Abstract
Until recently, the study of microbial diversity has mainly been limited to descriptive approaches, rather than predictive model-based analyses. The development of advanced analytical tools and decreasing cost of high-throughput multi-omics technologies has made the later approach more feasible. However, consensus is lacking as to which spatial and temporal scales best facilitate understanding of the role of microbial diversity in determining both public and environmental health. Here, we review the potential for combining these new technologies with both traditional and nascent spatio-temporal analysis methods. The fusion of proper spatio-temporal sampling, combined with modern multi-omics and computational tools, will provide insight into the tracking, development and manipulation of microbial communities.
Introduction
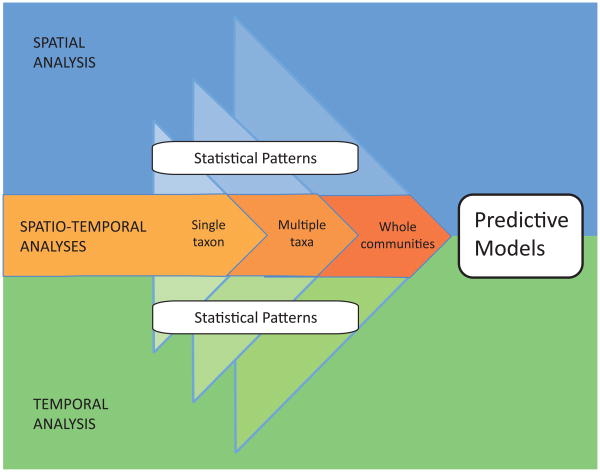
Microorganisms are ultimately responsible for the health of all other organisms. Communities of microbes decompose materials, recycle nutrients, break down pollutants, and aid in the digestion of food in “higher” animals -- they are the “ubiquitous janitors of the Earth” (1). Microbes can also cause disease, destroy our food sources, and degrade our structures (as reviewed in (1)). Early studies characterized a limited snapshot of microbial diversity. However, to predict or manipulate microbially mediated processes, we must understand spatial and temporal patterns of diversity at multiple levels, Figure 1.
Figure 1. Improving our understanding of the variability of microbial communities.
Spatial and temporal studies ranging in scale from that of individual taxa to communities have allowed us to detect patterns of distribution. However, in order to be able to fully understand the nature and ranges of variability in microbial systems, research focusing on improving our ability to predict whole communities across heterogenous space and time is much needed. Building upon existing studies and tools of analysis will help us move from simply describing statistical patterns to developing better predictive models. The accumulation of spatio-temporal studies for microbial communities across different ecosystems will provide essential information about the expected scales of variability, allowing for better biological and ecological interpretations of deviations from normal ranges of variability.
Because of the astounding diversity of microbial communities(2), the ability to characterize their fine-scale temporal and spatial variation has only become achievable within the past 5 years. Next-generation “-omics” technologies such as high-throughput amplicon, metagenome, and metatranscriptome sequencing (3, 4) allow collection of thousands to millions of sequences at prices the average researcher can now afford. Recent research using next-generation technologies has focused on temporal and spatial associations(5-8) of microbial communities, concentrating on clustering (e.g. microbial communities from different body habitats cluster separately(9)) or correlations (e.g. soil bacterial diversity is correlated with pH(10)) However, unifying theories linking overall spatial and temporal variation in microbial community structure to system functional processes have remained elusive. Under similar environmental conditions, microbial communities can have strikingly different composition and function(11, 12); conversely, different community configurations can function similarly(13, 14).
Here, we discuss how community statistics from ecology can increase understanding of microbial community structure-function relationships. This review covers recent advances in molecular, culture-independent studies, and highlights advances in statistical methods for detecting and explaining spatially and/or temporally explicit patterns in microbial community composition. We conclude that combined spatial and temporal studies that exploit inexpensive DNA sequencing, together with improved bioinformatics tools and large-scale automated sampling techniques, will aid in understanding and predicting both the temporal and spatial patterning of microbial communities, and the processes that underlie these patterns.
