Abstract
It is known that the liver undergoes size increase and differentiation simultaneously during the postnatal period. Cells in the liver undergo a period of well-controlled proliferation to achieve the adult liver-to-body weight ratio. The postnatal liver growth is also accompanied by simultaneous hepatic differentiation. However, the mechanisms of liver size regulation and differentiation are not completely clear. Herein we report that yes-associated protein (Yap), the downstream effector of the Hippo Kinase signaling pathway, plays a role in liver size regulation and differentiation during the postnatal liver growth period. Postnatal liver growth was studied in C57BL/6 mice over a time course of postnatal days (PND) 0-30. Analysis of nuclear Yap by Western blot indicated peak Yap activation between PND15–20, which coincided with increased cyclin D1 expression and liver cell proliferation. Analysis of postnatal liver development in Yap+/− mice revealed a significant decrease in the liver-to-body weight ratio compared with Yap+/+ mice at PND15 and -30. Yap+/− mice exhibited a significant decrease in postnatal liver cell proliferation, but no change in apoptosis was observed. Furthermore, global gene expression analysis of Yap+/− livers revealed a role of Yap in regulation of genes involved in bile acid metabolism, retinoic acid metabolism, ion transport, and extracellular matrix proteins. Taken together, these data indicate that Yap plays a role in both cell proliferation and possibly in hepatic differentiation during postnatal liver development.
Keywords: Hippo Kinase, caspase-3, proliferating cell nuclear antigen, Krt19, Nur77
it is known that the liver undergoes a remarkable maturation process following birth. This process occurs within the first 30 days in rodents but takes up to 5 years in humans (16). During this postnatal development, the liver acquires an adult gene expression program, and expression of several drug-metabolizing enzymes, transporters, and other enzymes involved in major metabolic processes is initiated (7, 10). Another striking feature of postnatal liver development is the increase in liver size due to extensive hepatic cell proliferation, which increases liver size to the adult liver weight-to-body weight ratio (1–3, 12). The postnatal liver development period is unique because the mechanisms involved in hepatic cell proliferation and hepatic differentiation operate simultaneously during this period. The exact signaling involved in coordinated regulation of the differentiation program and proliferation program is not clear. It is known that several nuclear receptors, including constitutive androstane receptor (CAR), pregnane X receptor (PXR), and hepatocyte nuclear factor (HNF) 4α, are involved in stimulation of hepatocyte-specific gene expression during postnatal liver development (5, 6, 9). Previous studies have shown that Wnt/β-catenin signaling is involved in postnatal liver growth and size increase (1). However, a central mechanism that regulates both liver size increase and hepatic maturation has not been identified.
Recent studies have shown that the Hippo Kinase pathway plays a critical role in regulation of liver size (15, 18). Hippo Kinase signaling is highly conserved between species and is involved in organ size regulation in general (4). Hippo Kinase signaling is initiated by cell-cell adhesion, and the upstream regulators include Fat cadherin as well as NF2 protein (17). The downstream effector of this pathway is a transcriptional coactivator called yes-associated protein (Yap) (4). Yap binds to transcription factor TEAD and initiates promitogenic gene expression. Yap expression is high in rapidly dividing cells, and Yap is concentrated in the nucleus where it activates genes such as survivin and connective tissue growth factor. Yap activity is regulated by phosphorylation catalyzed by a serine/threonine kinase called large tumor suppressor-1 (Lats-1), which is in turn activated by another serine/threonine kinase called Mst. Mst phosphorylates and activates Lats, which in turn phosphorylates Yap at serine-127. Phosphorylated Yap is exported out of the nucleus and targeted for 14–3-3-mediated degradation. Degradation of Yap is known to reduce cell proliferation (15). However, whether Yap plays a role in hepatic differentiation and maintenance of liver homeostasis is currently not known.
We have investigated the role of Yap in postnatal liver development during postnatal days (PND) 0-30. Our studies indicate that, in addition to stimulating cell proliferation, Yap is also involved in hepatic differentiation. Furthermore, our studies have identified additional target genes of Yap that are critical for liver homeostasis.
MATERIALS AND METHODS
Animals.
C57BL/6 pregnant female mice were maintained in an American Association of Laboratory Animal Care-accredited animal facility at the University of Kansas Medical Center. The females were allowed to give birth, and pups (n = 5–6/time point) were killed at PND0, -5, -10, -15, -20, -25, and -30. Both male and female pups were used at PND0, -5, and -10, whereas only male pups were used at the rest of the time points. Yap+/− mice were obtained from Dr. Hiroshi Sasaki of the RIKEN Institute in Japan (14). These mice were originally developed by Drs. Sharon Milgram and Elizabeth Morin-Kensicki at the University of North Carolina (13). The Yap+/− male mice were bred with C57BL/6 female mice for at least six generations before using them in any experiments, and this breeding scheme was maintained throughout these studies. The Yap+/+ mice used in these studies are all from breeding of Yap+/− males with C57BL/6 female mice. Male Yap+/+ and Yap+/− mice (n = 5) were killed at PND15 and -30. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center based on the National Institutes of Health guidelines.
Mice were killed by cervical dislocation under isoflurane anesthesia, and livers were removed. A part of the liver was fixed in 10% neutral buffered formalin and processed to obtain paraffin-embedded sections. A piece of liver was quickly frozen in optimum cutting temperature medium and used to obtain fresh frozen sections. Nuclear and cytoplasmic protein extracts were prepared from freshly isolated livers using the NE-PER cytoplasmic and nuclear protein isolation kit (Pierce, Rockford, IL) at the time of death. Remaining liver tissue was snap-frozen in liquid nitrogen, stored at −80°C, and used to make total cell extracts.
Immunohistochemistry and immunofluorescence.
