Abstract
The mevalonate pathway is highly conserved and mediates the production of isoprenoids, which feed into biosynthetic pathways for sterols, dolichol, ubiquinone, heme, isopentenyl adenine, and prenylated proteins. We found that in Caenorhabditis elegans, the nonsterol biosynthetic outputs of the mevalonate pathway are required for the activity of microRNAs (miRNAs) in silencing their target mRNAs. Inactivation of genes that mediate multiple steps of the mevalonate pathway causes derepression of several miRNA target genes, with no disruption of the miRNA levels, suggesting a role in miRNA-induced silencing complex activity. Dolichol phosphate, synthesized from the mevalonate pathway, functions as a lipid carrier of the oligosaccharide moiety destined for protein N-linked glycosylation. Inhibition of the dolichol pathway of protein N-glycosylation also causes derepression of miRNA target mRNAs. The proteins that mediate miRNA repression are therefore likely to be regulated by N-glycosylation. Conversely, drugs such as statins, which inhibit the mevalonate pathway, may compromise miRNA repression as well as the more commonly considered cholesterol biosynthesis.
MicroRNAs (miRNAs) are ~22-nucleotide noncoding RNAs that repress gene expression posttranscriptionally. In animals, miRNAs cause repression by base pairing to the 3′ untranslated region of their target mRNAs, which contain perfect or near-perfect sequence complementarity to nucleotides 2–7 (the “seed region”) of miRNAs and mismatches and bulges in other parts of the miRNA–mRNA duplex (1). miRNAs are processed to their mature form in a well-understood pathway involving the RNase III proteins Drosha and Dicer; mature miRNAs then associate with Argonaute proteins to form the core miRNA-induced silencing complex (miRISC). However, the exact mechanism by which miRNAs repress target mRNA stability or translation is still an open question (2). In addition, genetic and biochemical analysis has implicated multivesicular bodies in the repression of target mRNAs by miRNAs (3, 4), underscoring the need to better understand how membrane trafficking functionally intersects with the miRNA pathway. Genetic screens for mutants that phenocopy well-known miRNA mutants are a promising avenue to identify other components in the miRNA repression of target mRNAs.
The development of the nematode Caenorhabditis elegans proceeds through four larval stages (L1–L4) that are separated by molts, followed by the reproductive adult stage. During development, heterochronic genes regulate the timing of cell fate specification in several tissues, with the result that heterochronic mutants exhibit disrupted synchronization between tissues of developmental timing events (5). lin-4 and let-7 miRNAs are C. elegans heterochronic genes (6, 7). These miRNAs are generated at specific stages to repress their target genes, which encode regulators of cell fate specification (8–10).
The mevalonate pathway is present in all higher eukaryotes and many bacteria and mediates the production of isoprenoids. The isoprenoids feed into a wide range of biosynthetic pathways: sterols, primarily cholesterol; dolichol, which serves as the lipid carrier of the oligosaccharide moiety destined for protein N-linked glycosylation; ubiquinone and heme A, which function in the electron transport chain; prenylated proteins; and isopentenyl adenine, which is present at position 37 of tRNAs that read codons starting with U (11). Cholesterol, the bulk product of the mevalonate pathway in humans and many other organisms, is important for membrane structure and steroid hormone synthesis. C. elegans possesses a functional mevalonate pathway but lacks all enzymes for the synthesis of sterols, suggesting that mevalonate in C. elegans is an important precursor for other biosynthetic pathways (12). Although it does not synthesize cholesterol itself, C. elegans requires exogenously supplied cholesterol for growth and development (13, 14).
Here we present a link between the nonsterol biosynthetic products of the mevalonate pathway and miRNA activity in C. elegans. Inactivation of genes that function in rate-limiting steps of the mevalonate pathway causes desilencing of miRNA targets and retarded heterochronic defects that resemble the effects of mutations in lin-4 and let-7 miRNAs. We show that among the downstream branches of the mevalonate pathway, the biosynthesis of dolichol for protein N-linked glycosylation is important for miRNA activity. Biological functions of protein N-glycosylation include facilitating protein folding and stability, intracellular targeting, intercellular recognition, and others (15). Earlier studies identified Argonaute as a peripheral membrane protein located on the Golgi and/or endoplasmic reticulum (ER) in several mammalian cell lines, and the protein was originally named for this: GERp95 (Golgi ER protein 95 kDa) (16, 17). Our analyses suggest that N-glycosylation is required for miRISC function, for example via regulating the sorting of miRISC to the cellular membrane compartment.
