Abstract
Most patients with familial pulmonary arterial hypertension (FPAH) carry mutations in the bone morphogenic protein receptor 2 gene (BMPR2). Yet carriers have only a 20% risk of disease, suggesting that other factors influence penetrance. Thrombospondin-1 (TSP1) regulates activation of TGF-β and inhibits endothelial and smooth muscle cell proliferation, pathways coincidentally altered in pulmonary arterial hypertension (PAH). To determine whether a subset of FPAH patients also have mutations in the TSP1 gene (THBS1) we resequenced the type I repeats of THBS1 encoding the TGF-β regulation and cell growth inhibition domains in 60 FPAH probands, 70 nonfamilial PAH subjects, and in large control groups. We identified THBS1 mutations in three families: a novel missense mutation in two (Asp362Asn), and an intronic mutation in a third (IVS8+255 G/A). Neither mutation was detected in population controls. Mutant 362Asn TSP1 had less than half of the ability of wild-type TSP1 to activate TGF-β. Mutant 362Asn TSP1 also lost the ability to inhibit growth of pulmonary arterial smooth muscle cells and was over threefold less effective at inhibiting endothelial cell growth. The IVS8+255 G/A mutation decreased and/or eliminated local binding of the transcription factors SP1 and MAZ but did not affect RNA splicing. These novel mutations implicate THBS1 as a modifier gene in FPAH. These THBS1 mutations have implications in the genetic evaluation of FPAH patients. However, since FPAH is rare, these data are most relevant as evidence for the importance of TSP1 in pulmonary vascular homeostasis. Further examination of THBS1 in the pathogenesis of PAH is warranted.
Keywords: genetics, endothelium, vascular smooth muscle, matricellular proteins
pulmonary arterial hypertension (PAH) is a progressive disease with a high mortality rate (47). PAH is characterized by remodeling of distal pulmonary arterioles due to smooth muscle hypertrophy and endothelial proliferation. PAH displays familial transmission in a subset, termed familial PAH (FPAH). Heterozygous mutations in bone morphogenic protein receptor 2 (BMPR2) are the causative genetic event in over 50% of FPAH kindreds (43). Yet relatives within a kindred who are mutation carriers have only a 20% chance of developing disease, implying that environmental or genetic modifiers influence disease risk (51). Although various BMPR2 mutations occur in FPAH, few idiopathic PAH patients have BMPR2 mutations, and BMPR2 mutations do not occur in PAH associated with scleroderma (34, 50). BMPR2 is a member of the TGF-β superfamily, which encompasses a large number of genes whose products are TGF-β isoforms, related ligands, ligand activators, receptors, and signaling partners.
Thrombospondin-1 (TSP1) is a large secreted glycoprotein encoded by the gene THBS1 (Fig. 1) (27, 37). TSP1 activates latent TGF-β by structurally disrupting the noncovalent sequestration of TGF-β by latency-associated peptide (LAP) (60). Posttranslational activation is the rate-limiting step in TGF-β1 signaling (10). TSP1 is consistently expressed in the quiescent lung of rodents and humans (10, 67), where TSP1 protein is mostly detectable in epithelium, basement membranes, and the intercellular matrix, whereas RNA expression is more diffuse (35, 68). TSP1 expression is typically induced in many other cells during chronic lung disease (14, 67), including in smooth muscle cells and fibroblasts in pulmonary hypertension (PH) (6). Basal TSP1 expression in lung, although not high, is similar in level to that of proteins such as plasminogen-activator inhibitor 1 (59). Constitutive levels of TSP1 protein also appear to have a central role in creating the lung scaffolding network needed for cell differentiation (62).
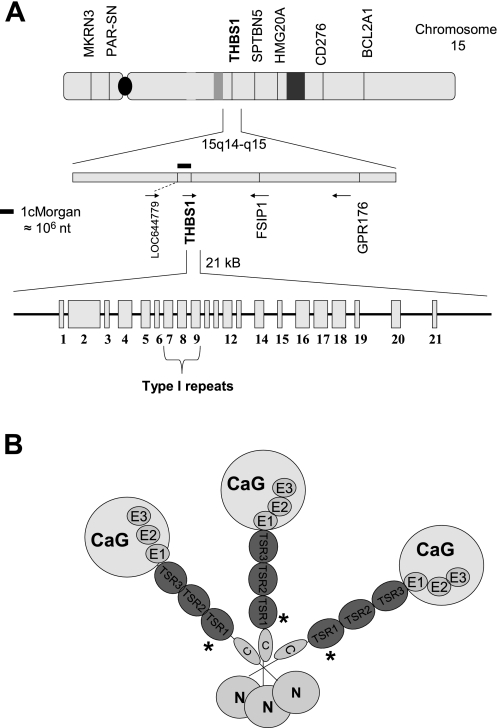
Fig. 1.