Four-micrometer-thick paraffin sections were used for proliferating cell nuclear antigen (PCNA) immunohistochemistry as previously described (1). Slides were viewed under an Olympus BX51 microscope equipped with a DP-71 camera and imaging system. Three slides per time point, each from a separate mouse, were used to count 1,000 cells/slide for PCNA-positive cells to obtain the percentage of PCNA-positive cells.
For immunofluorescence of Yap, 4-μm-thick fresh frozen sections were obtained, fixed in 4% formaldehyde, and blocked with 5% normal donkey serum in PBS. Slides were incubated in goat anti-Yap antibody or CK-19 antibody (1:50 in PBS; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Sections were treated with Alexa-fluoro 594-conjugated donkey anti-goat secondary antibody (1:500 in PBS with 2% normal donkey serum) for 1 h and mounted in Prolong Gold mounting medium with DAPI. TUNEL assay was performed on paraffin-embedded liver sections using an In Situ Cell Death Detection Kit, TMR red (catalog no. 2–156-792; Roche Applied Sciences, Indianapolis, IN) as per the manufacturer's protocol. Sections were visualized under an Olympus BX51 microscope equipped with a DP-71 camera and imaging system.
Protein isolation and Western blotting.
Cytoplasmic and nuclear extracts were prepared from fresh liver tissues using the NEPER Nuclear and Cytoplasmic Protein Extraction Reagents (Pierce).
Total protein was isolated from livers using RIPA buffer (1% SDS, 20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1% Triton X-100, 0.25% sodium deoxycholate, and protease and phosphatase inhibitors at a concentration of 1:100) using glass homogenizers. Protein concentrations of nuclear, cytoplasmic, and total protein lysates were determined using the bicinchoninic acid protein assay reagents (BCA method) (Pierce). Nuclear and cytoplasmic extracts (3–5 μg) and total protein lysates (50 μg) were separated by electrophoresis on 4–12% NuPAGE Bis-Tris gels with MOPS buffer (Invitrogen, Carlsbad, CA) and then transferred to Immobilon-P membranes (Millipore, Bedford, MA) in NuPAGE transfer buffer containing 20% methanol. Membranes were stained with Ponceau S to verify loading and transfer efficiency. Membranes were probed with primary and secondary antibodies in Tris-buffered saline-Tween 20 containing either 5% nonfat milk or 5% BSA depending on the antibody used. All antibodies used for Western blot (both primary and secondary) were purchased from Cell Signaling Technologies (Danvers, MA). Signal was visualized by incubating the blots in SuperSignal West Pico chemiluminescence substrate (Pierce) and exposure to X-ray film (MidSci, St. Louis, MO). Blots were scanned, and densitometry was performed using the UN-SCAN-It software (Silk Scientific, Orem, UT).
Caspase-3 assay.
DEVD-Afc was used as a substrate to determine caspase-3 activity. Cytoplasmic extracts (20 μg protein) from the livers of Yap+/+ and Yap+/− mice at PND15 and -30 were mixed with caspase-3 assay buffer [20 mM PIPES, 100 mM NaCl, 1 mM EDTA (0.5 M, pH 8.0), 0.1% CHAPS, 10% sucrose, pH adjusted to 7.2] along with 2 mM DEVD-Afc in a 96-well plate and incubated over a period of 3 h. Fluorescence was measured at an excitation wavelength of 400 nm and emission wavelength of 500 nm every hour on a Tecan Infinite M200 plate reader.
Gene array analysis.
Gene expression analysis for determining the global changes in gene expression of Yap+/− mouse livers was carried out using the Affymetrix Mouse430_2.0 gene chip. Livers were pooled (100 mg/mouse) from three individuals each for Yap+/+ and Yap+/− for RNA isolation using the TRIzol Reagent with the Phase Lock Gel Heavy protocol. The microarrays were background corrected and normalized, and the gene level was summarized using the Robust Multichip Average procedure (8). The resulting log (base 2)-transformed signal intensities were used for ascertaining differentially expressed genes. Fold changes were calculated by transforming the differences in log intensity values to the linear scale between Yap+/− and Yap+/+. All computations were performed in Matlab (R2009b; The MathWorks, Natick, MA) and the Partek Genomic suite (version 6.5; Partek, St. Louis, MO). The gene expression data have been submitted to the GEO database (GSE31281).
Gene set enrichment analysis (GSEA) was carried out using Ingenuity Pathways Analysis [IPA, version 7.6; Ingenuity Systems (www.ingenuity.com)]. Genes with an absolute fold change ≥1.5 (resulting in 590 genes of which 283 were upregulated and 307 downregulated) were selected for analysis. IPA identifies significant networks, functions, and canonical pathways associated with a set of genes based on information gathered in the Ingenuity Pathway Knowledge Base (IPKB). The IPKB is an extensive repository of information on genes and gene products that interact with each other. The significance (P value) of the identified networks, functions, and canonical pathways was calculated using the right-tailed Fisher's Exact Test. This test measures the significance of the overlap between the input genes and the genes in a particular category in the IPKB, reflecting the statistical significance of the network, function, or canonical pathway with respect to the input genes and the reference genes (all genes in the IPKB identified with the particular network, function, or canonical pathway).
To identify the subset of genes that may be directly regulated by Yap, the gene expression data were compared with published Yap-TEAD ChIP-sequencing data (11).
Real-time PCR.
mRNA levels of various genes were quantified in mouse livers using TaqMan-based Real-Time PCR gene expression assay (Applied Biosystems). Total mRNA was isolated from livers using TRIzol reagent (Invitrogen) and converted to cDNA, and Real-Time PCR was conducted on the Applied Biosystems Prism 7300 Real-time PCR instrument using TaqMan Gene Expression Assays as previously described (1).
