Abstract
The RMI subcomplex (RMI1/RMI2) functions with the BLM helicase and topoisomerase IIIα in a complex called the “dissolvasome,” which separates double-Holliday junction DNA structures that can arise during DNA repair. This activity suppresses potentially harmful sister chromatid exchange (SCE) events in wild-type cells but not in cells derived from Bloom syndrome patients with inactivating BLM mutations. The RMI subcomplex also associates with FANCM, a component of the Fanconi anemia (FA) core complex that is important for repair of stalled DNA replication forks. The RMI/FANCM interface appears to help coordinate dissolvasome and FA core complex activities, but its precise role remains poorly understood. Here, we define the structure of the RMI/FANCM interface and investigate its roles in coordinating cellular DNA-repair activities. The X-ray crystal structure of the RMI core complex bound to a well-conserved peptide from FANCM shows that FANCM binds to both RMI proteins through a hydrophobic “knobs-into-holes” packing arrangement. The RMI/FANCM interface is shown to be critical for interaction between the components of the dissolvasome and the FA core complex. FANCM variants that substitute alanine for key interface residues strongly destabilize the complex in solution and lead to increased SCE levels in cells that are similar to those observed in blm- or fancm-deficient cells. This study provides a molecular view of the RMI/FANCM complex and highlights a key interface utilized in coordinating the activities of two critical eukaryotic DNA-damage repair machines.
Keywords: DNA recombination, X-ray crystallography
Cells have evolved numerous corrective pathways to ensure fidelity and stability of their genomes. Deleterious mutations in genes encoding DNA-repair proteins can lead to several different diseases, many of which share common features such as formation of unusual chromosomal structures in affected cells and cancer predisposition in afflicted individuals. Fanconi anemia (FA), an autosomal recessive disease caused by mutations in any of 15 genes involved in the repair of DNA interstrand crosslinks (ICLs), is an example of such a disease (1–3). FA patients suffer developmental abnormalities, progressive bone marrow failure, and a high incidence of cancer. FA cell lines are hypersensitive to agents that induce ICLs, exhibiting high levels of chromosomal instability as exemplified by radial chromosomes or, in some cases, by high levels of sister chromatid exchange (SCE) in mitotic cells. A core complex of eight FA proteins, (FANCA, B, C, E, F, G, L, and M), cooperate in DNA-damage recognition and in catalyzing monoubiquitination of the FANCD2/FANCI heterodimer in response to DNA damage. Ubiquitinated FANCD2/FANCI in turn recruits downstream factors to facilitate DNA repair (2).
Bloom syndrome (BS), similar to FA, is an autosomal recessive DNA-repair deficiency disease that exhibits chromosomal instability. BS results from mutation of a single gene, BLM, which encodes one of five human RecQ-family DNA helicases (4). BS patients exhibit proportional dwarfism, sun-sensitivity, and a broad predisposition to cancer (5, 6). BS cells display elevated levels of SCEs, which is consistent with the proposed role for the BLM helicase in the nonrecombinogenic repair of double-Holliday junction (dHJ) DNA structures that can arise during repair. To catalyze this activity, BLM functions within a complex called the “dissolvasome” that also includes topoisomerase IIIα (Top3α) and the RecQ-mediated genome instability, or RMI, subcomplex (RMI1/RMI2 heterodimer) (7–13). The dissolvasome is thought to drive HJ branch migration (catalyzed by BLM) and DNA cleavage of the resulting hemi-catenated DNA (catalyzed by Top3α) in a reaction called “dissolution” (14). The RMI subcomplex stimulates these activities and stabilizes the dissolvasome (7, 8, 10).
Several studies have highlighed molecular connections between the FA core complex and the BLM dissolvasome. The initial finding linking the two complexes was identification of an ICL-induced supercomplex called BRAFT (BLM, replication protein A, FA, and Top3α) that included components of both complexes (15, 16). A subsequent study identified a direct link between a conserved 34 amino acid motif from FANCM (called MM2, for FANCM motif 2) and the C-terminal OB-fold of RMI1 (RMI1 OB2) and Top3α (Fig. 1A) (17). FANCM is a member of the FA core complex that contains an N-terminal helicase domain and a degenerate C-terminal endonuclease domain; the MM2 sequence resides in the linker that connect these two domains (17, 18). Consistent with a connection between FANCM and BS, FANCM-deficient cells are characterized by high levels of SCEs, which is distinct from most other FA complementation groups (19) but similar to BS-derived cells. Deletion of MM2 from FANCM or substitution of two Phe residues in the FANCM MM2 sequence (Phe1232 and Phe1236) with Ala abrogates its binding to RMI1 and results in a DNA-damage-dependent increase in SCE formation (17). These observations lead to a model in which FANCM is responsible for coordinating the activities of the FA complex and the BLM dissolvasome at damaged sites in the genome.