Foundations of Modern Microbial Community Ecology
Advances in sequencing technology and analytic tools to quantify differences between communities have enabled the recent boom in microbial diversity studies. Limitations in sequencing cost and coverage have largely been overcome with next generation sequencing technologies that directly read single-nucleotide additions to DNA strands (454 and Illumina)., Analytic obstacles due to issues of species identification were conceptually harder to overcome. For example, the traditional definition of “species-level” Operational Taxonomic Units (OTUs) at 97%, although useful for an overview of diversity, is problematic because it assumes that rates of evolution and inherent genetic variation are equivalent across species, and excludes phylogenetic information contained in sequence data(15-17). Phylogenetic community distance metrics such as UniFrac(18) reduce the impact of arbitrary OTU thresholds and often provide better community clustering than taxon-based methods, because even an imperfect phylogenetic tree gives a far better picture of relationships among microbes than does the “phylogeny” where all taxa are equally related implicitly assumed by taxon-based approaches(19).
Spatial studies of microbial diversity
Understanding how microbial communities vary at different spatial scales is important because diversity hotspots and deserts can be identified, correlations with environmental factors can be detected, and hypotheses about dispersal limitation or stochasticity of community assembly can be tested. These issues of spatial scaling can be critical in downstream applications; for example, it is essential to know the scale at which microbial diversity varies when designing a bioprospecting sampling campaign to maximize the diversity of organisms or genes surveyed(20-22).
Central to the investigation of microbial spatial patterns is one of the most-contested hypothesis in microbial ecology: “everything is everywhere, but the environment selects” (23); i.e. that local environmental conditions, or ‘filters’, select for community assemblages that can best exploit or survive. Although several important environmental filters have been well-studied (e.g. (18, 24-27), historical filters (i.e. traces of past events or community states) are also important in structuring communities (28, 29). Thus, a primary goal of spatial analysis techniques should be to measure the separate contribution of history (e.g. community assembly and patterns of microbial dormancy) and environment as a function of the spatial configuration of the dataset. Ultimately, spatial pattern analysis improves our ability to predict microbial diversity hotspots, and where microbes and their associated functions are abundant in a given habitat.
Indeed, there are some strongly predictive patterns in spatial microbial diversity. For example, taxa-area analysis (regression of taxa observed vs. sampling area) has been adopted from island biogeography(30). As with macroorganisms, larger areas harbor more microbial lineages, and the taxa-area relationship has been referred to as one of the only ‘laws’ of ecology(31). In contrast, other spatial patterning of microbial alpha diversity deviates from patterns observed for macroorganisms. For example, the well-established pattern in macroorganisms that diversity decreases with latitude seems to apply to marine (32)but not soil microbes (10), and significant relationships between elevation and microbial diversity have not been observed (33).
As noted above, phylogenetic beta diversity has been widely applied: it has been shown to vary systematically at scales ranging from between continents(17, 27, 34) to within a single hand or computer keyboard(35). Whether communities of similar composition cluster together in space can be detected using variograms, which graphically represent the relationship between two distance matrices(7, 17); the statistical significance of these associations can explored with the Mantel test(36). These representations also allow researchers to fit models that explicitly determine the range of spatial autocorrelation (37) and produce input for spatially explicit prediction algorithms, such as kriging or spatial-partial regression(38), which explain the overall patterns of variation and faithfully interpolate community structure at sites not directly observed.