Statistical analysis.
To determine differences between two groups, Student's t-test was performed. Difference between groups was considered statistically significant at P < 0.05.
RESULTS
Increase in liver size during postnatal liver development.
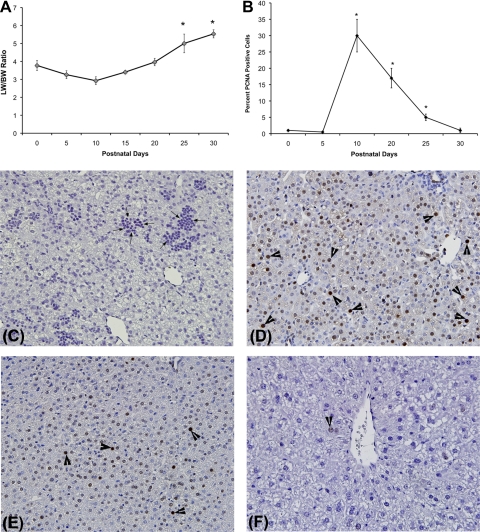
An increase in liver and body weight was observed during the postnatal period, resulting in increased liver-to-body weight ratio between PND0 and PND30. The liver weight increased substantially from PND10 to PND25 and reached the adult (PND90 or 3 mo of age) liver-to-body weight ratio (Fig. 1A) by PND25–30. The increase in liver weight was due to increased cell proliferation as demonstrated by the PCNA analysis. Marked increase in hepatic cell proliferation was observed between PND10 to PND25 (Fig. 1, B–F).
Fig. 1.
Increase in liver size during postnatal liver period. A: changes in liver weight (LW)-to-body weight (BW) ratios during postnatal days (PND) 0–30 in C57BL/6 mice (n = 5–6/time point). *Significant difference at P < 0.05 compared with PND0. B: percent of cells in cell cycle as measured by proliferating cell nuclear antigen (PCNA) immunohistochemistry. *Significant difference at P < 0.05 compared with PND0. C–F: representative photomicrographs of PCNA immunohistochemistry at PND0 (C), PND10 (D), PND20 (E), and PND30 (F). All images at ×400 magnification. Arrows in C point to hematopoietic cells, and arrowheads in D, E, and F show cells in S-phase.
Yap activation during postnatal liver development.
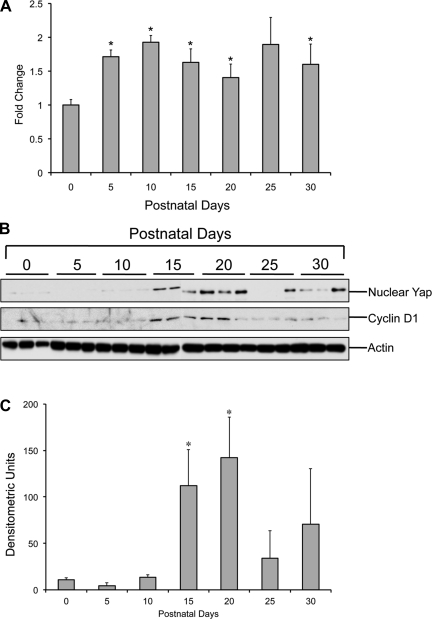
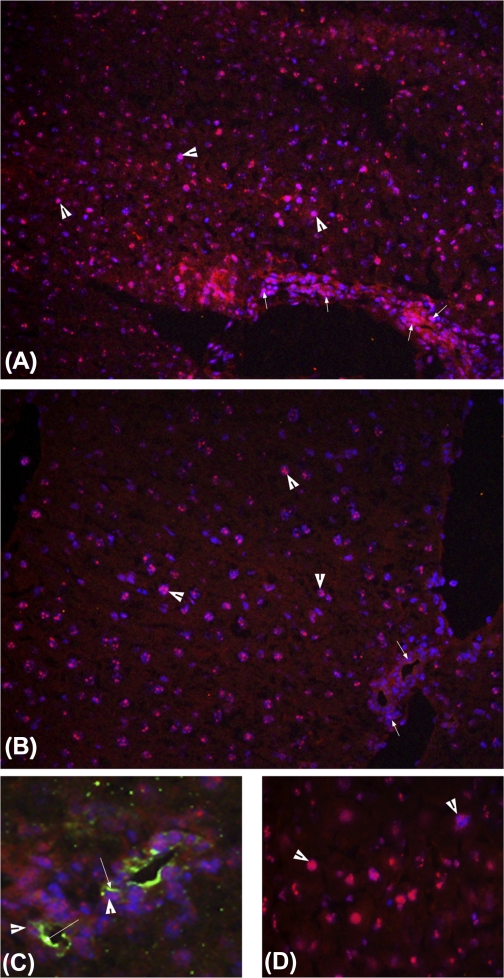
Real-Time PCR analysis for Yap showed a biphasic increase during the postnatal period (Fig. 2A). Yap mRNA increased after birth with a twofold increase at PND10 followed by a decline at PND20 and increased again (1.8-fold compared with PND0) at PND25, which remained unchanged at PND30. Western blot analysis of nuclear Yap (Fig. 2, B and C) showed substantial increase in nuclear Yap levels at PND15, which further increased at PND20. Immunofluorescence staining of Yap corroborated the Western blot data (Fig. 3). At PND20, extensive Yap staining was observed in both hepatocyte and biliary cell nuclei (Fig. 3, A and C). At PND30, substantial cytoplasmic Yap staining was observed along with nuclear staining in hepatocytes (Fig. 3B). Coimmunofluorescence staining of Yap and CK-19 indicated that Yap is also expressed in biliary epithelial cells (Fig. 3C).
Fig. 2.