Fig. 1.
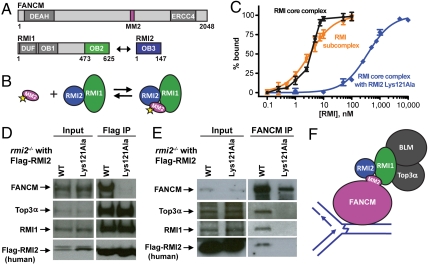
RMI2 Lys121Ala is defective in binding FANCM in vitro and in vivo. (A) Schematic diagram of the domain structures of human FANCM, RMI1, and RMI2. The location of the MM2 portion of FANCM is shown in pink, and interacting OB domains comprising the RMI core complex are shown in green (RMI1 OB2) and blue (RMI2 OB3). Additionally, FANCM contains a DEAH helicase domain and degenerate ERCC4-like domain. RMI1 also contains a domain of unknown function, DUF, and an OB domain that interacts with Top3α and BLM (OB1). (B) Scheme for the FP assay used to test binding in C. The MM2 peptide is FITC-labeled as indicated with the yellow star. (C) RMI2 Lys121Ala is defective in binding to MM2. Normalized binding curves for the RMI core complex (black), RMI subcomplex (orange), and RMI core complex with RMI2 Lys121Ala (blue) are shown. Experiments were done in triplicate and average FP value was plotted with one standard deviation of the mean shown as error. (D) RMI2 Lys121Ala is defective in FANCM binding in vivo. Protein input before coimmunoprecipitation (left), and the results of the coimmunoprecipitation (right), shows experiments with Flag-tagged RMI2 complemented into rmi2-/- chicken DT40 cells, followed by Western to probe for the protein designated to the left of the gel. (E) The RMI subcomplex acts as a bridge between FANCM and Top3α. Experiments were performed as in D, except FANCM was immunoprecipitated. The cell line is noted above each lane and to the left the protein probed for in Western is designated. (F) Model of the FANCM/BLM dissolvasome interaction. FANCM at a site of DNA damage (depicted as an ICL) recruits the dissolvasome through the RMI subcomplex.
X-ray crystal structures of a protease-resistant “RMI core complex” comprising RMI1 OB2 bound to RMI2 have recently been reported (12, 13). Sequence changes at the RMI1/RMI2 interface generally destabilize the dissolvasome and increase cellular SCEs levels (12). However, one unusual interface variant, RMI2 Lys121Ala, supports normal dissolvasome assembly but leads to SCE levels similar to those observed in rmi2-/- cells (12). Based on this result and the report that MM2 variants display SCE levels that are similar to those of BS cells (17), we hypothesized that FANCM binds at the RMI1/RMI2 interface and that RMI2 Lys121 is critical for proper formation of the MM2 binding site. In support of this hypothesis, we show that RMI subcomplex variants that include RMI2 Lys121Ala are defective in binding to a FANCM MM2 peptide in vitro and to full-length FANCM in vivo. Interestingly, the Top3α/FANCM interaction is no longer observed with the Lys121Ala RMI2 sequence change, supporting a model in which the interaction between Top3α and the FA core complex is mediated by the RMI/FANCM interface. To better understand this important molecular interface, we have determined the X-ray crystal structure of the FANCM MM2 peptide bound to the RMI core complex. The structure shows that FANCM MM2 binds to both RMI1 and RMI2 at a site involving RMI2 Lys121. The complex is formed through an extensive hydrophobic “knobs-into-holes” packing arrangement in which hydrophobic side chains from FANCM MM2 project into hydrophobic pockets on the RMI core complex. These MM2 “knob” residues include Phe residues previously identified as having roles in RMI subcomplex binding (17) and several additional residues from FANCM. Sequence changes to the hydrophobic knob residues in FANCM abrogate binding to the RMI core complex in vitro and in vivo, leading to increases in cellular SCE formation. Collectively, these studies define the key molecular surface that links the FA core complex to the BLM dissolvasome and allow a deeper understanding of the mechanisms that connect these two diseases.
Results
A Role for RMI2 Lys121 in Linking the Dissolvasome to FANCM.
