σ over background in blue) calculated with coefficients |Fobs| - |Fcalc| and phases from the final refined model with the coordinates of McC7 deleted prior to one round of refinement. (B) Stereoview showing the active site features of the MccF-S118A in complex with DSA. Superimposed is a difference Fourier electron density map (contoured at 2.5σ) calculated as above. (C) Stereoview showing the active site features of the MccF-S118A in complex with ESA. Superimposed is a difference Fourier electron density map (contoured at 2.5σ over background) calculated as above.
Although π-stacking interactions can provide extensive surface area for van der Waals contact, such interactions result in electrostatic repulsion as the delocalized π-electrons result in a concentration of negative electrostatic potential. However, the electrostatic potential of heterocyclic adenine and guanine nucleobases of DNA/RNA differ markedly from that of homocyclic conjugated systems such as benzene. Calculations of the electrostatic potential map of adenine illustrates that the negative charge is concentrated on N1, N3, and N7, whereas C8 and N9 have significant positive potential (12). Similar studies on the electrostatic potential of tryptophan suggest a relatively greater negative potential on the six-member ring of the indole side chain than on the five-member heterocycle (13). In the MccF cocrystal structures, the side chain of Trp186 and the adenine ring of the substrate are oriented to minimize unfavorable electrostatic interactions between the two heterocyclic π-electron systems (Fig. S2).
The phosphoramidate that links the adenine and the aspartate of McC7 also interacts with the peptide backbone atoms of the enzyme. The carbonyl oxygen atom of the amide bond of McC7 is positioned 3.1- and 2.8-Å away from the backbone amide nitrogen atoms of Asp119 and Gly92 residues, respectively. One of the phosphate oxygen atom is positioned 2.9-Å away from the Gly91 amide nitrogen. These interactions would stabilize the oxyanion of the tetrahedral acyl-enzyme intermediate formed upon attack of the Ser118 hydroxyl at the carbonyl (10). No other amino acid side chains are positioned to favorably interact with the oxygen atom. Unexpectedly, the propylamine appendage to the phosphate that increases the biological activity of McC7 (14) is fully extended and points outward from the active site, where there are no interactions with the terminal primary amine.
Because we could not determine the kinetic parameters for McC7 hydrolysis by MccF due to both acid lability of the phosphoramidate and inability to achieve sufficient HPLC separation of reaction products from unreacted substrate, we utilized DSA as a mimic for processed McC7. In order to confirm that the protein-ligand interactions with DSA are similar to those observed for processed McC7, we first determined the 1.5-Å resolution cocrystal structure of inactive Ser118Ala MccF in complex with DSA (Fig. 3B). Briefly, the interactions at the P1 site of the sulfate oxygen atoms and the stabilization of the oxyanion transition state are essentially identical to those observed in the processed McC7 cocrystal structure. The carboxylate oxygen atoms of the aspartate side chain at P1′ are also similarly engaged by Asn220 and Lys247 (Fig. 3B). The similarities in the two cocrystal structures validate the use of DSA as a substrate mimic of processed McC7 for kinetic analyses.
As noted above, MccF can also utilize ESA as a substrate, albeit with approximately eightfold decrease in catalytic efficiency compared to DSA. This substrate preference is reflected by a turnover number for DSA that is an order of magnitude higher, whereas the Km for the two substrates is comparable. In order to further examine the basis for difference in catalytic efficiency with the two substrates, we determined the 1.3-Å resolution cocrystal structure of Ser118Ala MccF in complex with ESA (Fig. 3C). Although the interactions at the P1 site are essentially identical to those in cocrystal structures with processed McC7 and DSA, the interactions with the glutamate side chain at P1′ are distinct. Notably, the glutamate side chain is tilted toward Lys247 (Oϵ1 and Oϵ2 to Nζ distances of 3.2 and 2.8 Å, respectively), with an additional interaction between Oϵ2 with Nδ2 of Asn220 (2.6 Å). As a consequence, MccF engages the larger ESA side chain with fewer interactions than either processed McC7 or DSA.
Structure-Function Analysis of Site-Specific MccF Mutants.