Changes in yes-associated protein (Yap) expression during postnatal period. A: quantification of Yap mRNA using Real-Time PCR during PND0–30 (n = 5/group). *Significant difference at P < 0.05 compared with PND0. B: representative Western blots of Yap in nuclear extracts and cyclin D1 in total cell extracts from the C57BL/6 mice livers (n = 3) during PND0–30. C: densitometric analysis of nuclear Yap Western blot (shown in B).
Fig. 3.
Immunofluorescence staining of Yap during postnatal period. Staining was performed on sections from 5 individual mice. Representative images are shown. Frozen liver sections from PND20 (A and C) and PND30 (B and D) were used for detection of Yap and CK-19 as described in materials and methods. Red color stains for Yap (all photographs) and green color for CK-19 (in C). DAPI (blue) was used as nuclear counterstain. A and B: images at ×400 magnification; C and D: images at ×600 magnification. Arrowheads point to nuclear Yap staining in all images. Smaller arrows in A and B point to biliary epithelium cells. Larger arrows in C point to CK-19 staining.
Partial deletion of Yap results in decreased liver size.
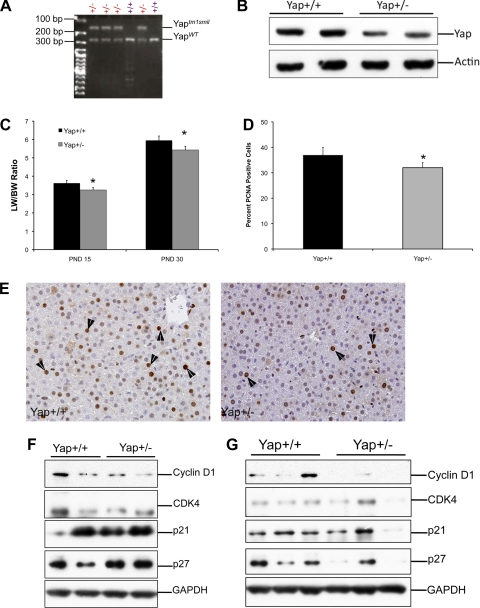
To further study the role of Yap in postnatal liver development, we determined changes in liver size and cell proliferation in the livers of Yap+/− mice. Previous studies have demonstrated that Yap−/− mice are embryonic lethal (13). Yap+/− mice were generated on a C57BL/6 background and identified by PCR-based genotyping (Fig. 4A). Western blot analysis of Yap showed a 50% decrease in Yap protein expression (Fig. 4B). Liver weight-to-body weight ratio analysis indicated a moderate but statistically significant decrease in total liver weight (Fig. 4C) at both PND15 and PND30. PCNA immunohistochemistry revealed that the decrease in liver weights of Yap+/− mice is due to decreased cell proliferation (Fig. 4, D and E). To determine the mechanism behind decreased proliferation in Yap+/− livers, we studied the expression of promitogenic cell cycle protein cyclin D1 and CDK4 along with cell cycle inhibitors p21 and p27 at PND15 and -30. The Western blot analysis indicates a significant decrease in cyclin D1 expression at PND15 and -30 in Yap+/− livers (Fig. 4, F and G). No difference in CDK4, p21, and p27 protein expression was observed. Previous studies have indicated that apoptosis plays an important role in organ size regulation, and Yap has been implicated in stimulating apoptosis. We studied apoptosis in Yap+/+ and Yap+/− livers at PND15 and -30 by caspase-3 activity and TUNEL assay, both of which indicated no change in apoptosis in the livers of Yap+/− mice during postnatal liver development (Fig. 5, A and B).
Fig. 4.
Decreased postnatal liver weights in Yap+/− mice. A: PCR analysis used to identify Yap+/− and Yap+/+ mice (n = 5). B: Western blot analysis of Yap using total cell extracts from livers of Yap+/+ and Yap+/− mice at PND30. C: bar graph showing liver-to-body weight ratios in Yap+/+ and Yap+/− mice livers at PND15 and PND30. *Significant difference at P < 0.05. D: percent of PCNA-positive cells in the livers of Yap+/+ and Yap+/− mice at PND15. *Significant difference at P < 0.05. E: representative photomicrographs of paraffin-embedded liver sections of Yap+/+ and Yap+/− livers stained for PCNA. Arrowheads point to cells in S-phase. Western blot analysis of cyclin D1, CDK4, p21, and p27 at PND15 (F) and PND30 (G) using total cell extracts of Yap+/+ and Yap+/− mice.
Fig. 5.
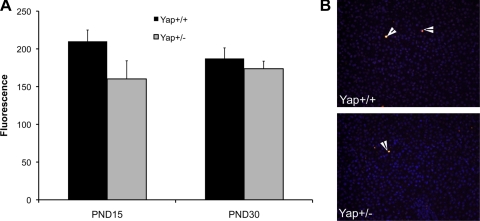
No change in apoptosis in Yap+/− mice. A: caspase-3 activity measured in cytoplasmic extracts obtained from livers of Yap+/+ and Yap+/− at PND15 and PND30 (n = 5). B: representative photomicrographs of TUNEL assay performed on frozen section of Yap+/+ and Yap+/− livers at PND15. Arrowheads point to cells in apoptosis.
Incomplete hepatic differentiation in Yap+/− mice.