Previous studies have shown that the C-terminal OB domain of RMI1 (RMI1 OB2) interacts both with the MM2 region of FANCM and with RMI2 (Fig. 1A) (7, 8, 12, 13, 17). We have confirmed the RMI/FANCM interaction in vitro by demonstrating that a glutathione S-transferase (GST)-MM2 fusion protein forms a complex with the RMI core complex (RMI1 OB2 in complex with RMI2) using analytical size-exclusion chromatography (Fig. S1). To assess the stability of the RMI/MM2 complex, MM2 peptide was purified, labeled with fluorescein (producing F-MM2) and used in a fluorescence polarization (FP) assay (Fig. 1 B and C). F-MM2 was bound by the RMI subcomplex (full-length RMI1 in complex with RMI2) with an apparent dissociation constant (Kd,app) of ≤ 4.5 nM. Because the F-MM2 concentration is the same as the Kd,app, we could only establish an upper limit for the binding affinity. F-MM2 binding by the RMI core complex was very similar to that observed with the full-length RMI subcomplex, indicating that the RMI core complex is sufficient for binding to MM2 (Fig. 1C).
Previous results have shown that the RMI2 variant Lys121Ala exhibits no defect in coimmunoprecipitation with other members of the BLM dissolvasome but shows increases in cellular SCE levels (7, 12). We therefore tested a possible role for Lys121 from RMI2 in MM2 binding using the FP assay. An RMI core complex that included the RMI2 Lys121Ala variant bound F-MM2 with a Kd,app of 360 ± 20 nM, which is ≥80-fold weaker than the wild-type RMI core complex (Fig. 1C). Because the structure of the RMI core complex is unaltered by the Lys121Ala sequence change (12), this result strongly supports a role for Lys121 in FANCM MM2 binding. To confirm the importance of RMI2 Lys121 in the context of full-length FANCM, Flag-tagged wild-type RMI2 or RMI2 Lys121Ala were expressed in rmi2-/- DT40 cells and immunoprecipitated; coprecipitation of FANCM was detected by Western blotting. Consistent with a role for RMI2 Lys121 in FANCM binding, FANCM coimmunoprecipitated with wild-type Flag-RMI2 but not with Flag-RMI2 Lys121Ala (Fig. 1D). Top3α and RMI1 coimmunoprecipitated with both Flag-tagged RMI2 proteins as has been observed previously (7, 12). These data indicate that the Lys121Ala sequence change in RMI2 specifically abrogates binding to FANCM and identify a role for RMI2 in FANCM binding.
With its selective disruption of the FANCM/dissolvasome interaction, the RMI2 Lys121Ala variant is a useful tool for determining whether FANCM interacts directly with Top3α, as has been suggested (17), or instead requires interaction with the RMI subcomplex to associate with Top3α and other dissolvasome components. To determine whether interaction between FANCM and Top3α depends on the RMI subcomplex, FANCM was immunoprecipitated from DT40 cells and FANCM, RMI1, and Top3α levels were assessed by Western blotting. Top3α and the RMI subcomplex coimmunoprecipitated with FANCM in extract from wild-type cells but not rmi2-/- cells (Fig. S2), indicating that RMI2 is essential for the association between the BLM dissolvasome and FANCM. We next performed coimmunoprecipitations in rmi2-/- cells complemented with RMI2 or the RMI2 Lys121Ala variant in order to selectively disrupt the RMI/FANCM interaction (Fig. 1E). RMI1 and Top3α coimmunoprecipitation with FANCM was observed in cells complemented with wild-type RMI2 but not in cells complemented with RMI2 Lys121Ala (Fig. 1E; longer exposures of experiments failed to reveal the presence of RMI1, Flag-RMI2, and Top3α in the cells complemented with RMI2 Lys121Ala as well). These results indicate that the RMI/FANCM interaction is necessary for FANCM interaction with the Top3α protein (Fig. 1F).
X-Ray Crystal Structure of FANCM MM2 Bound to the RMI Core Complex.