In order to investigate the role of active site residues of MccF, we generated several single point substitutions at residues that are engaged in interactions with the substrate in the cocrystal structures described above. As the interactions at the P1 site are identical for ESA and DSA, ESA was used for kinetic measurements for the MccF P1 site mutant enzymes. In the structures, the catalytic loop residue Trp186 is the primary determinant for substrate recognition at the P1 site, where it engages in π-stacking interactions with the substrate adenine ring. Mutation of Trp186 to Phe results in a fourfold increase in the Km for ESA hydrolysis (Table 1). The 1.50-Å resolution cocrystal structure of Trp186Phe MccF in complex with adenosine monophosphate reveals that the Phe side chain also engages the adenine ring through π-stacking interactions (Fig. S3). The lower substrate affinity of the Trp186Phe mutant must be caused by decreased π-stacking interaction. Notably, mutation of Trp186 to Ala resulted in a complete loss of activity, likely as a result of the abolition of the π-stacking interactions with the substrate nucleotide. The 2′ hydroxyl of the substrate nucleotide interacts with Arg246. Substitution of this residue to Ala did not alter the kcat of the enzyme, but instead led to a fivefold increase in the Km for the substrate.
At the P1′ site, the carboxylate side chains of the substrates interact with Lys247 and Asn220. Replacement of Lys247 with alanine led to over 30-fold reduction in the catalytic efficiency toward ESA. The substitution of Asn220 to alanine resulted in only a twofold increase in Km and no change in kcat for ESA, leading to twofold decrease in the catalytic efficiency of the enzyme for ESA hydrolysis. The effect of this mutation on kcat/Km is insignificant and can be explained by compensatory interactions of both substrate oxygen atoms of ESA with Lys247 in the mutant protein. This observation is further exemplified by a much larger decrease in kcat for DSA hydrolysis for the Asn220 to alanine mutation (and an eightfold reduction in the catalytic efficiency) and a lesser decrease in kcat for the Lys247 to alanine mutation. The two carboxylate oxygen atoms of ESA are skewed toward the Lys247 side chain, but are nearly equidistant to the Asn220 and Lys247 side chains for DSA. Hence the Asn220 to alanine mutation cannot be compensated by binding of both carboxylate oxygen atoms to Lys247 side chain for DSA. However, substitution of both Asn220 and Lys247 to alanine led to complete loss of activity for both DSA and ESA. Crystal structure of the Ser118Ala/Asn220Ala/Lys247Ala triple mutant enzyme in the apo form confirmed that this loss in activity is not due to rearrangements at the P1 site, but rather due to loss in interactions at the P1′ site (Fig. S4).
Alteration of MccF Substrate Scope Through Active Site Mutations.
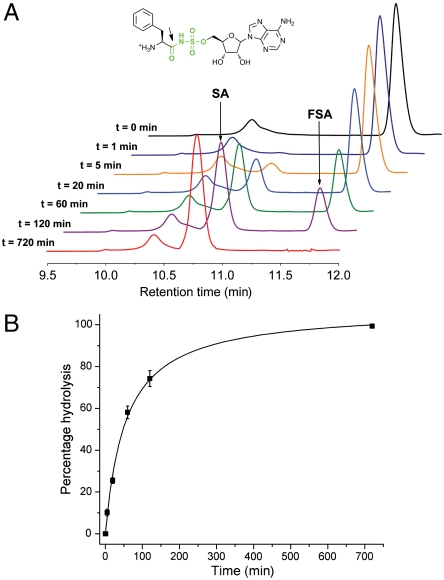
As shown by our combined biochemical and structural biological data, the stringent specificity of MccF for acidic side chains is dictated mainly by the presence of two residues, Asn220 and Lys247, which contact the carboxylate oxygen atoms of processed McC7 and its analogues. We hypothesized that alterations at either or both of these residues could result in a change in substrate scope of MccF toward substrates that contain aromatic residues. We generated four single mutants and four combinations of double mutants in which each or both of these residues were mutated to either leucine or phenylalanine to test the activity of resultant variants for hydrolysis of FSA. With the exception of Asn220Leu/Lys247Leu, the mutant proteins were either insoluble or prone to aggregation as judged by analytical size exclusion chromatography. We carried out time-resolved HPLC analysis of Asn220Leu/Lys247Leu MccF-catalyzed reaction with FSA as a substrate and observed that this mutant could indeed hydrolyze FSA, albeit at a rate much slower than that observed with the wild-type enzyme and DSA (Fig. 4 A and B). Electrospray ionization mass spectrometry analysis confirmed that the site of cleavage of FSA was at the sulfamoyl amide bond (Fig. S5). Notably, the rate of FSA cleavage by either wild-type MccF or a soluble double alanine mutant Asn220Ala/Lys247Ala, which were used as controls, was negligible under similar experimental conditions, demonstrating that hydrophobic aminoacyl-sulfamoyl adenosines are only accepted as substrates when hydrophobic side chains are introduced at the P1′ recognition site of the enzyme.