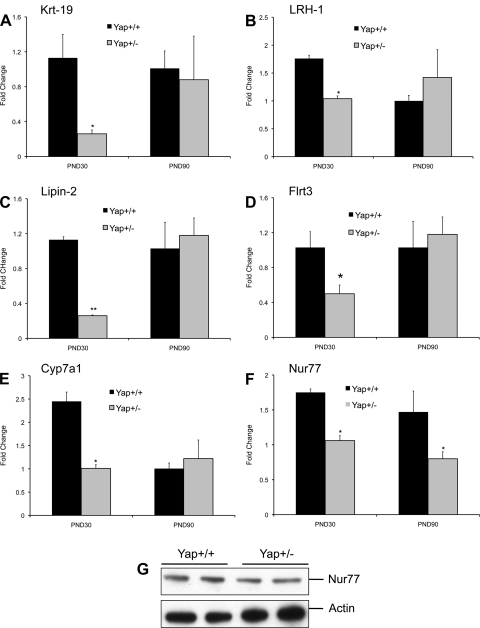
To study the global gene expression changes in the livers of Yap+/− mice, we performed a microarray analysis using an Affymetrix Mouse430_2.0 chip. The data indicate 22 genes were upregulated (Table 1), and 56 genes were downregulated more than twofold in the Yap+/− mice compared with Yap+/+ mice (Table 2). The upregulated genes include enzymes such as aspargine synthetase and glucokinase; proteins involved in lipid metabolism and signaling such as lipin1 and fatty acid binding protein 5; and nonprotein coding mRNA H19. The downregulated genes included a number of genes involved in hepatic differentiation, including many cytochrome P-450 enzymes involved in endobiotic metabolism such as Cyp7a1, Cyp26a1, and Cyp2c39; transporters such as Slc26a3 (a chloride ion exchanger), Slc9a3 (a sodium/hydrogen pump also called NHE-3), and Slc25a25 (calcium-dependent mitochondrial solute carrier); nuclear receptors such as NR5A2 (LRH-1) and NR4A1 (Nur77); and a variety of other enzymes, including cathepsin E, glucose-6-phosphatase, and arginosuccinate synthetase 1. To confirm the gene array data, we performed Real-Time PCR analysis for several genes, including Krt19 (cytokeratin-19), Lipin 2, LRH-1, Flrt3, Cyp7a1, and Nur77 (Fig. 6, A–F) at PND30 and PND90 (3 mo of age). The Real-Time analysis corroborated the gene array data. However, except Nur77, all other genes were expressed at similar levels in Yap+/+ and Yap+/− mice at PND90. The decrease in Nur77 was further confirmed by Western blot analysis at PND30 (Fig. 6G). These data indicate that the Yap expression resulting from the single functional copy of Yap gene in the Yap+/− mice is sufficient to regulate the majority of Yap target genes. To determine which of these genes may be direct targets of Yap, we compared the gene expression data with published ChIP-sequencing data. The results indicate that 26 of the 76 differentially expressed (up or down) genes in Yap+/− livers have a putative Yap (TEAD) binding site (Table 3).
Table 1.
Upregulated genes in Yap+/− mice
| Gene Symbol | Gene Title | Fold Change | Representative Public ID |
|---|---|---|---|
| Asns | Asparagine synthetase | 3.08 | AV212753 |
| S100a8 | S100 Calcium binding protein A8 (calgranulin A) | 3.00 | NM_013650 |
| H19 | H19 fetal liver mRNA | 2.89 | NM_023123 |
| Id2 | Inhibitor of DNA binding 2 | 2.84 | AK013239 |
| Lpin1 | Lipin 1 | 2.76 | AK014526 |
| S100a9 | S100 Calcium binding protein A9 (calgranulin B) | 2.70 | NM_009114 |
| EG624219 | Predicted gene; EG624219 | 2.69 | BM122014 |
| BC023105 | CDNA sequence BC023105 | 2.60 | BC023105 |
| Hes1 | Hairy and enhancer of Split 1 (Drosophila) | 2.45 | BC018375 |
| Agfg2 | Arfgap with FG repeats 2 | 2.37 | BC003330 |
| Serpina7 | Serine (or cysteine) peptidase inhibitor-member 7 | 2.36 | BB222737 |
| Gdf15 | Growth differentiation factor 15 | 2.23 | NM_011819 |
| Lepr | Leptin receptor | 2.19 | U42467 |
| Gfra2 | Glial celllLine-derived neurotrophic factor family receptor-α2 | 2.16 | BB284482 |
| Chka | Choline kinase-α | 2.16 | BB546429 |
| Fabp5 | Fatty acid binding protein 5 | 2.11 | BC002008 |
| Gck | Glucokinase | 2.11 | L38990 |
| Asns | Asparagine synthetase | 2.10 | BC005552 |
| Chka | Choline kinase-α | 2.08 | BB129366 |
| Slc25a32 | Solute carrier family 25; member 32 | 2.06 | AI662800 |
| Hspa1b | Heat shock protein 1B | 2.05 | M12573 |
| Zbtb16 | Zinc finger and BTB domain containing 16 | 2.03 | BQ174973 |
Yap, yes-associated protein.
Table 2.