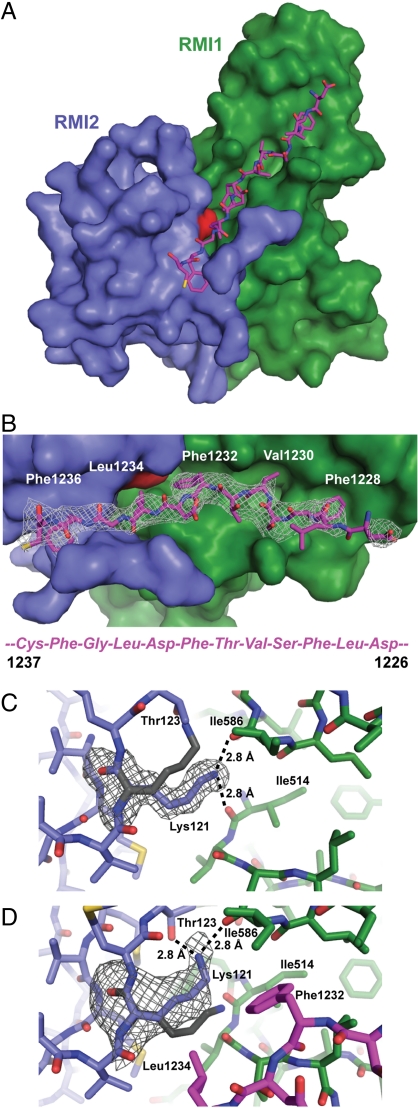
A crystallographic approach was taken to better define the interaction between FANCM and the RMI core complex. Crystals of the RMI core complex bound to the FANCM MM2 peptide diffracted to 3.3-Å resolution, and the structure of the ternary complex was determined using molecular replacement (Table S1 and Fig. 2A). Fo - Fc electron density maps revealed the presence of a single FANCM MM2 peptide (residues 1226–1237) bound to a surface comprised of elements from both RMI1 and RMI2 (Fig. 2B). Additionally, electron density for the N-terminal region of RMI2 (residues 13–16) was observed in the ternary structure; this region was not resolved in earlier structures of the apo RMI core complex (12, 13) (Fig. 2B and Fig. S3). These residues form a series of apparent hydrogen bonding interactions with the MM2 peptide.
Fig. 2.
The structure of the RMI core complex bound to the MM2 peptide. (A) The overall structure of the RMI core complex with bound MM2 peptide. MM2 residues 1226–1237 are shown in pink. RMI1 is shown in green, RMI2 in blue, and RMI2 Lys121 is highlighted in red. (B) A closeup of the interface between the RMI core complex and the MM2 peptide. The peptide is shown with the C terminus on the left with the corresponding residues shown below. Also shown is an Fo - Fc electron density map (2σ) for the MM2 peptide. The same color scheme is used as in A. (C) Shown in green (RMI1) and blue (RMI2) is the 1.55-Å resolution apo X-ray crystal structure (PDB ID code 3MXN) with Fo - Fc electron density map (2σ) for Lys121. Lys121 (gray) for the MM2-bound structure shows the alignment of the Cα at Lys121. (D) The same is shown as in C except the MM2-bound structure is shown in color with Lys121 in gray from the apo structure. The Fo - Fc electron density map (2σ) is shown for the MM2-bound Lys121.
The MM2/RMI interface is primarily defined by a knobs-into-holes packing arrangement in which alternating hydrophobic side chains from the MM2 peptide project into hydrophobic pockets on the RMI1/RMI2 surface (Fig. 2B). The MM2 peptide is highly extended in the complex, docking a total of 5 hydrophobic knob residues onto the RMI core complex. Each of the MM2 knob residues is highly conserved among FANCM proteins, and Phe1232 and Phe1236 are known to be critical for FANCM binding to the RMI subcomplex (17). Two of the hydrophobic MM2 residues, Phe1232 and Leu1234, dock into a pocket formed at the RMI1/RMI2 interface formed by RMI1 residues 514–517, 585, and 586 and RMI2 residues 90 and 120–121. The remaining MM2 knob residues bind to either RMI1 (Phe1228 and Val1230) or RMI2 (Phe1236).
Consistent with its importance in mediating the RMI/FANCM interaction, RMI2 residue Lys121 helps to form the hydrophobic pocket where MM2 residues Phe1232 and Leu1234 bind (Fig. 2 A and B). In structures of the apo RMI core complex, the ε-amino group of the Lys121 side chain is bound to the main chain carbonyl groups of residues 514 and 586 from RMI1 (Fig. 2C) (12, 13). However, in the ternary complex with MM2, the Lys121 side chain is altered and its ε-amino group is bound to the side chain of RMI2 Thr123 and the carbonyl of RMI1 residue 586 (Fig. 2D). This change is accompanied by structural changes in adjacent residues (120–124) in RMI2 (Fig. 2 C and D and Fig. S4). This overall movement appears to enhance the hydrophobicity of the binding pocket to better accommodate the MM2 peptide. These rearrangements suggest that Lys121 is important for conformational changes in the RMI subcomplex that help stabilize formation of the RMI/MM2 complex. Moreover, the hydrophobic portion of the Lys121 side chain is in proximity to Phe1232 (3.5 Å) and Leu1234 (3.3 Å) from the MM2 peptide, indicating a possible direct role in stabilizing the complex through hydrophobic contacts.
Assessing the Importance of RMI/FANCM Interactions.