A: THBS1 gene, including chromosomal neighbors. All exons (rectangles) and introns (lines) are depicted. Exons 7–9 (type I repeats) were resequenced for this project. Direction of gene translation is per arrows. B: trimeric THBS1 protein (TSP1). N, globular region N (NH2-terminal); C, procollagen homology domain; TSR1, 2, and 3, thrombospondin type I repeats; E1, 2, 3, EGF homology (type II) repeats; CaG, calmodulin homology (type III) repeats; G, globular domain (COOH-terminal). The location of the missense mutation found in TSR1 is designated with an asterisk (*). Exon sizes are relative to each other but are exaggerated for display.
In TSP1-null mice, latent TGF-β1 remains abundant in lung but active TGF-β1 is scarce. These mice display a principal phenotype of an inflammatory pneumonitis with secondary features that include pulmonary vascular remodeling and elevated right ventricular pressures (10, 38, 53). Pulmonary endothelial secretion of TSP1 increases with stretch, and TSP1 inhibits growth of pulmonary vascular smooth muscle cells (52). TGF-β1 activation by TSP1 and its potent inhibitory effects on pulmonary vascular cell proliferation localize to three type I repeats (TSR 1–3). Among the most potent natural inhibitors of endothelial cell proliferation and angiogenesis, TSR are highly conserved among vertebrates (30). These endothelial inhibitory effects are independent of TGF-β activation domains within the TSR (20), instead localizing to WXXW repeats and CD36/β1 integrin activation regions (11, 16, 20, 33, 63). Binding and sequestration of endothelial mitogens such as VEGF and bFGF occur at WXXW domains, thereby decreasing binding of these cytokines to their endothelial receptors and slowing proliferation, whereas CD36 and β1-integrin signaling cause endothelial cell apoptosis and inhibition of migration. TSP1 proteins are 420- to 450-kDa trimers; thus each trimer has 9 TSR (Fig. 1).
We hypothesized that a subset of FPAH patients with BMPR2 mutations would also have mutations in the THBS1 gene as genetic modifier events disrupting key pathways in PAH pathogenesis. Because TSR 1–3 are critical domains for TSP1 effects relevant to PAH, we resequenced these regions in probands from 60 FPAH kindreds, in patients with nonfamilial PAH, and in control populations. After finding novel THBS1 mutations, we evaluated them for effects on latent TGF-β1 activation, on inhibition of lung vascular cell growth, and on transcription factor binding.
MATERIALS AND METHODS
Human Subjects
Probands from 60 FPAH families, 70 subjects with PAH of various causes, and subjects from large control groups were screened for THBS1 mutations (Table 1). Research was approved by University of Colorado and Vanderbilt University Institution Review Boards, and written consent was provided. Anonymous DNA sequence data from sepsis control subjects and additional healthy control subjects in the regions of THBS1 analysis were available from a previous project approved by the Medical College of Wisconsin institutional review board. Sepsis subjects were used as a control group afflicted with multiple non-PAH comorbidities; this population was previously described (71).
Table 1.
Demographics of human subjects
| Group | N | % Female | % Caucasian |
|---|---|---|---|
| FPAH | 60 | 73 | 94 |
| PAH | 70 | 90 | 96 |
| Healthy controls | 650 | 45 | 37 |
| Sepsis controls | 144 | 44 | 78 |
PAH, pulmonary arterial hypertension; FPAH, familial PAH. Nonfamilial patients (PAH) had the following disease associations: idiopathic (50), congenital heart (3), scleroderma (12), diet drug (3), connective tissue (2).
Materials
All cytokines were from R&D Systems. All restriction enzymes were from New England Biolabs. All oligonucleotide primers were synthesized by Operon except where indicated. All cell culture reagents, plastics, and chemicals were from Fisher unless otherwise specified.