Downregulated genes in Yap+/− mice
| Gene Symbol | Gene Title | Fold Change | Representative Public ID |
|---|---|---|---|
| Dmbt1 | Deleted in malignant brain tumors 1 | −119.351 | NM_007769 |
| Cyp2c39 | Cytochrome P-450; family 2; subfamily C; polypeptide 39 | −29.7689 | NM_010003 |
| Slc26a3 | Solute carrier family 26; member 3 | −14.2758 | BB118542 |
| Ctse | Cathepsin E | −11.3218 | NM_007799 |
| Cyp26a1 | Cytochrome P-450; family 26; subfamily A; polypeptide 1 | −8.55307 | NM_007811 |
| Myom2 | Myomesin 2 | −7.62812 | AV241307 |
| Abhd1 | Abhydrolase domain containing 1 | −7.49449 | NM_021304 |
| Cox6b2 | Cytochrome c oxidase subunit Vib polypeptide 2 | −7.24847 | AV013496 |
| Muc3 | Mucin 3; intestinal | −7.00953 | AF027131 |
| B4 galnt2 | β-1;4-N-acetyl-galactosaminyl transferase 2 | −5.77575 | AI593864 |
| Flrt3 | Fibronectin leucine rich transmembrane protein 3 | −4.75927 | BE945486 |
| Gadd45 g | Growth arrest and DNA damage-inducible 45 γ | −4.55873 | AK007410 |
| Car9 | Carbonic anhydrase 9 | −4.15615 | AJ245857 |
| EG432681 | Predicted gene; EG432681 | −3.97624 | BQ128572 |
| Krt19 | Keratin 19 | −3.73817 | NM_008471 |
| G6pc | Glucose-6-phosphatase; catalytic | −3.73163 | NM_008061 |
| Cd24a | CD24a antigen | −3.66397 | BB560574 |
| Reg1 | Regenerating islet-derived 1 | -3.41103 | NM_009042 |
| Nr4a1 | Nuclear receptor subfamily 4; group A; member 1 | −3.29199 | NM_010444 |
| Muc1 | Mucin 1; transmembrane | −3.19237 | NM_013605 |
| Lgals2 | Lectin-galactose-binding; soluble 2 | −3.07931 | NM_025622 |
| Cyp7a1 | Cytochrome P-450; family 7; subfamily A; polypeptide 1 | −3.02401 | NM_007824 |
| Pik3ap1 | Phosphoinositide-3-kinase adaptor protein 1 | −2.99077 | BI684288 |
| Cyp4a12a | Cytochrome P-450; family 4; subfamily A; polypeptide 12a | −2.95464 | BC025936 |
| Pde4b | Phosphodiesterase 4B; camp specific | −2.66652 | BM246564 |
| Tspan8 | Tetraspanin 8 | −2.63999 | BC025461 |
| Clic6 | Chloride intracellular channel 6 | −2.63663 | BQ176424 |
| Slc26a3 | Solute carrier family 26; member 3 | −2.61996 | NM_021353 |
| D4Wsu53e | DNA segment; Chr 4; Wayne State University 53; expressed | −2.59026 | BE447520 |
| Myom2 | Myomesin 2 | −2.54276 | BB288010 |
| Lnx1 | Ligand of numb-protein X 1 | −2.53692 | BB131619 |
| Raet1a | Retinoic acid early transcript 1; α | −2.51838 | NM_009016 |
| Ctrb1 | Chymotrypsinogen B1 | −2.4229 | NM_025583 |
| Cd24a | CD24a antigen | −2.42084 | NM_009846 |
| Cyp7a1 | Cytochrome P-450; family 7; subfamily A; polypeptide 1 | −2.39345 | BB667338 |
| Ly6d | Lymphocyte antigen 6 complex; locus D | −2.38334 | NM_010742 |
| Ccrn4l | CCR4 carbon catabolite repression 4-like (Saccharomyces cerevisiae) | −2.35816 | AF199491 |
| Slu7 | SLU7 splicing factor homolog (S. Cerevisiae) | −2.34734 | BC025870 |
| Igh | Immunoglobulin heavy chain complex | −2.3189 | BC019425 |
| Agxt2l1 | Alanine-glyoxylate aminotransferase 2-like 1 | −2.30236 | BC022644 |
| Spink4 | Serine peptidase inhibitor; Kazal type 4 | −2.29513 | AV066321 |
| Shh | Sonic hedgehog | −2.29266 | AV304616 |
| Anxa13 | Annexin A13 | −2.24302 | BC013521 |
| Slc25a25 | Solute carrier family 25 | −2.19421 | BB186319 |
| Acta2 | Actin; α2; smooth muscle; aorta | −2.19323 | NM_007392 |
| Slc9a3 | Solute carrier family 9 (sodium/hydrogen exchanger); member 3 | −2.19082 | AV373598 |
| Lcn2 | Lipocalin 2 | −2.148 | X14607 |
| Igh | Immunoglobulin heavy chain complex | −2.13175 | NM_134051 |
| Dido1 | Death inducer-obliterator 1 | −2.08458 | BM224349 |
| Cd209 g | CD209 g antigen | −2.08145 | NM_027343 |
| Trim12 | Tripartite motif-containing 12 | −2.03762 | BM244351 |
| Ndufa12 | NADH dehydrogenase (ibiquinone) 1α subcomplex; 12 | −2.03668 | BC004633 |
| Fst | Follistatin | −2.03639 | BB444134 |
| Ass1 | Argininosuccinate synthetase 1 | −2.02596 | BM213298 |
| Lpin2 | Lipin 2 | −2.02257 | BB378708 |
| Nr5a2 | Nuclear receptor subfamily 5; group A; member 2 | −2.01462 | NM_030676 |
Fig. 6.
Gene expression change observed in Yap+/− livers. Gene array analysis was performed on pooled liver samples from Yap+/+ and Yap+/− mice (n = 3). mRNA expression of selected gene found to be downregulated in Yap+/− mice at PND30 using gene array analysis was confirmed using Real-Time PCR at PND30 and PND90 (3 mo). Bar graphs show fold change in mRNA expression of Krt19 (A), LRH-1 (B), Lipin-2 (C), Flrt3 (D), Cyp7a1 (E), and Nur77 (F). Gene expression of 18S mRNA was used to normalize the gene expression data. *Statistically significant difference between the groups at P < 0.05. **Statistically significant difference between the groups at P < 0.01. G: Western blot analysis of Nur77 in Yap+/+ and Yap+/− livers at PND30.
Table 3.