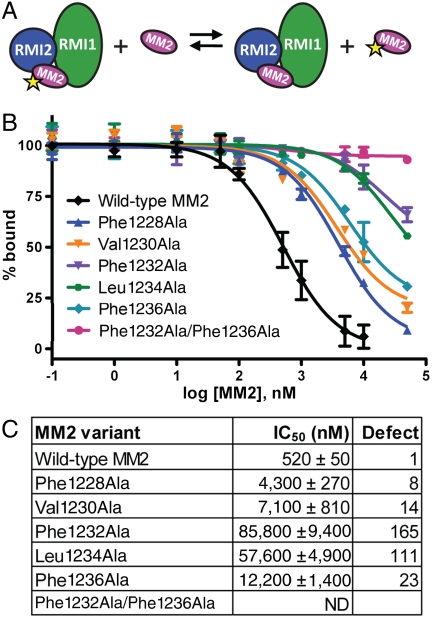
A competition peptide-binding assay was developed to test the effects of changes in the MM2 peptide sequence on the stability of the RMI/MM2 complex (Fig. 3A). In this assay, the RMI core complex was prebound to F-MM2 and unlabeled MM2 peptide variants were titrated into the RMI core complex/F-MM2 solutions. Concentration-dependent binding of the MM2 peptide variants to the RMI core complex was monitored as a decrease in FP that is observed as F-MM2 is liberated from the complex. Wild-type MM2 peptide dissociated F-MM2 with an IC50 of 520 ± 50 nM (Fig. 3 B and C). A panel of MM2 variants in which hydrophobic knob residues were individually or multiply substituted for alanine were then tested to determine the impact of the sequence changes on binding the RMI core complex. The most dramatic effects were observed with single Ala variants of Phe1232 (IC50 85,800 ± 9,400 nM) and Leu1234 (IC50 57,600 ± 4,900 nM) and with the double Phe1232/Phe1236 Ala variant (no competition observed) (Fig. 3 B and C). Because Phe1232 and Leu1234 dock in the hydrophobic pocket formed at the RMI1/RMI2 interface, these dramatic effects coupled with the strong effects of the RMI2 Lys121Ala variant on complex formation argue that this interaction is a key anchor point in the RMI1/RMI2/FANCM ternary complex. The strong defect of the double Phe1232/Phe1236 variant was consistent with previous results in which the same sequence change in MM2 resulted in a defect in FANCM coimmunoprecipitation with BLM and RMI1 (17). The single MM2 Phe1228, Val1230, and Phe1236 Ala variants were defective in the competition assay as well, but with more modest defects than were observed with the other peptides.
Fig. 3.
MM2 variants have defects in binding the RMI core complex. (A) The scheme used in the FP competition assay. Unlabeled MM2 or variants are tested for their ability to compete with FITC-MM2 for binding to the RMI core complex. (B) Normalized FP competition results with different MM2 variants. Experiments were done in triplicate and average FP value was plotted with one standard deviation of the mean shown as error. (C) Table showing the IC50 and competition defect for each MM2 variant.
To verify the competition binding results in a cellular context and with full-length FANCM, we performed coimmunoprecipitation experiments with FANCM variant proteins transiently transfected into HEK293 cells (Fig. S5). The results strongly paralleled the competition binding results, with the strongest defects observed for the Phe1232Ala and Leu1234Ala single site variants, and weaker defects for Phe1228Ala, Phe1236Ala, and Val1230Ala.
The RMI/FANCM Interaction Is Important for Suppressing Cellular SCEs.
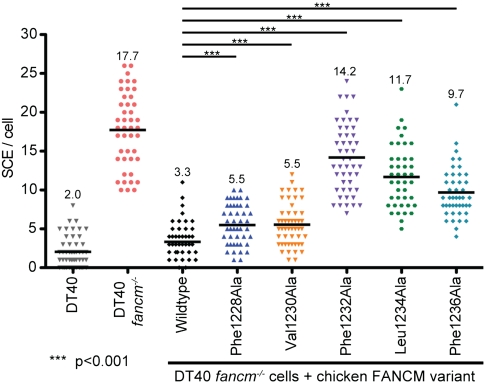
To determine the cellular effects of destabilizing the RMI/FANCM interaction, FANCM variants with MM2 sequence changes were stably expressed in fancm-/- DT40 cells and SCE levels were measured. In fancm+/+ DT40 cells, the mean number of SCEs per cell was 2.0 whereas in fancm-/- cells the mean increased to 17.7 SCEs/cell (Fig. 4). Expression of wild-type FANCM complemented the SCE phenotype in fancm-/- cells, reducing the mean to 3.1 SCEs/cell. The SCE levels of FANCM variants were then measured in the same assay. Expression of variants with the most severe in vitro binding defects, Phe1232Ala and Leu1234Ala, led to mean SCEs/cell levels of 14.2 and 11.7, respectively. Expression of FANCM variants with intermediate binding defects, Phe1236Ala, Val1230Ala, and Phe1228Ala, showed mean SCE/cell levels of 9.7, 5.5, and 5.5, respectively. Thus the defects of MM2 variants in RMI/FANCM complex binding paralleled the SCE increases that accompanied the MM2 sequence changes. This observation strongly supports a model in which the interaction between FANCM and the RMI subcomplex through the interface identified in the structure is critical for coordinated activity of the FA and BLM dissolvasome complexes in vivo.