PCR, DNA Sequencing, and RFLP
When necessary, reactions were optimized (MasterAmp PCR Optimization Kit, Epicentre). Amplifications were done with 20 ng of genomic DNA in 50 μl using 0.5 μl of Taq, 1 μl of 10 mM dNTP, 1 μl of 50 mM MgCl2, 5 μl of 10× PCR buffer without MgCl2, 1 μl of each 10 μM primer, and 39.5 μl molecular grade H2O. Reagents were from Invitrogen. Thermocycling was done on an MJ Dyad 96-well unit (Bio-Rad). Products were purified with ExoSAP-IT (USB). Primers and conditions are in Table 2. DNA resequencing was performed by using these primers at 10 μM, 50 ng of PCR products at 25 ng/μl, and Big-Dye chemistry on a 3730× sequencer (Applied Biosystems, ABI) at the UCD Barbara Davis Molecular Biology Core Facility. Genotype calls were by Consed software (University of Washington). For restriction fragment length polymorphism analysis (RFLP), PCR products were digested with BccI, and 20 μl of digestates were run on a 1% agarose gel for 45 min at 80 V. Band sizes (285 nt cuts into 62 nt and 223 nt) were determined by ethidium bromide staining with UV light. Two 362Asn carriers (1/kindred) and two noncarriers were tested.
Table 2.
Primer/probe sequences and reaction conditions
| Region | Product Size, nt | Sequence | Conditions |
|---|---|---|---|
| Resequencing | |||
| Thbs1Exon 7 | 677 | Fw: 5′-TTGCTTTTCACCCTCTGGC | Buffer E |
| Rv: 5′-CGGTAGAAATTGGCTCCTG | TSP(6–7) | ||
| Thbs1Exon 8–9 | 998 | Fw: 5′-TAACAGCAACCCATAAAAACAGC | |
| Rv: 5′-GCATCTTAGAGCAATCAAATCG | TSP(54) | ||
| Bmpr2Exon 9 | 336 | Fw: 5′-AGAATATGCTACGTTCTCTC | Buffer C |
| Rv: 5′-ACACTAGATAGCAATGAACTAAAGG | TSP(54)+10cycles | ||
| RT-PCR | |||
| Thbs1Exon 5–17 | 1,825 | Lt: GGAGTTCAGTACAGAAATAACG | RT(1) |
| Rt: ATCATCTTTGCAGGCATCACCTCG | |||
| Thbs1Exon 5–9 | 682 | Lt: GGAGTTCAGTACAGAAATAACG | RT(1) |
| Rt: GTTTTCTGTTACATCACCAACG | |||
| Expression Vector and Subcloning Vector Sequencing | |||
| BGH | N/A | Rv: 5′-TAGAAGGCACAGTCGAGG | N/A |
| T7 | N/A | Fw: 5′-GTAATACGACTCACTATAGGG | N/A |
| M13 | N/A | Fw: 5′-GTTGTAAAACGACGGCCAGT | N/A |
| Thbs1 Exon 7 | N/A | Fw: 5′-TCAACAACCGATGTGAGGGCTCC | N/A |
| Thbs1 Exon 13 | N/A | Fw: 5′-CAACAAGAACGCCAAGTGCAACTACC | N/A |
| Thbs1 Exon 18 | N/A | Rw: 5′-AGCCAAAGACAAATCCAGC | N/A |
| ChIP | |||
| Region Thbs1 | 318 | Fw: GAAAAACAAACTCACCCTCTT | Buffer B |
| IVS8 + 255 G/A | Rv: GACGACTACGTTTCTGTACC | Tsp(54) all ChIP | |
| MAZ (+) | 254 | Fw: 5′-GATCCTCTCTCGCTAATCTCC | Buffer F |
| Rv: 5′-TGCCTCTCGCTGGAATTAC | |||
| SP1 (+) | 283 | Fw: 5′-TAACTTCTGGACTATTTGCGGACTTTTTGG | Buffer H |
| Rv: 5′-GGGCGAGGGAGGAGAGAA | |||
| VMAT (−) | 186 | Fw: 5′-GCAGGCACCCTTTACCATAA | Buffer B |
| Rv: 5′-GGGAGGACAGAAGGAAAACC | |||
| Taqman Probes and Primers | |||
| Asp362Asn | 107 nt | Fw: 5′-CCCTCTAAAGAACAGCTTGTCCTAA | Per ABI |
| Rv: 5′-GTCCACTCGGACCATGGA | |||
| Probe 1: CAGCCATCGTCCGCA-VIC (WT) | |||
| Probe 2: CAGCCATTGTCCGCA-FAM (MUT) | |||
| Restriction Enzyme Validation (RFLP) | |||
| Asp362Asn | 285 nt | Fw: CAGCTTGTCCTAATCATCACG | Buffer B |
| Rv: CTTGGTGACGACAACAAGG | TSP 57 | ||
TSP1(6–7): lid 105°C; [95°C for 2:00, 95°C for 0:30, 56.7°C for 1:00, 55°C for 0:30] × 9 cycles; 40 cycles of [95°C for 0:30, 48.7°C for 0:30, 68°C for 1:00]; 72°C for 10:00; 4°C. TSP(54): lid 100°C; [94°C for 1:30, 94°C for 0:30, 54°C for 0:30; 68°C for 1:30] × 29 cycles, 72°C for 3:00; 4°C; TSP 57 — same as Ref. 54 except anneal is 57°C. RT(1): 50°C for 2:00, 95°C for 10:00; then 40 cycles of [95°C for 0:15, 60°C for 1:00]. FAM and VIC, dyes labeled onto probes. Site of mutation is underlined. WT, wild-type allele; MUT, mutant allele; Fw, forward; Rv, reverse; ChIP, chromatin immunoprecipitation; RFLP, restriction fragment length polymorphism analysis. T7, M13, and BGH are universal primers flanking the insert site in the pcDNA 3.1.