Results
hmgs-1/HMG-CoA Synthase Functions in the let-7 miRNA Pathway.
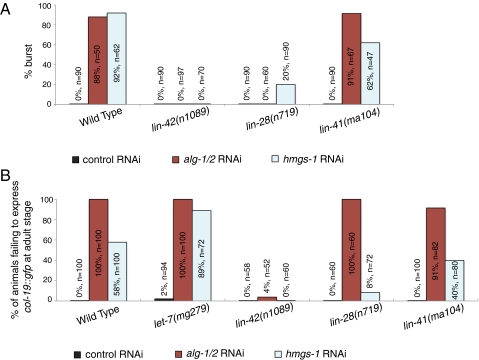
In C. elegans, let-7 miRNA regulates developmental timing events during the L4-to-adult transition. let-7 down-regulates its target genes, such as lin-41 and hbl-1, and this releases their repression of LIN-29, a key transcription factor that is required for the terminal specification of epidermal cells at the adult stage (9, 18, 19). Loss of let-7 activity either by mutation of the let-7 gene or inactivation of dcr-1/Dicer or alg-1/Argonaute, the core components in miRNA maturation and function, causes retarded heterochronic phenotypes in which larval developmental patterns are reiterated and adult-specific specializations do not occur (7, 20). To identify other proteins that act in the miRNA pathway, we screened for gene inactivations that enhance a weak let-7(mg279) reduction-of-function mutation (21). One of the strong hits from this screen, F25B4.6/hmgs-1, encodes the C. elegans ortholog of HMG-CoA synthase. Inactivation of hmgs-1 by RNAi causes let-7–like phenotypes in three independent assays: first, the burst-through-vulva phenotype characteristic of let-7 strong loss-of-function mutations (Fig. 1A); second, the failure to express a reporter gene for an adult-specific collagen, col-19::gfp (Fig. 1B); and third, the failure to produce alae, an adult-specific cuticle structure, at the nominal adult stage (Table S1).
Fig. 1.
Inactivation of hmgs-1 causes let-7–like phenotypes. (A) Shown are the percentage of animals that burst after the L4-to-adult molt upon treatment with control, alg-1/2, or hmgs-1 RNAi in the indicated genetic background. Inactivation of alg-1/2 or hmgs-1 causes bursting with high penetrance in the wild-type animals but lower penetrance in the lin-42(n1089) or lin-28(n719) loss-of-function mutants. (B) Inactivation of hmgs-1 causes animals to not express col-19::gfp in hyp7 cells at the adult stage. The penetrance of this phenotype is elevated in the let-7(mg279) mutant and decreased in the lin-42(n1089), lin-28(n719), or lin-41(ma104) loss-of-function mutants. Inactivation of alg-1/2 also causes animals to not express col-19::gfp in hyp7 cells; this phenotype is suppressed in lin-42(n1089).
Two lines of evidence further support that hmgs-1 functions in the let-7–regulated heterochronic pathway. First, although hmgs-1 inactivation causes relatively weak or incompletely penetrant retarded phenotypes on its own, these phenotypes are strongly enhanced in sensitized genetic backgrounds with compromised let-7 activity (Fig. S1 and Table S1). For example, upon hmgs-1 inactivation, 9% of wild-type animals, but 100% of alg-1(gk214) and 67% of ain-1(ku322) mutants, fail to produce adult-specific lateral alae (Table S1). These mutations in genes that encode the ALG-1/Argonaute protein or the AIN-1/ALG-1 interacting protein compromise miRNA function, and inactivation of hmgs-1 is strongly synergistic with these mutations. Second, the retarded phenotypes caused by hmgs-1 inactivation are suppressed by the loss of function of validated let-7 target genes. For example, the retarded phenotypes of hmgs-1 inactivation are completely suppressed by lin-42(n1089), partially suppressed by lin-28(n719), and weakly suppressed by lin-41(ma104), a hypomorphic mutation that causes only weak precocious phenotypes (Fig. 1 A and B). These data suggest that hmgs-1 functions in the let-7 pathway via the regulation of the activity of validated let-7 target genes lin-28, lin-41, and lin-42.
hmgs-1 Is Required for let-7 Family and lin-4 miRNA Silencing of Target Genes.