Genes changed in the livers of Yap+/− mice with putative Yap binding site
| Gene Symbol | Gene Title | Change in Yap+/− Livers |
|---|---|---|
| Krt19 | Keratin 19 | Repressed |
| Gdf15 | Growth differentiation factor 15 | Induced |
| Ctrb1 | Chymotrypsinogen B1 | Repressed |
| Ndufa12 | NADH dehydrogenase (ubiquinone) 1α subcomplex; 12 | Repressed |
| Clic6 | Chloride intracellular channel 6 | Repressed |
| B4 galnt2 | β-1;4-N-acetyl-galactosaminyl transferase 2 | Repressed |
| Hes1 | Hairy and enhancer of split 1 (Drosophila) | Induced |
| Pik3ap1 | Phosphoinositide 3-kinase adaptor protein 1 | Repressed |
| Zbtb16 | Zinc finger and BTB domain containing 16 | Induced |
| Ass1 | Argininosuccinate synthetase 1 | Repressed |
| Gck | Glucokinase | Induced |
| EG624219 | Predicted gene; EG624219 | Induced |
| Asns | Asparagine synthetase | Induced |
| Shh | Sonic hedgehog | Repressed |
| Pde4b | Phosphodiesterase 4B; cAMP specific | Repressed |
| Lnx1 | Ligand of numb-protein X 1 | Repressed |
| Cyp7a1 | Cytochrome P-450; family 7; subfamily A; polypeptide 1 | Repressed |
| Cyp26a1 | Cytochrome P-450; family 26; subfamily A; polypeptide 1 | Repressed |
| Tspan8 | Tetraspanin 8 | Repressed |
| Fst | Follistatin | Repressed |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α2 | Induced |
| Gadd45 g | Growth arrest and DNA damage-inducible 45 γ | Repressed |
| Nr5a2 | Nuclear receptor subfamily 5; group A; member 2 | Repressed |
| Myom2 | Myomesin 2 | Repressed |
| Id2 | Inhibitor of DNA binding 2 | Induced |
| Flrt3 | Fibronectin leucine-rich transmembrane protein 3 | Repressed |
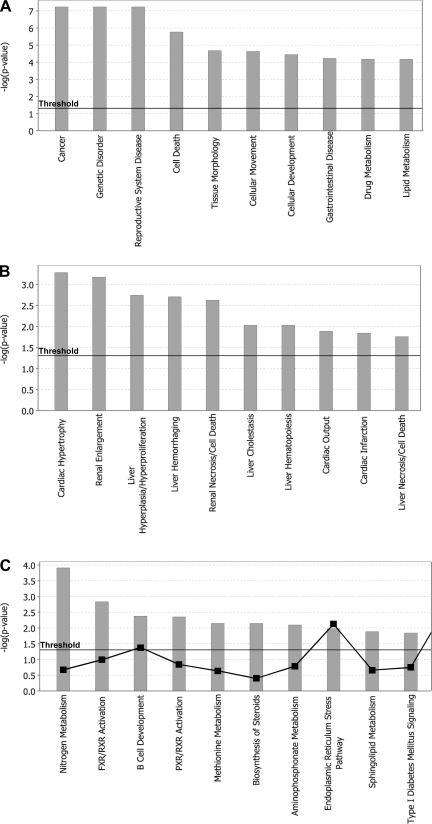
To determine the major metabolic, disease-related, and toxicologically important pathways affected by Yap, we performed gene set enrichment analysis (GSEA) using the IPA software. We selected genes with an absolute fold change ≥1.5. This criterion provided a total of 590 genes, out of which 283 were upregulated and 307 downregulated. The GSEA indicated that genes regulated by Yap are associated mainly with cancer, genetic diseases, and gastrointestinal diseases (Fig. 7A) and cardiac hypertrophy, renal and hepatic enlargement, and hepatic cell deaths (Fig. 7B). The IPA analysis also revealed that Yap regulates genes associated with many liver-specific physiological processes, including nitrogen metabolism, sphingolipid and methionine metabolism, and pathways coregulated by farnesoid X receptor and PXR (Fig. 7C).
Fig. 7.
Gene set enrichment analysis [Ingenuity Pathway Analysis (IPA)] of global gene expression changes in Yap+/− mice during postnatal days. Gene array analysis was performed on pooled liver samples from Yap+/+ and Yap+/− mice (n = 3). Global gene expression changes were determined in the livers of Yap+/− mice at PND30 using Affymetrix Mouse430_2.0 microarray. The data were subsequently analyzed using the IPA system. Bar graphs show the top 10 gene sets for the particular IPA category in the biological function group (A), toxicological function group (B), and canonical pathways involved in normal physiology (C).
DISCUSSION
Hippo Kinase pathway has been recognized as the major pathway regulating liver size and is implicated in the pathogenesis of hepatocellular carcinoma (HCC) (18). Yap, the downstream activator of the Hippo Kinase pathway, is an oncogene, and an increase in Yap activation has been observed in many cancers, including HCC (15). However, the role of the Hippo Kinase pathway and Yap in liver development and maturation has not been studied. Liver undergoes extensive metabolic maturation and size increase during the postnatal period, which is an excellent model to study growth and differentiation mechanisms simultaneously (1, 16). In these studies, we tested the hypothesis that Yap is involved in hepatic cell proliferation during postnatal liver development. Interestingly, our data suggest that Yap is not only involved in postnatal liver proliferation and size increase but also in hepatic maturation.