Although community-level measurements and associations provide useful tools for microbial biogeography, studies of individual taxa that are key players in an ecosystem are also critical: for example, knowing that cyanobacteria are early colonizers because they fix nitrogen and act as primary producers provides a level of insight that cannot be obtained purely from patterns of similarity and difference at the whole community level. Recently employed methods that refine overall spatial patterns in microbial communities include subdividing analyses by taxon (ranging from species to phylum-level), which can be used as inputs for niche modeling (7, 39), and using ordination techniques to identify indicator taxa (these include SAMOVA, DFA, and SIMPER), Table 1. These methods allow the user to identify an individual clade or consortium, and, when applied to metagenomic data, specific genes, that affect function (e.g. the ability of gut Bacteroidetes in Japanese people to degrade polysaccharides found in seaweed, presumably horizontally transferred from marine Bacteroidetes consumed along with sushi(40)). These newer methods share similar goals and often-statistical methodologies, but detailed comparisons on the same data have not yet been performed and there is not yet consensus about their strengths and weaknesses (41).
Table 1. Different types of spatial and temporal analysis, and their applications.
This table highlights the deep connections between spatial and temporal studies, and the frequent applicability of techniques developed for one type of analysis to the other. Further spatial and temporal sampling in projects such as the Earth Microbiome Project will likely highlight additional deep connections.
| Original use (developed for) |
Analysis type | Analytical method / tool | Information provided when applied in spatial studies | Information provided when applied in temporal studies |
|---|---|---|---|---|
| Spatial studies | Correlative | Variograms | Indication of spatial autocorrelation | Identification of the appropriate time scale for sampling |
| Spatial studies | Correlative | Taxa-accumulation plots | An indication of how many more taxa are detected by sampling larger areas (i.e. taxa-area curves) | An indication of how many more taxa are detected by sampling additional time points |
| Spatial studies | Predictive | Kriging | Interpolation of data at locations missing direct observation (e.g. continuous chloropleth maps) | Interpolation of data at time points missing direct observation |
| Temporal studies | Correlative | Frequency transformations | NA (or not yet determined?) | Identification of cyclical or periodic fluctuations in community structure/composition over time |
| Both | Correlative | Ordination methods | Geographic structuring of taxa/communities based on a metric of similarity | Temporal structuring of taxa/communities based on a metric of similarity |
| Both | Correlative | Network analyses | Association or co-occurrence patterns of taxa and/or communities over space | Association or co-occurrence patterns of taxa and/or communities over time |
| Both | Predictive | Niche modeling | Prediction of expected taxon/community distributions in space, indication of important environmental filters | Forecasting of taxon/community distributions with changing conditions over time, assessment of niche conservation |
Temporal studies of microbial diversity
Like spatial studies, temporal studies can identify taxa shared at different times, correlations with environmental conditions that affect the communities, and the relative contribution of different processes, including stochastic processes and priority effects, to community structure (14, 42). However, unlike spatial studies, temporal studies have provided greater insight into processes. Recent studies in different environments have shown that some communities exhibit cyclical patterns (26), others exhibit a monotonic trajectory (e.g. the development of the gut microbial community from the newborn to the adult (43)), and others remain relatively stable over time (e.g. the human mouth community (44), or the hypolimnion of stratified bog lakes(45)).
Temporal studies are now moving beyond observing temporal patterns using ordination or other visualizations of community associations. A growing toolbox of descriptive, non-parametric statistics for temporal microbial dynamics exists. These techniques include correlation networks and analysis of community rate of change over time, and can be used for exploring temporal patterns, then relating these patterns to specific biotic or abiotic drivers. For example, temporal associations between commonly occurring OTUs in aquatic microbial time series have be uncovered using correlation-based networks analyses, such as local similarity analysis (46-48). Specifically, different ecosystems sharing a regional or climactic driver were shown to exhibit similar changes in their microbial communities (e.g. (46, 49)). In addition, aquatic bacterial communities correlate far better with phytoplankton communities than with physical and chemical properties of the system (50). There are also non-parametric methods that specifically test for temporal structure such as cycles, trajectories, and serial objects (e.g. (36, 51)). Finally, measuring rates of change in the whole microbial community or in specific taxa over time allows comparisons of these rates across ecosystems or experimental treatments(52).