TaqMan Genotyping
Asp362Asn genotyping in controls was done using a custom TaqMan assay (Table 2) made with File-Builder 3.0 software (ABI), with 10% of all samples reassessed by sequencing. All reagents were from ABI. PCR was as follows: 95°C for 10:00; then 40 cycles of 0:15 at 92°C, 1:00 at 60°C. Reactions were performed in 25 μl by using 0.5 μl of 20× SNP genotyping assay mix, 12.5 μl of Master Mix, 3 μl of genomic DNA at 8 ng/μl, and 9 μl water. Allele calling was performed on an ABI 7300 platform. Undetermined calls were repeated; if needed the subject's DNA was sequenced for genotype. On each plate, DNA from two 362Asn carriers was used to improve genotype calls as a positive control. An assay for IVS8+255 G/A failed construction, so genotyping of non-PAH controls for this was performed only by sequencing.
RNA and Protein Isolation
After washing, cells were lysed and total RNA was isolated using a kit (Qiagen), quantified by UV spectroscopy at OD 260/280, then aliquoted and stored at −70°C. Protein was isolated from cells by a lysis/recovery kit (Active Motif) or from media by a column method (Qiagen) according to manufacturer guidelines. Protein concentration was determined by Bradford assay. Proteins were aliquoted and frozen at −70°C before assays.
Western Blotting
Twenty-microgram aliquots of total protein (100 ng for recombinant TSP1) and 20 μl of 2× reducing loading buffer were loaded into wells of a 7.5% Tris·HCl acrylamide gel (Bio-Rad). All proteins were reduced by boiling 10 min. Platelet TSP1 (100 ng; Athens Biotechnology) was a positive control. Gels were run at 40 V for 1 h and 100 V for 1 h, then transferred at 250 mA for 2 h at 4°C onto 0.2 μm nitrocellulose in transfer buffer (600 ml H2O, 3 g Tris·HCl, 14.4 g glycine, 200 ml MeOH). After washing (0.1% Tween-TBS), membranes were blocked with 3% nonfat dried milk (nfdm) in Tween-TBS overnight at 4°C with rotation. After washing, the membrane was incubated with a mouse anti-human TSP1 antibody (Abnova; H00007057) at 1 μg/ml in Tween-TBS-3% nfdm for 2 h at 22°C. After four washes, secondary antibody was applied (1:5,000 rabbit anti mouse-horseradish peroxidase; Bio-Rad) in Tween-TBS-3% nfdm for 2 h at 22°C. Blots were washed, incubated in chemiluminescent solution, then imaged onto X-ray film.
RT-PCR
Using 5 μg of RNA, cDNA was made with oligo(dT) primers (SuperScript III, Invitrogen); 5 μl was used for PCR. Amplification of the THBS1 exon 7–9 region was done with primers in exon 5 (forward) and exons 9 and 17 (reverse) (Table 2). Ten microliters of products were run on a 1% agarose gel and ethidium bromide stained with UV light. Bands were eluted from gels (Qiaquick, Qiagen) and 50 ng of cDNA was sequenced bidirectionally with the primers to detect potential splicing events. THBS1 cDNA sequence was compared with an NCBI reference (X04665).