Fig. 4.
Cleavage of hydrophobic sulfamoyl adenosine by mutant forms of the MccF enzyme. (A) Time-dependent cleavage of substrate FSA (Inset) by MccF-N220L/K247L mutant enzyme demonstrated by separation of substrate and product peaks by HPLC. (B) Percentage hydrolysis of FSA plotted against reaction time for MccF-N220L/K247L enzyme.
Discussion
McC7 is a Trojan horse antibiotic that, following processing, mimics an aminoacyl–adenylate intermediate to target and inhibit aspartyl tRNA synthetase. Because unwanted, premature processing of McC7 within the producer cells could result in toxicity, the producing organisms utilize the MccF immunity determinant to hydrolyze the amide bond that connects the peptide and nucleotide moieties of McC7. In this study, we present biochemical, kinetic, and structural biological characterization of the hydrolysis of processed McC7 and its synthetic analogs by MccF. Three independently functional domains can be identified in the structure of MccF: The catalytic nucleophile and residues necessary for the stabilization of the oxyanion intermediate are provided by the amino terminal domain, and the carboxy-terminal domain provides residues necessary for catalysis and recognition of the acidic side chain of the substrate.
The principal determinant for substrate binding is provided by Trp186, located in a unique catalytic loop, which stacks with the nucleotide adenine ring of McC7. This catalytic loop is absent in the structurally homologous ld-carboxypeptidases. The lack of a cocrystal structure of the ld-carboxypeptidase with bound substrate precludes a comparison of the substrate binding sites of the two enzymes, though the absence of the catalytic loop provides a possible explanation for the reported inability of ld-carboxypeptidase to detoxify McC and its analogs (8, 9). We propose that the presence of the catalytic loop can be used to distinguish proteins with MccF-like activity from ld-carboxypeptidases that have been misannotated in sequence databases (Fig. S1). Analysis of MccF homologs’ catalytic loop sequences reveals that hydrophobic residues corresponding to Trp186 are conserved, suggesting that these proteins should also interact with adenylated targets. Curiously, many MccF homologs are encoded by genes that are not part of recognizable McC7-like biosynthetic operons. The intracellular function of these proteins remains to be determined.
MccF has nearly 10-fold greater turnover number for the hydrolysis of aspartyl sulfamoyl adenylate as compared to that of glutamyl sulfamoyl adenylate but the Km values for the two substrates are comparable. This reflection for substrate preference through a change in kcat and not in Km is reminiscent of earlier work with serine proteases in which synthetic substrates designed with altered binding interactions at the P1′ and subsequent downstream binding subsites have led to nonsignificant changes in the Km for these enzymes, while displaying large variations in the kcat (15–18). For MccF, the order of magnitude increase in kcat for DSA as compared to ESA can be attributed to the differential mode of stabilization of the acyl-enzyme transition state (formed after the departure of the adenine sulfonamide) in the active site. Because the aspartate chain makes more contacts with the enzyme than the glutamate side chain, the activation energy for the formation of this transition state would be lower, leading to an increase in the rate of hydrolysis of DSA. It should also be noted that substitutions at the P1 site, Trp186Phe and Arg246Ala, result in much larger alterations in the Km values for ESA hydrolysis and much less significant changes in kcat values.