An extensive legacy of aquatic timeseries observations exists due to routine limnological and marine sampling efforts, and timeseries are now becoming available in other systems such as wastewater treatment (e.g. (53, 54)), host-associated systems(44), and air (e.g. (55)). The accumulation of more of these rich time series datasets for microbial communities across different ecosystems will likely provide essential the first baseline for the expected nature and scale of variability in microorganisms. An added benefit will be improved detection of responses to disturbance events in microbial communities outside of the expected variation, such as those observed after episodic typhoon events in a sub-tropical lake (56)or in the human gut after antibiotic treatment(57). Such studies of response to perturbation are critical for developing an understanding of factors that lead to resilience in different communities, and for predicting microbial responses to a changing planet.
Combining spatial and temporal studies of microbial communities towards predictive models
Combining spatial or temporal series can reveal key features of a system. For example Caporaso et al. (44) combined both spatial and temporal components in a microbial study to assess the variation of microbial communities of the human microbiome. The addition of extensive temporal sampling to a previous study(9) led to a novel perspective: although each spatially explicit location on the human body retains a compositional difference from other locations, the communities within each location shift over time. That is, each location retains only a small ‘temporal core’ of species-level phylotypes within a community over time.
Analysis tools developed for spatial studies can sometimes be applied to temporal studies, or vice versa. For example, in(58), variograms were used to identify the temporal scales at which E. coli concentrations increased within a watershed. Similarly, wavelet analysis, a method to find the dominant periodic phenomena in a time series by decomposing those signals on a local timescale, was used in an in vitro gut microbial community analysis which revealed the strongest population cycles in Bacteroidetes and Firmicutes(59). Creative applications of techniques developed for one type of study to others will likely yield additional benefits in future, as will merging the two types of studies. For example, samples taken along a spatial gradient that vary due to time, known as chronosequences, e.g. ecosystem succession (60), glacial recession(61), spoil heap development (62) inherently have both spatial and temporal components. However, spatial and temporal patterning are conflated in such studies; techniques such as niche modeling could potentially assist in resolving these issues. Similarly, studies that track timeseries at multiple sites or in multiple subjects will be essential for understanding factors that affect community dynamics as well as structure. Describing different resolutions of microbial community spatial variability, from microns to continents, and temporal variability, from hours to decades, will inform prediction of dynamics and responses to novel events.
Conclusion
Our review has described issues concerning the description of microbial communities and their relevant impact on the systems in which they reside. We have also described the use of existing and nascent analytical tools. In the past, studies of spatial and temporal dynamics of microbial communities have been limited to descriptions, rather than predictive models (7, 22). The advent of high-throughput multi-omics tools and the decreasing cost of automated sampling equipment enables adoption by the scientific community. Eventually, model-based approaches will be a key goal of future studies of microbial communities, and will allow us to test and appropriately apply what we think we have learned.
Highlights.
Microbial community studies with spatial or temporal axes have become more common due to the democratization of sequencing technologies
Current analysis tools allow us to statistically test our hypothesis but give few insights into causal relationships or the development of predictive models
There are multiple scales of temporal and spatial dynamics in microbial communities that have yet to be described in many systems, but will inform prediction.