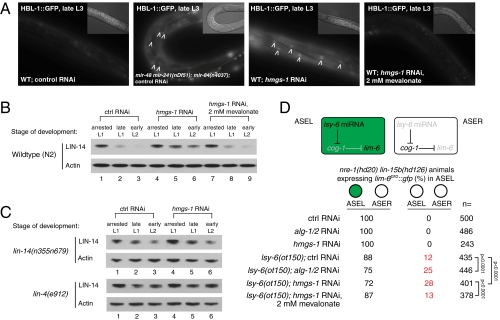
To ask more directly whether hmgs-1 functions in the miRNA pathway, we assayed whether miRNA target mRNAs become derepressed upon inactivation of this gene. We focused on the genetically verified targets of the let-7 family and lin-4 miRNAs. The hunchback factor hbl-1 (hunchback like) is silenced synergistically by the let-7 family of miRNAs (mir-48, mir-241, and mir-84) during the L2-to-L3 stage transition (22). Knocking down hmgs-1 by RNAi prevents down-regulation of hbl-1::gfp at the L3 stage. This resembles the phenotype caused by mutations in the let-7 family of miRNAs (Fig. 2A). We assayed whether the silencing of lin-14 by lin-4 miRNA during the late L1 stage is dependent on hmgs-1. LIN-14 protein levels become derepressed at the late L1 stage by approximately twofold comparing hmgs-1 RNAi-treated to stage-matched control animals, and the derepression is still apparent at the L2 stage (Fig. 2B). To ask whether the desilencing of lin-14 upon inactivation of hmgs-1 is due to reduced lin-4 miRNA repression of lin-14 via its 3′ untranslated region (3′ UTR), we analyzed the down-regulation of lin-14 in the lin-14(n355) gain-of-function mutant, which lacks all of the sites in the lin-14 3′ UTR that are complementary to lin-4 and let-7 and its paralogs (23, 24). lin-14 is not further desilenced when hmgs-1 is inactivated in the lin-14(n355) mutant background (Fig. 2C, Upper). lin-14 is also not further desilenced in the lin-4(e912) null mutant upon inactivation of hmgs-1 (Fig. 2C, Lower). This indicates that the derepression of lin-14 after hmgs-1 inactivation is dependent on the lin-14 3′ UTR and lin-4 miRNA. Together, these results show that hmgs-1 is required for silencing of lin-14 by lin-4 miRNA.
Fig. 2.
Inactivation of hmgs-1 causes desilencing of miRNA target genes. (A) Inactivation of hmgs-1 causes defects in the down-regulation of hbl-1::gfp at the L3 stage. This resembles the phenotype caused by mutations in the let-7 family of miRNAs: mir-48, mir-241, and mir-84. The desilencing of hbl-1::gfp upon hmgs-1 RNAi is rescued by supplementation with 2 mM mevalonate. Images were captured using the same exposure settings and processed identically. Arrowheads point to the desilenced hbl-1::gfp in the nuclei of hyp7 cells. (Insets) Nomarski images. (B and C) Immunoblots. Actin was probed as a control for even loading. (B) Inactivation of hmgs-1 causes defects in the down-regulation of lin-14, the target of lin-4 miRNA, at the late L1 stage (lane 5) and early L2 stage (lane 6). Mevalonate supplementation rescues this phenotype (lanes 8 and 9). (C) Inactivation of hmgs-1 does not further desilence lin-14 in the lin-14(n355n679) mutant lacking the lin-14 3′ UTR or the lin-4(e912) null mutant (compare lanes 5 and 6 to lanes 2 and 3). (D) lsy-6 miRNA is expressed in the ASEL but not ASER neuron in the wild type. It is required for ASEL specification, as judged by the expression of lim-6pro::gfp in ASEL, which is promoted by down-regulation of cog-1 by lsy-6. In a sensitized genetic background with a weak allele of lsy-6, ot150, inactivation of alg-1/2 or hmgs-1 significantly enhanced the ASEL specification defect. Supplementing 2 mM mevalonate rescued the phenotype of hmgs-1 inactivation. Brackets indicate statistically significant difference as judged by a two-tailed χ2 test.