Fig. 4.
FANCM variants with defects in binding increase SCEs. The number of SCEs per cell in chicken DT40 cells are shown as a histogram in which each point in a column is the number of SCE events in a single cell. The mean level of SCEs/cell is displayed as a black line and numerically at the top of the column. Statistical significance is indicated with *** (p < 0.001). “DT40” denotes wild-type DT40 cells, “fancm-/-” denotes DT40 cells lacking FANCM, “wildtype” denotes fancm-/- cells complemented with FANCM and the remainder denote the FANCM variants that have been substituted.
Discussion
Coordination of BLM dissolvasome and FA core complex activities is critical for suppressing potentially harmful SCE events that can arise from DNA-damage repair processes. Here, we have defined the molecular interface that mediates this coordination, formed between the RMI subcomplex from the BLM dissolvasome and the FANCM protein from the FA core complex. These studies identify a previously unrecognized role for RMI2 in binding FANCM and show that the RMI/FANCM interface is required for the proper cellular assembly of FANCM with the BLM dissolvasome. Our results support a model in which the RMI/FANCM interaction provides the primary direct link between the BLM dissolvasome and the FA core complex to preserve genomic stability (Fig. 1F, model).
Structure of the RMI/FANCM Interface.
Our structural and biochemical observations show that interaction between FANCM and the RMI core complex depends heavily upon docking of conserved hydrophobic side chains from FANCM into hydrophobic pockets on the RMI1/RMI2 surface (Fig. 2). This knobs-into-holes packing arrangement utilizes an extended surface presented by both RMI1 and RMI2, with a key anchor site for FANCM binding located at a pocket formed at the interface between the two RMI proteins. Sequence changes to either the RMI1/RMI2 pocket (RMI2 Lys121) or in two FANCM knob residues (Phe1232 or Leu1234) that pack at this three-protein interface dramatically destabilize the ternary complex (Figs 2 and 3 and Fig. S5). The importance of this central site in RMI/FANCM complex stability is consistent with previously identified roles for FANCM Phe1232 in RMI1 binding and for FANCM Phe1232 and RMI2 Lys121 in the joint activities of the FA and BLM dissolvasome complexes (7, 12, 17). In addition to this central interface position, several other important interactions involving FANCM residues Phe1228, Val1230, and Phe1236 were identified. These residues dock into hydrophobic pockets on RMI1 or RMI2 that are adjacent to the central anchor site. These secondary pockets stabilize the ternary complex and are likely to be important for the specificity of peptide binding.
Insights into the Architecture of the BLM Dissolvasome/FA Core Complex Superstructure.
Our findings point to a primary role for the RMI/FANCM interface in mediating complex formation between the BLM dissolvasome and the FA core complex. An earlier study that identified the FANCM MM2 region as the RMI1 interaction site also demonstrated binding between an extended polypeptide including the MM2 region and purified Top3α in vitro (17). However, immunoprecipitation experiments from cell lysates showed that the Flag-tagged RMI1 OB2 domain could coprecipitate with RMI2 and FANCM but not Top3α (17), leaving the role of the possible Top3α/FANCM interaction in directly mediating interaction between the larger BLM dissolvasome and the FA core complex unclear. Our results using the RMI2 Lys121Ala variant, which disrupts RMI1/RMI2 interaction with FANCM but not with components of the BLM dissolvasome (Fig. 1D), help clarify the relative importance of the RMI/FANCM and possible Top3α/FANCM interfaces in higher order complex formation. Top3α coimmunoprecipitated with FANCM from cells expressing wild-type RMI2 but not from cells lacking RMI2 or expressing the RMI2 Lys121Ala variant (Fig. 1E and Fig. S2). Thus, a direct interaction between the RMI proteins and FANCM is required for coimmunoprecipitation of Top3α with FANCM, highlighting the primacy of the RMI/FANCM interface in higher order complex formation. A direct interaction between FANCM and Top3α in a supercomplex formed through the RMI/FANCM interface is not excluded by these findings, but it appears to require the RMI/FANCM interface to form in cells.
Possible Regulation of RMI/FANCM Complex Assembly.