Future spatial or temporal analysis must use both spatial and temporal components with model-based algorithms to properly assess and predict microbial community divergence between health states
Acknowledgments
We thank Diana Nemergut for helpful comments and suggestions. The work described in this review was supported by the National Institutes of Health, the Bill and Melinda Gates Foundation, the Crohns and Colitis Foundation of America, the Colorado Center for Biofuels and Biorefining and the Howard Hughes Medical Institute. Ashley Shade is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Colwell RR. Microbial diversity: the importance of exploration and conservation. J Ind Microbiol Biotechnol. 1997 May;18(5):302–7. doi: 10.1038/sj.jim.2900390. [DOI] [PubMed] [Google Scholar]
- 2.DeLong EF, Pace NR. Environmental diversity of bacteria and archaea. Syst Biol. 2001 Aug;50(4):470–8. [PubMed] [Google Scholar]
- 3.Green JL, Bohannan BJ, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008 May 23;320(5879):1039–43. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 4.DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009 May 14;459(7244):200–6. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- 5.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003 Aug 15;301(5635):976–8. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 6.Paerl RW, Turk KA, Beinart RA, Chavez FP, Zehr JP. Seasonal change in the abundance of Synechococcus and multiple distinct phylotypes in Monterey Bay determined by rbcL and narB quantitative PCR. Environ Microbiol. 2011 Sep 29; doi: 10.1111/j.1462-2920.2011.02594.x. [DOI] [PubMed] [Google Scholar]
- **7.King AJ, Freeman KR, McCormick KF, Lynch RC, Lozupone C, Knight R, et al. Biogeography and habitat modelling of high-alpine bacteria. Nat Commun. 2010;1:53. doi: 10.1038/ncomms1055. This paper combines high-throughput sequencing with spatial interpolation (kriging) and comparison of physical/chemical with spatial gradients to uncover ecological drivers of distributional patterns across soil communities. [DOI] [PubMed] [Google Scholar]
- 8.Boucher D, Jardillier L, Debroas D. Succession of bacterial community composition over two consecutive years in two aquatic systems: a natural lake and a lake-reservoir. FEMS Microbiol Ecol. 2006 Jan;55(1):79–97. doi: 10.1111/j.1574-6941.2005.00011.x. [DOI] [PubMed] [Google Scholar]
- 9.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009 Dec 18;326(5960):1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. 2006 Jan 17;103(3):626–31. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006 Feb;4(2):102–12. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 12.Balser TC, Firestone MK. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry. 2005;73(2):395–415. [Google Scholar]
- 13.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009 Jan 22;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14288–93. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009 Jul;19(7):1141–52. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2010 Oct;7(10):813–9. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robeson MS, King AJ, Freeman KR, Birky CW, Jr, Martin AP, Schmidt SK. Soil rotifer communities are extremely diverse globally but spatially autocorrelated locally. Proc Natl Acad Sci U S A. 2011 Mar 15;108(11):4406–10. doi: 10.1073/pnas.1012678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008 Jul;32(4):557–78. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010 Jan;4(1):17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivaji S, Pratibha MS, Sailaja B, Hara Kishore K, Singh AK, Begum Z, et al. Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles. 2011 Jan;15(1):1–22. doi: 10.1007/s00792-010-0333-4. [DOI] [PubMed] [Google Scholar]
- 21.Lacap DC, Barraquio W, Pointing SB. Thermophilic microbial mats in a tropical geothermal location display pronounced seasonal changes but appear resilient to stochastic disturbance. Environ Microbiol. 2007 Dec;9(12):3065–76. doi: 10.1111/j.1462-2920.2007.01417.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Kang S, Schadt CW, Garten CT., Jr Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7768–73. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Malley MA. The nineteenth century roots of ‘everything is everywhere’. Nat Rev Microbiol. 2007 Aug;5(8):647–51. doi: 10.1038/nrmicro1711. [DOI] [PubMed] [Google Scholar]