An assessment of the rate enhancement for adenylated substrate hydrolysis by MccF requires knowledge of the uncatalyzed rate for the cleavage of an amide with a phosphoramide leaving group. Although, to our knowledge, such rates have yet to be determined, limits for the rate can be estimated to be between the uncatalyzed rate for the hydrolysis of an sd-peptide (2 × 10-10 s-1) (19) and the rate for hydrolysis of a methyl phosphate dianion (2 × 10-20 s-1) (20). Hence, the rate enhancement (kcat/knon-cat) for the MccF hydrolysis of DSA can be estimated to be between 3.5 × 1010- and 3.5 × 1020-fold. This enhancement is comparable to that for the hydrolysis of peptide substrates by serine proteases, such as trypsin and chymotrypsin, that demonstrate roughly 1011-fold enhancement over the uncatalyzed reaction rate (10).
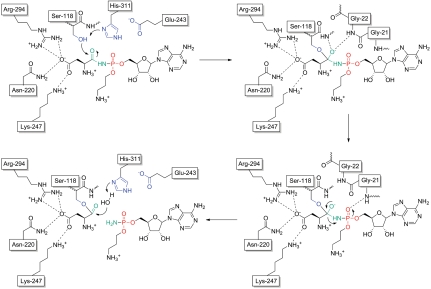
The cocrystal structure of MccF in complex with processed McC7 presents a mechanistic challenge for the protonation of the α-amine of the departing carboxy terminus following cleavage of the scissile peptide bond. In serine proteases, the pKa of the catalytic triad histidine side has been determined to be close to 7.0 (21) and consistent with its role in protonating the departing α-NH2 of the peptide, which would have a pKa of greater than 9.0. However, in the case of a phosphoramide leaving group of McC7, the pKa of the α-NH2 would be close to 4.5 (22), making protonation by the catalytic histidine side chain (His311) difficult. The McC7 α-NH2 can only be protonated by either a glutamate or aspartate (pKa lower than 4.5), and no candidate carboxylate is present in the vicinity of the departing amine. However, the lone pair of electrons on the α-amine of adenosine phosphoramine are in conjugation with the phosphate oxygen atom, which in turn, is within hydrogen binding distance with the backbone amide nitrogen of Gly91 (Fig. 5). Hence, we propose that, in substrates of MccF, the α-amine does not need to be protonated to be a leaving group, because the developing negative charge on the α-amine nitrogen can then be delocalized by resonance conjugation to the significantly more electronegative phosphate oxygen atom.
Fig. 5.
Putative mechanism for McC7 hydrolysis by MccF. Hydrogen bond interactions are shown as dashes. MccF catalytic triad residue side-chain atoms are colored blue, the scissile peptide bond atoms are colored green, and the atoms of the phosphomoiety of the phosphoramidate bond are colored red.
Experimental Procedures
Protein Expression, Crystallization, and Structure Determination.
The expression and purification of recombinant MccF from Escherichia coli has been described previously (8). For crystallization, an additional size exclusion chromatography (Superdex 75 16/60; GE Healthcare) was added at the end of purification. MccF wild-type enzyme structure was solved using single wavelength anomalous diffraction dataset collected on selenomethionine derivatized protein crystals and was subsequently used as a search model for structure determination of the substrate cocrystal structures and structures of the mutant enzymes. Detailed information is provided in the SI Materials and Methods. Relevant data collection and refinement statistics are provided in Table S1.
MccF Enzyme Kinetics.
Kinetics for the hydrolysis of ESA by MccF was monitored by a continuous coupled assay to detect the formation of glutamate. Kinetics for the hydrolysis of DSA and FSA were determined by discontinuous separation of the reactant and products by HPLC. Detailed assay and HPLC conditions are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors are grateful to Drs. Keith Brister, Joseph Brunzelle, and the staff at Life Sciences Collaborative Access Team (Argonne National Laboratory) for facilitating data collection. We thank Gaston Vondenhoff and Arthur Van Aerschot (Rega Institute, Belgium) for providing us with aminoacyl-sulfamoyl adenylates used in this work. This work was supported, in part, by a Dynasty Foundation Fellowship to A.T., a Russian Foundation for Basic Research Grant to A.M., and a Rutgers University Technology Commercialization Fund Grant and Russian Academy of Sciences Molecular and Cellular Biology Program Grant to K.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes: MccF wild type, 3TLA; MccF S118A Apo, 3TLG; MccF S118A McC7, 3TLC; MccF S118A DSA, 3TLB; MccF S118A ESA, 3TLE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114224109/-/DCSupplemental.