The RMI subcomplex forms a very stable interaction with the isolated MM2 peptide (Kd,app of ≤ 4.5 nM) that could imply the proteins form a ternary complex under all cellular conditions. However, previously published data indicate that the BLM dissolvasome and FA core complexes only colocalize under conditions in which replication fork stalling is triggered, with colocalization being MM2-dependent (17). The mechanisms that regulate RMI/FANCM complex formation are presently unclear, but the interface could be regulated in a manner that facilitates conditional coordination of the activities of the complexes. One possibility is that FANCM and/or the RMI subcomplex undergo conformational changes that subsequently allow complex formation. Except for the highly conserved MM2 sequence, the central portion of FANCM is generally poorly conserved and predicted to be unstructured (Fig. 1A) (17). It could be that FANCM folding occludes the MM2 sequence until it is triggered to expose the element through modification or ligand (DNA) binding to allow interaction with the dissolvasome. Conversely, regulation could occur through changes to the RMI subcomplex. Regulated assembly of the RMI subcomplex is not likely because it appears to be an obligatory heterodimer; the hydrophobic surface area between RMI1 and RMI2 is extensive and cellular RMI1 and RMI2 protein levels are dependent on each other (7, 12). However, full-length RMI1 is structurally dynamic, and there is a possibility that the RMI1 structure is altered upon response to a cellular trigger in such a way that exposes the FANCM binding site (Fig. 1A) (12).
A Model for Complex Coordination and SCE Suppression.
A theme that emerges from this work and previous studies is that depletion of proteins in either the BLM dissolvasome or the FA core complex, or disruption of protein interfaces that stabilize each complex or link the two, lead to similar increases in SCE levels (Fig. 4 and (7, 11, 12, 19, 20). Taken together, these data suggest that the RMI/FANCM interface is critical in bridging the BLM dissolvasome and the FA core complexes and that proper association of these two complexes is vital in SCE suppression. This indicates that the BLM dissolvasome and the FA core complex are highly interdependent and may have roles beyond ICL repair.
A model in which SCEs can result from improper coordination between the BLM dissolvasome and the FA core complex during repair of stalled replication forks is supported by these findings. In this model, FANCM binds to a damaged site in the genome and recruits the BLM dissolvasome through its interaction with the RMI subcomplex (Fig. 1F). FANCM could act to remodel the replication fork, possibly in coordination with BLM and Top3α (20–24). Furthermore, FANCM interacts with a large number of proteins that mediate many DNA-repair pathways. It may be that FANCM binds to stalled replication forks that form as a consequence of many types of DNA damage and in certain cases recruits the BLM dissolvasome to aid in replication fork remodeling, whereas other FA proteins may only be recruited by FANCM when the damage is an ICL. Additional interactions among proteins implicated in FA and BS (e.g., BLM and FANCJ, a helicase that acts downstream of the FA core complex) could also aid in the process of replication fork remodeling, as FANCJ-deficient cells also show increases in SCEs (25). Failure to properly orchestrate the activities of these complexes would result in an accumulation of SCE events. Although we have identified the primary molecular and functional basis underlying the RMI/FANCM interaction, how other FANCM interactions mediate genome stability remains to be determined.
Materials and Methods
Purification of the RMI Complexes.
The RMI core complex, RMI core complex with RMI2 Lys121Ala, and RMI subcomplex were purified as described previously (12).
Purification of the MM2 Peptide.
The different MM2 constructs that were used along with detailed purification methods can be found in SI Text. Briefly, cDNA encoding the FANCM MM2 peptide (residues 1218 to 1251) was cloned into a modified pGEX4T-1 plasmid (Amersham) to express a glutathione S-transferase (GST)-MM2 fusion protein in which GST and MM2 are separated by atobacco etch virus (TEV) protease cleavage sequence.. Rosetta 2 (DE3) Escherichia coli cells transformed with pLysS (Novagen) and the appropriate GST-MM2 vector were grown and GST-MM2 overexpression was induced with isopropyl β-D-thiogalactopyranoside. Cells were harvested with centrifugation and frozen at -80 °C. For MM2 isolation, cells were resuspended, lysed by sonication, and centrifuged. The soluble lysate was purified using Glutathione Sepharose 4B resin (Amersham), followed by TEV cleavage, and finally resolved on a Sephacryl S-100 column (Amersham). The MM2 peptides that were used in crystallization or labeled with FITC were further purified using reverse-phase HPLC and their masses confirmed with MALDI mass spectrometry. MM2 peptide variants for competition assays were not HPLC purified; these were dialyzed against 10 mM Na2CO3 (pH 9.3), lyophilized, and resuspended in water.
FITC Labeling of MM2 (F-MM2).