- 24.Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011 Jan;13(1):135–44. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. Global patterns in belowground communities. Ecol Lett. 2009 Nov;12(11):1238–49. doi: 10.1111/j.1461-0248.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- **26.Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2011 Aug 18; doi: 10.1038/ismej.2011.107. This 6 year marine timeseries shows which bacterial taxa have strong seasonal patterns as well as seasonal patterns in diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011 Sep;5(9):1414–25. doi: 10.1038/ismej.2011.24. This study tracks both bacterial and eukaryotic marine taxa, showing that the use of correlation networks among taxa and among environmental parameters detects keystone species and reveals that the abundance of many bacterial taxa correlates better with protist taxa than with physical/chemical parameters, suggesting that top-down control of this ecosystem is critical. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Bahl J, Lau MC, Smith GJ, Vijaykrishna D, Cary SC, Lacap DC, et al. Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat Commun. 2011 Jan 25;2:163. doi: 10.1038/ncomms1167. This study shows that at fine scales of taxonomic resolution the timing of adaptive radiation in microbes can be detected, suggesting that short-read sequencing techniques that give a broad view of microbial diversity will need to be complemented with whole-genome methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt SK, Lynch RC, King AJ, Karki D, Robeson MS, Nagy L, et al. Phylogeography of microbial phototrophs in the dry valleys of the high Himalayas and Antarctica. Proc Biol Sci. 2011 Mar 7;278(1706):702–8. doi: 10.1098/rspb.2010.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Colloquium paper: microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci U S A. 2008 Aug 12;105(1):11505–11. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoener TW, Spiller DA, Losos JB. Natural restoration of the species-area relation for a lizard after a hurricane. Science. 2001 Nov 16;294(5546):1525–8. doi: 10.1126/science.1064396. [DOI] [PubMed] [Google Scholar]
- 32.Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7774–8. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, et al. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011 Apr;92(4):797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- 34.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009 Aug;75(15):5111–20. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010 Apr 6;107(14):6477–81. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967 Feb;27(2):209–20. [PubMed] [Google Scholar]
- 37.Franklin RB, Mills AL. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol. 2003 Jun 1;44(3):335–46. doi: 10.1016/S0168-6496(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 38.Luc A. Under the hood: Issues in the specification and interpretation of spatial regression models. Agricultural Economics. 2002;27(3):247–67. [Google Scholar]
- 39.Bru D, Ramette A, Saby NP, Dequiedt S, Ranjard L, Jolivet C, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2011 Mar;5(3):532–42. doi: 10.1038/ismej.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010 Apr 8;464(7290):908–12. doi: 10.1038/nature08937. This paper presents the intriguing conjecture that some factors, such as diet, that are geographically variable can affect not only the short-term set of genes in host-associated microbial communities by promoting the growth of specific microbes but might also lead to horizontal transfer of genes into the microbiome. [DOI] [PubMed] [Google Scholar]
- *41.Dale MRT, Dixon P, Fortin MJ, Legendre P, Myers DE, Rosenberg MS. Conceptual and mathematical relationships among methods for spatial analysis. Ecography. 2002;25(5):558–77. This paper shows that despite potential dispersal limitations, both source and sink populations of diatoms in a Danish fjord and the sea that drains it remained essentially constant over a century, resisting potential cross-colonization. This study emphasizes the importance of examining systems over appropriate timescales. [Google Scholar]
- 42.Harnstrom K, Ellegaard M, Andersen TJ, Godhe A. Hundred years of genetic structure in a sediment revived diatom population. Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):4252–7. doi: 10.1073/pnas.1013528108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011 Mar 15;108(1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011 May 30;12(5):R50. doi: 10.1186/gb-2011-12-5-r50. This paper demonstrates the value of comparing day-to-day variability, in this case across four body sites of two individuals, to the framework provided by a larger reference population. Specifically, it shows the extent to which each individual's microbiota remains distinct, and relates intra- to inter-individual variability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shade A, Jones SE, McMahon KD. The influence of habitat heterogeneity on freshwater bacterial community composition and dynamics. Environ Microbiol. 2008 Apr;10(4):1057–67. doi: 10.1111/j.1462-2920.2007.01527.x. [DOI] [PubMed] [Google Scholar]