References
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 3.Tao J, Schimmel P. Inhibitors of aminoacyl-tRNA synthetases as novel anti-infectives. Expert Opin Invest Drugs. 2000;9:1767–1775. doi: 10.1517/13543784.9.8.1767. [DOI] [PubMed] [Google Scholar]
- 4.Ataide SF, Ibba M. Small molecules: Big players in the evolution of protein synthesis. ACS Chem Biol. 2006;1:285–297. doi: 10.1021/cb600200k. [DOI] [PubMed] [Google Scholar]
- 5.Reader JS, et al. Major biocontrol of plant tumors targets tRNA synthetase. Science. 2005;309:1533. doi: 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 6.Stefanska AL, et al. A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J Antibiot. 2000;53:1346–1353. doi: 10.7164/antibiotics.53.1346. [DOI] [PubMed] [Google Scholar]
- 7.Kazakov T, et al. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J Bacteriol. 2008;190:2607–2610. doi: 10.1128/JB.01956-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tikhonov A, et al. The mechanism of microcin C resistance provided by the MccF peptidase. J Biol Chem. 2010;285:37944–37952. doi: 10.1074/jbc.M110.179135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korza HJ, Bochtler M. Pseudomonas aeruginosa ld-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow. J Biol Chem. 2005;280:40802–40812. doi: 10.1074/jbc.M506328200. [DOI] [PubMed] [Google Scholar]
- 10.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 11.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 12.Boehr DD, et al. Analysis of the π-π stacking interactions between the aminoglycoside antibiotic kinase APH(3′)-Illa and its nucleotide analogs. Chem Biol. 2002;9:1209–1217. doi: 10.1016/s1074-5521(02)00245-4. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty DA. Cation-π interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 14.Metlitskaya A, et al. Maturation of the translation inhibitor microcin C. J Bacteriol. 2009;191:2380–2387. doi: 10.1128/JB.00999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumann WK, Bizzozero SA, Dutler H. Specificity of alpha-chymotrypsin. Dipeptide substrates. FEBS Lett. 1970;8:257–260. doi: 10.1016/0014-5793(70)80280-0. [DOI] [PubMed] [Google Scholar]
- 16.Baumann WK, Bizzozero SA, Dutler H. Kinetic investigation of the alpha-chymotrypsin-catalyzed hydrolysis of peptide substrates. The relationship between peptide-structure N-terminal to the cleaved bond and reactivity. Eur J Biochem. 1973;39:381–391. doi: 10.1111/j.1432-1033.1973.tb03136.x. [DOI] [PubMed] [Google Scholar]
- 17.Thompson RC, Blout ER. Elastase-catalyzed amide hydrolysis of tri-and tetrapeptide amides. Biochemistry. 1973;12:66–71. doi: 10.1021/bi00725a012. [DOI] [PubMed] [Google Scholar]
- 18.Sachdev GP, Fruton JS. Kinetics of action of pepsin on fluorescent peptide substrates. Proc Natl Acad Sci USA. 1975;72:3424–3427. doi: 10.1073/pnas.72.9.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith RM, Hansen DE. The pH profile for the hydrolysis of a peptide bond. J Am Chem Soc. 1998;120:8910–8913. [Google Scholar]
- 20.Lad C, Williams NH, Wolfenden R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc Natl Acad Sci USA. 2003;100:5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachovchin WW, et al. Catalytic mechanism of serine proteases: Reexamination of the pH dependence of the histidyl 1J13C2-H coupling constant in the catalytic triad of alpha-lytic protease. Proc Natl Acad Sci USA. 1981;78:7323–7326. doi: 10.1073/pnas.78.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamcsik MP, et al. Protonation of phosphoramide mustard and other phosphoramides. J Med Chem. 1993;36:3636–3645. doi: 10.1021/jm00075a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.