We observed an increase in Yap activation coinciding with increased cell proliferation in C57BL/6 mice. Furthermore, we observed decreased liver size in Yap+/− mice secondary to decreased cell proliferation. These data indicate that Yap plays a critical role in postnatal liver size increase. This observation is in agreement with previous reports that ectopic expression of Yap stimulates cell proliferation and hepatomegaly in the liver (4). The moderate decrease in liver-to-body weight ratios and cell proliferation in the Yap+/− mice can be explained by the fact that the Yap+/− mice are heterozygous and still maintain ∼50% of Yap protein expression. Similarly, other pathways such as Wnt/β-catenin signaling have also been implicated in postnatal liver growth, which may compensate for the loss of Yap-mediated cell proliferation (1). The decrease in cell proliferation in Yap+/− mice may be associated with a significant decrease in cyclin D1 expression, but the exact mechanism by which Yap regulates cyclin D1 levels during postnatal liver growth is unknown.
Interestingly, we also observed a significant increase in Yap activation during the late postnatal period at PND30, during which hepatocyte proliferation decreases and hepatocyte differentiation is completed. A gene array analysis on the livers of Yap+/+ and Yap+/− mice at PND30 indicated a significant decrease in liver-specific gene expression in Yap+/− livers. These data suggest that Yap is also involved in hepatocyte maturation. Our analysis further revealed that 26 of the 76 differentially expressed genes in Yap+/− mice have a putative Yap-TEAD binding site indicating these genes may be directly regulated by Yap. This observation is supported by a recent study where hepatocyte-specific deletion of Yap resulted in decreased liver function and increased hepatocyte turnover (17). The gene array data indicate that Yap regulates genes involved in a variety of hepatocyte-specific functions, including retinoic acid metabolism, bile acid metabolism, and nitrogen metabolism. Furthermore, a significant decrease in Krt19 gene expression was observed in Yap+/− mice, indicating a role of Yap in regulating expression of CK-19, a protein involved in biliary epithelium development. These data indicate that Yap activation occurs in mature hepatocytes as well as biliary epithelium cells and is involved in regulation of cell-specific gene expression.
It is known that Yap is a transcriptional coactivator and stimulates gene expression after forming a dimer with transcription factors such as TEAD. Our data raise the possibility that Yap may regulate liver-specific gene expression by cooperating with liver-specific transcription factors such as HNF4α, HNF1β, and other nuclear receptors such as CAR and PXR. The exact mechanism behind regulation of many of the specific genes that were downregulated in Yap+/− livers remains to be studied. However, the gene expression data clearly support the role of Yap in hepatic maturation.
Our results indicate that, apart from regulating organ size via control of cell proliferation, the Hippo Kinase signaling pathway also contributes to cell differentiation. This is plausible because the Hippo Kinase pathway is activated by cell-cell contact, which is necessary for epithelial cell maturation. Thus, the role of Yap in postnatal liver growth and maturation suggests that the Hippo Kinase signaling pathway and Yap respond not only to stimuli that regulate organ size but also to stimuli that induce cell differentiation. The Hippo Kinase pathway and Yap may in fact be the switch between cell proliferation and cell maturation.
GRANTS
This research was supported in part by the American Cancer Society Institutional Research Pilot Grant and National Institutes of Health-COBRE Pilot project (5 P20 RR-021940-03) to U. Apte. The KUMC-Microarray Facility is supported by the Kansas University-School of Medicine, KUMC Biotechnology Support Facility, the Smith Intellectual and Developmental Disabilities Research Center (HD-02528), and the Kansas IDeA Network of Biomedical Research Excellence (RR-016475).
DISCLOSURES
No conflicts of interest are declared by the authors.
ACKNOWLEDGMENTS
We thank Dr. Steve Hart (University of Kansas Medical Center) for the comparative gene expression and ChIP-Seq analysis. We thank Dr. Elizabeth Morin-Kensicki (University of North Carolina at Chapel Hill) and Dr. Hiroshi Sasaki (RIKEN Institute) for kindly providing Yap+/− mice. We thank the Kansas University Medical Center (KUMC)-Microarray Facility for generating array data sets.
REFERENCES
- 1. Apte U, Zeng G, Thompson MD, Muller P, Micsenyi A, Cieply B, Kaestner KH, Monga SP. beta-Catenin is critical for early postnatal liver growth. Am J Physiol Gastrointest Liver Physiol 292: G1578–G1585, 2007 [DOI] [PubMed] [Google Scholar]
- 2. Behrens A, Sibilia M, David JP, Mohle-Steinlein U, Tronche F, Schutz G, Wagner EF. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J 21: 1782–1790, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dalu A, Mehendale HM. Efficient tissue repair underlies the resiliency of postnatally developing rats to chlordecone + CCl4 hepatotoxicity. Toxicology 111: 29–42, 1996 [DOI] [PubMed] [Google Scholar]
- 4. Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science 281: 692–695, 1998 [DOI] [PubMed] [Google Scholar]
- 6. Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 23: 2–7, 2008 [DOI] [PubMed] [Google Scholar]
- 7. Hart SN, Cui Y, Klaassen CD, Zhong XB. Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab Dispos 37: 116–121, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data (Anstract). Nucleic Acids Res 31: e15, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kakizaki S, Yamazaki Y, Takizawa D, Negishi M. New insights on the xenobiotic-sensing nuclear receptors in liver diseases–CAR and PXR. Curr Drug Metab 9: 614–621, 2008 [DOI] [PubMed] [Google Scholar]
- 10. Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62: 1–96, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, Ding S, Guan KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Limaye PB, Bhave VS, Palkar PS, Apte UM, Sawant SP, Yu S, Latendresse JR, Reddy JK, Mehendale HM. Upregulation of calpastatin in regenerating and developing rat liver: role in resistance against hepatotoxicity. Hepatology 44: 379–388, 2006 [DOI] [PubMed] [Google Scholar]
- 13. Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Pan D. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walthall K, Cappon GD, Hurtt ME, Zoetis T. Postnatal development of the gastrointestinal system: a species comparison. Birth Defects Res B Dev Reprod Toxicol 74: 132–156, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol 20: 638–646, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]