FITC was reacted with approximately 0.5 mM MM2 peptide (with the sequence GKWEDIFDCSRDLFSVTFDLGFCSPDSDDEILEHTSD, where the underlined sequence is that of FANCM) at a 15-fold excess for 1 h [in 100 mM Na2CO3 (pH 9.3), in the dark, at room temperature]. Extensive dialysis against 10 mM Na2CO3 (pH 9.3) removed unreacted FITC. The calculated degree of labeling was approximately 0.8 label/MM2 peptide.
Fluorescence Polarization Assays.
FP was measured at 25 °C using a Panvera Beacon 2000 system (Invitrogen) with 490 nm excitation and 535 nm emission wavelengths for three replicates; the average FP value was plotted with one standard deviation of the mean shown as error. For direct binding experiments, dilutions of the RMI subcomplex, RMI core complex, or RMI core complex with RMI2 Lys121Ala (in 10 mM Tris pH 8.8, 1 mM DTT) were incubated for over 50 min at room temperature with 4–4.5 nM F-MM2 before measurement. Lower amounts of probe could not be tested due to high signal-to-noise. For competition experiments, 4–5 nM F-MM2 was preincubated for 50 min with 100 nM RMI core complex prior to the addition of MM2 variants; FP was measured after an additional 50 min incubation. IC50 values are the concentrations of unlabeled peptides necessary for 50% inhibition of F-MM2 binding. Data were fitted with a single binding-site model using the GraphPad Prism software package; lower baseline values were fixed at 27 mA to allow IC50 determination in cases where 100 % dissociation was not observed with the peptide concentrations tested.
RMI/MM2 Complex Crystallization and Structure Determination.
RMI core complex (5 g/L, in 10 mM Tris pH 8.8, 1 mM DTT) was mixed at a 1∶1 ratio with MM2 peptide (sequence GHMEDIFDCSRDLFSVTFDLGFCSPDSDDEILEHTSD) in the same buffer. The complex was mixed with mother liquor (200 mM Na2SO4, 5% polyethylene glycol 3350) at a 1∶1 (vol) ratio and crystals were formed by hanging drop vapor diffusion over a 6-mo period. Crystals were transferred to a cryoprotectant solution (200 mM Na2SO4, 10% polyethylene glycol 3350, 30% ethylene glycol) and flash-frozen in liquid nitrogen.
X-ray diffraction data were indexed and scaled using HKL2000 (26). The structure of the RMI/MM2 complex was determined by molecular replacement using the apo RMI core complex structure (12) as a search model in the program Phaser (27) followed by rounds of manual fitting using Coot (28) and refinement using REFMAC5 (29). Coordinate and structure factor files have been deposited in the Protein Data Bank (PDB ID code 4DAY).
Coimmunoprecipitation and SCE Assays.
For coimmunoprecipitation with wild-type and RMI2 Lys121Ala mutant, the Flag-tagged human RMI2 variants were stably expressed in chicken DT40 rmi2-/- cells. Whole-cell extracts were prepared as described previously (30). Coimmunoprecipitation experiments were performed using anti-FLAG M2-agarose (Sigma) as described (30).
For coimmunoprecipitation with FANCM, experiments were performed using anti-chicken FANCM antibody as described (20, 30). Immunoblotting shows levels of FANCM and Top3α in whole-cell lysates from various DT40 cells (wild-type, rmi2-/- cells, and rmi2-/- cells complemented with human WT or Lys121Ala RMI2).
In order to measure SCE levels, chicken FANCM variants were stably expressed in chicken DT40 fancm-/- cells. The SCE frequency was measured as previously described (12).
Supplementary Material
Acknowledgments.
We thank Advanced Photon Source staff (LS-CAT beamline) and Ken Satyshur for assistance with data collection and members of the Keck Lab for critical reading of this manuscript. We thank members of the Denu lab for their assistance with peptide purification and mass spectrometry. This work was funded by a grant from the National Institutes of Health (NIH) (GM068061, J.L.K.) and by the Intramural Research Program of the National Institute on Aging (Z01 AG000657-08, W.W.). K.A.H. was supported in part by an NIH training grant in Molecular Biosciences (GM07215).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117279109/-/DCSupplemental.
Data deposition: Model coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4DAY).
References
- 1.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: Cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Bachrati CZ, Hickson ID. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.German J. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 7.Xu D, et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin J, et al. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoadley KA, et al. Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome. Structure. 2010;18:1149–1158. doi: 10.1016/j.str.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, et al. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure. 2010;18:1159–1170. doi: 10.1016/j.str.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 15.Meetei AR, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 17.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi Anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Meetei AR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37:4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Z, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh TR, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 23.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 24.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhasini AN, et al. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Meth Enzy. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 30.Guo R, Xu D, Wang W. Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods. 2009;48:72–79. doi: 10.1016/j.ymeth.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.