EMSA
Biotin-labeled oligonucleotides (oligos) centered on IVS8+255 G/A were tested for binding to nuclear extract proteins (NE) and recombinant proteins. An unbiased in silico analysis of the region of this mutation was performed by using Accelrys Gene v2.5 software to identify putative transcription factor binding sites created or altered by this mutation. Positive and negative control oligos for transcription factors were as published (4, 26, 44). All oligos are shown in Table 3. Only sense oligos were biotin labeled. Single-stranded oligos were diluted in Tris-EDTA (TE; 10 mM Tris, 1 mM EDTA in deionized water) to 150 μM; pairs were mixed in equimolar amounts and then annealed at 95°C for 10 min. To remove free biotin, double-stranded oligos were dialyzed against a 20-kDa membrane (Slide-A-Lyzer, Pierce) for four rounds of 1 h each in 100 ml of TE buffer pH 8.0 at 4°C. All oligo concentrations were verified by UV spectrophotometry and adjusted with deionized water. Unlabeled oligos were added for competition assays 15 min before addition of labeled oligos. EMSA was performed by use of a kit (LightShift, Pierce). Incubations were performed for 30 min on ice with 1 μg of NE, 100–200 ng of recombinant human SP1 protein (Promega), or 50–200 ng of recombinant MAZ protein (H00004150-Q01, Abnova) and with 8.5 ng/μl of labeled oligos. Tests of oligo binding to recombinant SP1 and MAZ were done in lieu of supershifts because azides in antibody solutions interfere with biotin chemistry. NE from cells (106 cells, passages 2–5) were prepared using a commercial kit (Active Motif) and quantified. After incubation, the reaction volumes were electrophoresed on 5% Tris-borate-EDTA acrylamide gels, transferred to membranes, incubated with anti-horseradish peroxidase antibody, washed, and detected on X-ray film. Experiments were repeated at least four times. Band densities were determined with NIH Image J software, and variables were normalized to vehicle values for each condition.
Table 3.
EMSA oligonucleotides
| Oligonucleotide | DNA Sequence |
|---|---|
| THBS1 IVS8 + 255 | Sense: 5′-AGAGTCTATGACAAGGGAGGGATTTGAAA |
| Common allele | Antisense: 5′-TTTCAAATCCCTCCCTTGTCATAGACTCT |
| THBS1 IVS8 + 255 | Sense: 5′-AGAGTCTATGACAAGAGAGGGATTTGAAA |
| Mutant allele | Antisense: 5′-TTTCAAATCCCTCTCTTGTCATAGACTCT |
| MAZ | Sense: 5′-GAAAAAGAAGGGAGGGGAGGGATC |
| Positive control | Antisense: 5′-GATCCCTCCCCTCCCTTCTTTTTC |
| MAZ | Sense: 5′-GAAAAAGAAGGGAATTCAGGGATC |
| Negative control | Antisense: 5′-GATCCCTGAATTCCCTTCTTTTTC |
| SP1 | Sense: 5′-AGGGAATGGGGGCGGGATGAGGGCCT |
| Positive control | Antisense: 5′-AGGCCCTCATCCCGCCCCCATTCCCT |
| SP1 | Sense: 5′-AGGGAATGGATTCATTATGAATTCCT |
| Negative control | Antisense: 5′-AGGAATTCATAATGAATCCATTCCCT |
Transcription factor binding sites are underlined. Positive control oligos contain a strong consensus element for binding; negative control oligos contain sequences known to minimize binding. Sense oligos were constructed with a 5′-TEG biotin label.
Site-Directed Mutagenesis of THBS1 cDNA
A pGEM4 plasmid containing human THBS1 cDNA was a kind gift of Dr. Joanne Murphy-Ullrich. The insert was sequence verified (comparison NCBI X04665) then was expanded in Escherichia coli and purified with spin columns (Bio-Rad). Ten micrograms of DNA were cut in the multicloning site region with HindIII (5′) and KpnI (3′) to isolate the THBS1 insert, which was then purified by running on a 1% agarose gel followed by elution (QIAquick, Qiagen) of the 4.3-kb cDNA fragment. This insert was directionally cloned into the expression vector pcDNA3.1 (Invitrogen) by digestion of the vector with KpnI and SalI (and of the insert with SalI) followed by subsequent fragment ligation with Fast Link ligase (Epicentre) to make pcDNA3.1(THBS1wt). After sequence verification, the 362Asn mutation was introduced by site-directed mutagenesis (QuikChange II XL, Stratagene) to make pcDNA3.1(THBS1mut) by following manufacturer guidelines and using HPLC-purified primers (sense 5′-GGC CCA GCG ACT CTG CGG ACA ATG GaT GGT CTC CAT GGT CCG AG, antisense CTC GGA CCA TGG AGA CCA tCC ATT GTC CGC AGA GTC GCT GGG CC, mutant allele underlined; an engineered C to T silent mutation 5 nt 3′ to native mutation in lower case was needed to generate a FokI site, GGATG, for clone screening). The Asp362Asn allele, lack of cloning artifacts, and orientation were verified by sequencing with primers T7, BGH, and THBS1 (exons 7 and 13, forward; exon 18, reverse; Table 2).