- 46.Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2011 Sep 1; doi: 10.1038/ismej.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics. 2006 Oct 15;22(20):2532–8. doi: 10.1093/bioinformatics/btl417. This paper demonstrates how, in a lake ecosystem, detailed timeseries analysis can reveal associations with specific ecosystem events including ice formation, mixing, and phytoplankton blooms. It also demonstrates that many taxa share responses to the same events. [DOI] [PubMed] [Google Scholar]
- 48.Shade A, Chiu CY, McMahon KD. Differential bacterial dynamics promote emergent community robustness to lake mixing: an epilimnion to hypolimnion transplant experiment. Environ Microbiol. 2010 Feb;12(2):455–66. doi: 10.1111/j.1462-2920.2009.02087.x. [DOI] [PubMed] [Google Scholar]
- 49.Crump BC, Peterson BJ, Raymond PA, Amon RM, Rinehart A, McClelland JW, et al. Circumpolar synchrony in big river bacterioplankton. Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21208–12. doi: 10.1073/pnas.0906149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. Synchrony in aquatic microbial community dynamics. ISME J. 2007 May;1(1):38–47. doi: 10.1038/ismej.2007.6. [DOI] [PubMed] [Google Scholar]
- 51.Clarke KR, Somerfield PJ, Airoldi L, Warwick RM. Exploring interactions by second-stage community analyses. Journal of Experimental Marine Biology and Ecology. 2006;338(2):179–92. [Google Scholar]
- 52.Shade A, Read JS, Welkie DG, Kratz TK, Wu CH, McMahon KD. Resistance, resilience and recovery: aquatic bacterial dynamics after water column disturbance. Environ Microbiol. 2011 Oct;13(10):2752–67. doi: 10.1111/j.1462-2920.2011.02546.x. [DOI] [PubMed] [Google Scholar]
- 53.Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):4158–63. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He S, McMahon KD. ‘Candidatus Accumulibacter’ gene expression in response to dynamic EBPR conditions. ISME J. 2011 Feb;5(2):329–40. doi: 10.1038/ismej.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowers RM, Sullivan AP, Costello EK, Collett JL, Jr, Knight R, Fierer N. Sources of Bacteria in Outdoor Air across Cities in the Midwestern United States. Appl Environ Microbiol. 2011 Sep;77(18):6350–6. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones SE, Chiu CY, Kratz TK, Wu JT, Shade A, McMahon KD. Typhoons initiate predictable change in aquatic bacterial communities. Limnol Oceanogr. 2008;53(4):1319–26. [Google Scholar]
- *57.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011 Mar 15;108(1):4554–61. doi: 10.1073/pnas.1000087107. This paper demonstrates the importance of inter-subject variability not just in terms of the static state of the microbiota but also in terms of its capacity to vary on perturbation, in this case with administration of repeated doses of antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Schilling KE, Zhang YK, Hill DR, Jones CS, Wolter CF. Temporal variations of Escherichia coli concentrations in a large Midwestern river. Journal of Hydrology. 2009;365(1-2):79–85. [Google Scholar]
- 59.Trosvik P, Rudi K, Straetkvern KO, Jakobsen KS, Naes T, Stenseth NC. Web of ecological interactions in an experimental gut microbiota. Environ Microbiol. 2010 Oct;12(10):2677–87. doi: 10.1111/j.1462-2920.2010.02236.x. This is one of the few studies to apply wavelet analysis to microbial ecology, examining changes in a simplified community of gut microbes in vitro. [DOI] [PubMed] [Google Scholar]
- **60.Banning NC, Gleeson DB, Grigg AH, Grant CD, Andersen GL, Brodie EL, et al. Soil microbial community successional patterns during forest ecosystem restoration. Appl Environ Microbiol. 2011 Sep;77(17):6158–64. doi: 10.1128/AEM.00764-11. This chronosequence study demonstrates that soil bacterial communities, but not fungal communities, may follow a predictable pattern of succession during forest regrowth after disturbance events such as mining. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, et al. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol. 2009 Dec;47(6):673–81. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- 62.Urbanova M, Kopecky J, Valaskova V, Sagova-Mareckova M, Elhottova D, Kyselkova M, et al. Development of bacterial community during spontaneous succession on spoil heaps after brown coal mining. FEMS Microbiol Ecol. 2011 Jun 27; doi: 10.1111/j.1574-6941.2011.01164.x. [DOI] [PubMed] [Google Scholar]