hmgs-1 Acts in miRNA Pathways in Other Cell Types as Well.
hmgs-1 also modulates the activity of miRNAs whose functions are unrelated to developmental timing. lsy-6 is an miRNA that regulates the specification of the taste neurons ASE left (ASEL) and ASE right (ASER) which, even though they are bilaterally symmetric, express distinct patterns of receptor genes based on the asymmetric activity of lsy-6 miRNA (25, 26). Specifically expressed in less than 10 neurons including ASEL but not ASER, lsy-6 down-regulates the cog-1 transcription factor only in ASEL, thus distinguishing the gene expression profile of ASEL from ASER (25, 26). The ASEL neuron of lsy-6(ot71) null mutants fails to down-regulate cog-1 and adopts the ASER pattern of gene expression as a result. On the other hand, animals bearing a hypomorphic allele of lsy-6, ot150, display the ASEL specification defect with incomplete penetrance. Inactivation of genes that are key for miRNA activity, for example nhl-2, significantly enhances the ASEL fate specification defect in the lsy-6(ot150) but not in the wild-type background (27). We asked whether knocking down hmgs-1 causes an ASEL specification defect by scoring lim-6pro::gfp, an ASEL-specific reporter. To enhance the efficiency of RNAi in neurons, we crossed the lim-6pro::gfp reporter into the RNAi-hypersensitive nre-1(hd20) lin-15b(hd126) mutant background (28). RNAi was initiated at the L3 stage of lsy-6(ot150); nre-1(hd20) lin-15b(hd126) parental (P0) animals, and lim-6pro::gfp was scored in the progeny. Twenty-eight percent of hmgs-1 RNAi-treated animals (n = 401), compared with 12% of control RNAi-treated animals (n = 435), showed the ASEL specification defect (Fig. 2D). However, the progeny of nre-1(hd20) lin-15b(hd126) animals with the wild-type lsy-6 gene did not show any ASEL specification defect upon inactivation of hmgs-1. Similar results were obtained by knocking down alg-1/2 by RNAi. This result supports a requirement for hmgs-1 for the efficient down-regulation of cog-1 by lsy-6 miRNA.
Taken together, these observations suggest that hmgs-1 modulates the function of many and perhaps all miRNAs in multiple tissues, and at multiple stages during development.
hmgs-1 Acts Downstream of miRNA Biogenesis and Loading of ALG-1/Argonaute.
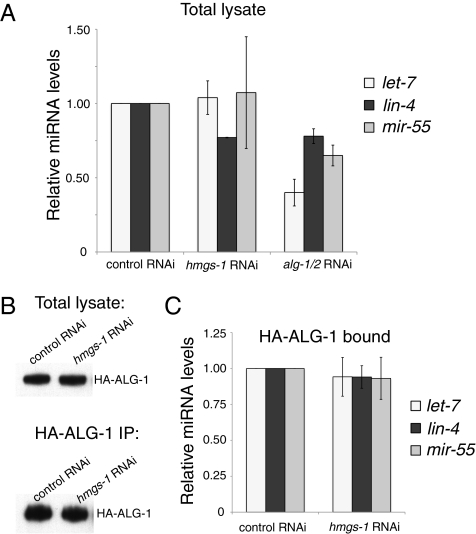
We asked whether hmgs-1 is required for miRNA biogenesis/accumulation or activity. To distinguish between these possibilities, we first assayed the mature miRNA levels by real-time PCR. The levels of let-7, lin-4, and mir-55 all remained unchanged upon knocking down hmgs-1 (Fig. 3A) despite the fact that the targets of let-7 and lin-4 became derepressed. We also assayed the let-7 level in the let-7(mg279) mutant, which has a reduced level of mature let-7 miRNA resulting from defects in the splicing and processing of the let-7 transcript (29). Inactivation of hmgs-1 did not reduce the level of mature let-7 even in this sensitized genetic background (Fig. S2). In contrast, knocking down alg-1/2 caused a significant reduction of miRNA levels, consistent with its role in miRNA biogenesis and stability.
Fig. 3.
hmgs-1 acts downstream of miRNA biogenesis/accumulation and loading of ALG-1. (A) Shown are the mature miRNA levels in total worm lysate, determined by real-time PCR. The miRNA levels are reduced upon alg-1/2 inactivation but remain unchanged upon hmgs-1 inactivation. (B and C) HA-ALG-1 was immunoprecipitated from animals treated with control or hmgs-1 RNAi, and the level of HA-ALG-1–bound miRNAs was determined. (B) Equal amounts of HA-ALG-1 were purified from control and hmgs-1 RNAi-treated animals. Shown is the Western blot of HA-ALG-1 in total lysate (Upper) and from HA-ALG-1 IP (Lower). (C) Relative levels of let-7, lin-4, and mir-55 bound by HA-ALG-1; they remain unchanged upon hmgs-1 inactivation. In A and C, for each miRNA, the result is shown relative to its level in animals treated with control RNAi. The mean and SD were calculated from three biological replicates. Error bars represent SEM.
We also surveyed whether hmgs-1 regulates the protein level and/or cellular localization of the core miRNA cofactors. We monitored ALG-1/Argonaute and AIN-1/ALG-1 interacting protein. Neither the overall expression level nor the subcellular localization of GFP::ALG-1 or AIN-1::GFP was altered upon knocking down hmgs-1 (Fig. S3).
To assay whether hmgs-1 regulates the competence of ALG-1/Argonaute in loading miRNAs, we purified miRISC from synchronized L4-stage alg-1(gk214) mutants rescued with an HA-ALG-1 single-copy construct. HA-ALG-1–bound let-7, lin-4, and mir-55 levels remained unchanged upon inactivation of hmgs-1 (Fig. 3 B and C), indicating ALG-1 loading is unaltered. We also found that the guide:passenger strand ratio of these miRNAs remained unchanged when hmgs-1 was inactivated (Fig. S4). Together, these results position hmgs-1 downstream of miRISC loading and duplex unwinding. It suggests that one or multiple downstream steps, for example the competence of miRISC in finding and silencing its target mRNAs, are dependent on hmgs-1.
Noncholesterol Output of the Mevalonate Pathway Modulates miRNA Activity.
Humans and some other organisms have two forms of HMG-CoA synthase: the cytosolic form, which acts in the mevalonate pathway, and the mitochondrial form, which functions in the production of ketone bodies during starvation. C. elegans bears just the hmgs-1 ortholog of HMG-CoA synthase, which is predicted to be cytosolic. Therefore, we hypothesized that the isoprenoid output of the mevalonate pathway has a role in miRNA activity. Three strands of evidence support this hypothesis, as follows.
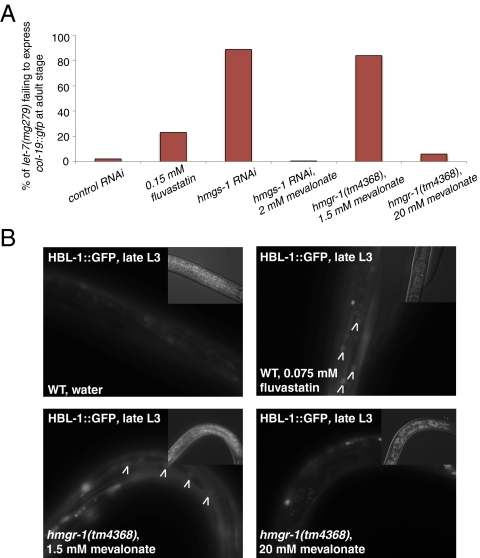
First, we reasoned that if the retarded phenotypes caused by hmgs-1 inactivation are due to reduced biosynthetic outputs of the mevalonate pathway, then supplementing mevalonate, the downstream product of HMG-CoA synthase, should rescue these phenotypes. Indeed, mevalonate supplementation completely rescued all retarded phenotypes caused by inactivation of hmgs-1 (Fig. 4A and Table S2) but did not rescue the retarded phenotypes induced, for example, by inactivation of the Argonaute gene alg-1/2 (Table S2). Mevalonate supplementation also rescued the desilencing of hbl-1::gfp and lin-14 in hmgs-1 RNAi-treated animals (Fig. 2 A and B). Furthermore, the ASEL specification defect upon knocking down hmgs-1 was also rescued by mevalonate (Fig. 2D). In contrast, supplementation to even 50 μg/mL cholesterol (the standard nematode growth medium contains 5 μg/mL cholesterol) did not rescue any of the retarded phenotypes caused by knocking down hmgs-1 (Table S2). This indicates that instead of cholesterol, other biosynthetic products of the mevalonate pathway modulate miRNA activity.
Fig. 4.
The mevalonate pathway modulates miRNA activity. (A) Inactivation of the mevalonate pathway by either application of fluvastatin, inactivation of hmgs-1 by RNAi, or mutation of hmgr-1, with a low level of mevalonate (1.5 mM) supplied in the medium, causes let-7(mg279) mutants to fail to express col-19::gfp in hyp7 cells. This phenotype can be rescued by supplementing mevalonate [2 mM for hmgs-1 RNAi and 20 mM for the hmgr-1(tm4368) mutant]. (B) Inactivation of the mevalonate pathway by application of fluvastatin or mutation of hmgr-1, with a low amount of mevalonate (1.5 mM) supplied in the medium, causes defects in the down-regulation of hbl-1::gfp (targeted by the let-7 family of miRNAs) at the L3 stage. However, hmgr-1(tm4368) animals growing on high mevalonate (20 mM) show wild-type down-regulation of hbl-1::gfp. Images were captured using the same exposure settings and processed identically. Arrowheads point to the desilenced hbl-1::gfp in the nuclei of hyp7 cells. (Insets) Nomarski images.
Second, HMG-CoA reductase (encoded by F08F8.2/hmgr-1) also acts in the miRNA pathway. HMG-CoA reductase is the rate-limiting enzyme that acts immediately downstream of HMG-CoA synthase in the production of mevalonate. The C. elegans hmgr-1(tm4368) mutant strain bears a 620-bp deletion that spans the first three exons, causing a likely null mutation in this gene. The homozygous hmgr-1(tm4368) mutants that segregate from a heterozygote arrest at the L1 stage. However, if mevalonate is added to the growth media to 20 mM final concentration, the homozygous hmgr-1(tm4368) mutants are viable and fertile. We found that hmgr-1(tm4368); let-7(mg279) mutants grown with low (1.5 mM) mevalonate failed to express col-19::gfp, a defect that was not observed when mevalonate was increased to 20 mM (Fig. 4A). In addition, hmgr-1(tm4368) mutants grown with 1.5 mM mevalonate failed to properly down-regulate hbl-1::gfp at the L3 stage, a phenotype that was also rescued by a higher concentration of mevalonate (Fig. 4B).
Third, we found that statins, cholesterol-lowering drugs that inhibit HMG-CoA reductase activity, can compromise let-7 activity. When fluvastatin was added to the growth medium, it caused let-7(mg279) animals to fail to express col-19::gfp at the adult stage (Fig. 4A). Fluvastatin also prevented the proper down-regulation of hbl-1::gfp at the L3 stage (Fig. 4B), similar to RNAi depletion of hmgs-1 or the hmgr-1(tm4368) mutation.
Together, the above results strongly indicate that in C. elegans, the nonsterol outputs of the mevalonate pathway modulate miRNA activity.
The Dolichol Pathway for Protein N-Linked Glycosylation Is Required for miRNA Activity.
Isoprenoids, the end products of the mevalonate pathway, feed into a wide range of downstream pathways in addition to the better-known synthesis of sterols (11). This includes protein prenylation; tRNA isopentenylation; and biosynthesis of ubiquinone, heme A, and dolichol (Fig. S5A). To further delineate which downstream biosynthetic output of the mevalonate pathway modulates miRNA activity, we inactivated genes corresponding to each step in the production and use of isoprenoids and surveyed the phenotypes. We screened for which of these gene inactivations caused let-7(mg279) animals to fail to express col-19::gfp (Table S3).
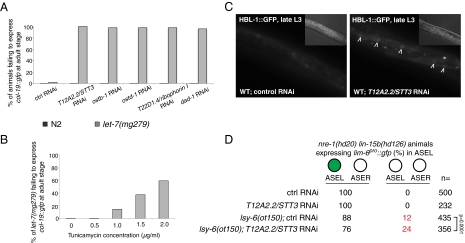
The gene inactivations that caused a let-7–like phenotype, the failure to up-regulate col-19::gfp expression at the adult stage, all correspond to proteins that act in the dolichol pathway for protein N-linked glycosylation (Table S3; for a diagram of the dolichol pathway, see Fig. S5B). The C. elegans oligosaccharyltransferase (OST) complex, which carries out protein N-glycosylation, has five subunits: the catalytic subunit T12A2.2/STT3 and four accessory subunits: T22D1.4/ribophorin I, OSTB-1, OSTD-1, and DAD-1. Depleting any of these subunits by RNAi caused a defect in col-19::gfp expression in the let-7(mg279) mutant but not the wild-type background (Fig. 5A), suggesting a strong genetic interaction with let-7. To further validate this result, we performed the same assay using tunicamycin. Tunicamycin is an antibiotic that blocks the reaction of UDP-GlcNAc and dolichol phosphate in the first step of the dolichol pathway and thus inhibits the synthesis of N-linked glycoproteins. When tunicamycin was added to the worm growth medium, it caused let-7(mg279) adult animals to fail to express col-19::gfp in a dose-dependent manner (Fig. 5B).
Fig. 5.
The dolichol pathway for protein N-glycosylation is required for miRNA activity. (A) Inhibiting OST activity by RNAi depletion of any of its five subunits causes defects in the nominal adult-stage up-regulation of col-19::gfp in hyp7 cells in a let-7(mg279) but not wild-type background. (B) Tunicamycin causes defects in the nominal adult-stage up-regulation of col-19::gfp in the let-7(mg279) mutant in a dose-dependent manner. (C) Inhibiting OST activity by RNAi depletion of its catalytic subunit, T12A2.2/STT3, causes defects in the down-regulation of hbl-1::gfp at the L3 stage. Images were captured using the same exposure settings and processed identically. Arrowheads point to the desilenced hbl-1::gfp in the nuclei of hyp7 cells. (Insets) Nomarski images. (D) RNAi depletion of T12A2.2/STT3 enhances the ASEL specification defect in the lsy-6(ot150) mutant but not wild-type genetic background. Brackets indicate a statistically significant difference as judged by a two-tailed χ2 test.
More directly, we found that inactivation of T12A2.2/STT3 disrupted down-regulation of the hbl-1::gfp reporter at the L3 stage (Fig. 5C). Furthermore, knocking down T12A2.2/STT3 also exacerbated the ASEL neuron specification defect in the lsy-6(ot150) mutant (Fig. 5D), similar to the inactivation of the mevalonate pathway.
Because N-glycosylation facilitates protein folding in the ER, blockage of N-glycosylation by RNAi, mutation, or drugs causes protein misfolding, which induces ER stress and the unfolded protein response (UPR). We asked whether the desilencing of miRNA target genes is an element of the unfolded protein response. We found that several gene inactivations that strongly induce ER UPR did not prevent proper down-regulation of hbl-1::gfp at the L3 stage (Fig. S6A). However, several gene inactivations that induce ER UPR did enhance the failure to express col-19::gfp in the let-7(mg279) mutant, and a mutation in the unfolded protein response factor xbp-1 (30) suppressed this defect (Fig. S6A). The xbp-1(zc12) mutation also suppressed the failure to express col-19::gfp induced by inactivation of T12A2.2/STT3, but not the failure to express col-19::gfp induced by inactivation of alg-1/2 or hmgs-1 (Fig. S6B). This suggests that the mevalonate pathway is required for miRNA activity for more than one reason: first, a relatively direct role of N-glycosylation in miRISC function possibly via regulating the sorting of miRISC to the appropriate cellular membrane compartment; and second, a relaying signaling cascade downstream of ER UPR in opposing miRNA activity (Fig. S6C).
Discussion
Here we report that the dolichol phosphate/protein N-glycosylation output of the mevalonate pathway is required in the miRNA repression of target mRNAs in C. elegans. Mechanistically, our analyses positioned HMG-CoA synthase downstream of miRISC loading and duplex unwinding. This suggests that one or multiple downstream steps, for example the competence of miRISC in finding and silencing its target mRNAs, are dependent on the isoprenoid output of the mevalonate pathway. Support for this idea also emerged from plant miRNA genetics, although that work points to cholesterol output (31).
Several studies have identified Argonaute as a peripheral membrane protein, and its membrane association might be crucial for its function (16, 17, 31). Therefore, our study suggests that N-linked glycosylation could have a relatively direct role in the regulation of assembly, sorting, or turnover of miRISC on the cellular membrane compartment. Meanwhile, disruption of N-glycosylation elicits the ER UPR signaling cascade, which also mildly opposes let-7 activity. However, it is unknown whether the ER UPR intersects a specific subset or a broad group of miRNAs.
Whereas the efficacy of statins to lower cholesterol and in this manner treat atherosclerosis is the general view of their molecular mechanism, the role of the mevalonate pathway in miRNA repression of target mRNAs suggests an miRNA axis in the treatment of heart disease with statins. There are tantalizing hints of the involvement of miRNAs in heart disease and the effects of statins on miRNAs. The mammalian cardiac miRNA miR-208 has been implicated in cardiac fibrosis: Deletion of miR-208 suppresses the high-blood-pressure induction of fibrosis by the heterochronic misexpression of the fetal myosin heavy-chain gene (32). Moreover, the mammalian miRNA miR-122 activates the replication of the hepatitis C virus (HCV) via complementary sequences in the 5′ UTR of the virus (33–36). Statins emerged from drug screens for inhibition of HCV replication and act via blocking the geranylgeranyl pyrophosphate output of the mevalonate pathway (37, 38), which possibly mediates the membrane localization of a particular viral replication protein or down-regulates mir-122 function in a manner similar to the inhibition of let-7 and lin-4 miRNA function in C. elegans we have found. Because statins are so broadly prescribed, it is important to determine whether the therapeutic activities or side effects of statin treatment are due to reduced miRNA activity.
Materials and Methods
C. elegans strains used in this study are listed in SI Materials and Methods. The bursting and alae assays were performed on carefully staged worms as previously described (21). Animals were scored as young gravid adults for expression of veIs13[col-19::gfp] in hyp7 cells. Fluvastatin (EMD) was dissolved in deionized water to produce a 100 mM stock solution, and then added to nematode growth media to final concentrations mentioned in the text. Cholesterol (in ethanol), mevalonic acid [mevalonolactone powder (Sigma), dissolved in deionized water], and tunicamycin (Sigma; dissolved in DMSO) were used similarly, with concentrations mentioned in the text. Details for feeding RNAi, LIN-14 Western blots, miRNA real-time PCR, and ALG-1 immunoprecipitation (IP) are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank G.R. laboratory members, in particular G. Hayes, for constructive suggestions and thoughtful critiques of the work; T. Montgomery for kindly providing the worm strain for ALG-1 IP; M. Newman for helping with mass spectrometry; and Y. Kang for help with sequencing RNAi clones. We thank F. Slack for the [alg-1p::gfp::alg-1] strain, and the Caenorhabditis Genetics Center and the Mitani laboratory for other worm strains. Z.S. is supported by an American Heart Association predoctoral fellowship. This work was supported by National Institutes of Health Grant R01-GM44619 (to G.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202421109/-/DCSupplemental.
References
- 1.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 2.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 4.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 10.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 12.Mörck C, et al. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:18285–18290. doi: 10.1073/pnas.0907117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 14.Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- 15.Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3(2):97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cikaluk DE, et al. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tahbaz N, Carmichael JB, Hobman TC. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem. 2001;276:43294–43299. doi: 10.1074/jbc.M107808200. [DOI] [PubMed] [Google Scholar]
- 18.Rougvie AE, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- 19.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 20.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott AL, et al. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wightman B, Bürglin TR, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 24.Hayes GD, Ruvkun G. Cambridge, MA: Harvard University; 2005. Control of developmental timing by microRNAs in C. elegans. PhD thesis. [Google Scholar]
- 25.Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 27.Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci USA. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 31.Brodersen P, et al. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:1778–1783. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 33.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 34.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda M, et al. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44(1):117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 36.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, et al. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci USA. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.