Synthetic TSP1 Peptides
Peptides with the Asp362Asn TSR1 mutation were synthesized as wild-type (DDG WSP WSE WTSC), mutant (DNG WSP WSE WTSC), and scrambled (WCP DSW ESD TSWG) (mutant allele underlined; Anaspec). Peptides were dissolved in DMSO at 1 μg/ml and frozen until use.
Production of rTSP1
The vectors pcDNA3.1(THBS1wt), pcDNA3.1(THBS1mut), or pcDNA3.1(empty) were transfected (1 μg) into adherent HEK cells (GIBCO) overnight in six-well plates with Lipofectamine 2000 (Invitrogen). Standard incubator conditions were 37°C, 5% CO2, and 21% O2. Media consisted of 10% FBS, DMEM with 4.5 g/l of glucose, and 2.5 mM l-glutamine, with antibiotics (PCN 50 U/ml, SM 50 μg/ml). The next day, selection of transfectants was begun with 0.5 μg/ml Geniticin (G418; GIBCO); a titration experiment established this dose as optimal to kill mock-transfected cells while letting transfected cells proliferate. After 10 passages of 1:20 splitting (95% of cells were discarded), high-expressing clones were identified by use of a human TSP1 ELISA (R&D Systems) to screen media, then expanded in T150 flasks. At confluence, flasks were washed with PBS and media were changed to 30 ml of serum-free (SF) media with 1 μM forskolin and 10 nM phorbol ester to stimulate CREB and NF-κB sites in the CMV promoter of pcDNA 3.1 (stimulation increased production 2- to 3-fold) (18). Ten T150 flasks were used for each genotype, and SF medium was harvested and changed once every 48 h four times. Media were centrifuged at 300 g to pellet cells, supernatants (SN) were filtered thorough 0.4 μM membranes, and 10 mM PMSF was added before freezing. Recombinant full-length TSP1 protein (rTSP1) was purified from thawed SN by centrifugal concentrators (100,000 molecular weight cutoff, 15–70 ml tubes, Millipore; 3,000 g) at 4°C to a volume of 2–5 ml, then washed three times (in concentrators) at 4°C with 15 ml of TBS-C2 (10 mM Tris·HCl, 0.15 M NaCl, 2 mM CaCl2, pH 7.4). Concentrated media (5 ml) was applied to a prewashed 1 ml heparin column (GE Life Sciences), washed with 5 ml of TBS-C2, then 5 ml of TBS-E (same as TBS-C2 except 0.7 M NaCl) was added to elute rTSP1 in fractions. Fractions were screened by ELISA (rTSP1 eluted in fractions 2–4), pooled, and concentration was also determined by ELISA. Correct rTSP1 size was verified by Western blot. Purity was verified by silver staining of 200 ng run on a 7.5% PAGE gel (Bio-Rad): purified rTSP1 was diluted in TBS-C2 and frozen. Average yields were 0.5 mg of TSP1/l of media. Cells transfected with pcDNA 3.1(empty) were antibiotic resistant but did not produce TSP1.
Latent TGF-β1 Activation
Mink lung epithelial cells (MLEC) were a gift of Dr. Daniel Rifkin. MLEC are a well standardized and sensitive assay for TGF-β activity. MLEC were grown at 37°C in 21% O2 and 5% CO2 in T-75 flasks as published (1). At subconfluence, cells were washed with PBS, trypsin harvested, and plated at 2 × 104 cells/well in 96-well plates. Four hours later, media were changed to include 0.3 nM human latent TGF-β1 (all wells) and log concentrations of synthetic TSP1 peptides (vehicle, 0.01–100 nM; wild-type, mutant 362Asn, or scrambled) or trimeric rTSP1 (wild-type or mutant 362Asn; 0–1 nM). All concentrations were in triplicate wells. After a 14-h incubation, then washing, cells were lysed with 20 μl/well of lysis buffer (Promega). Luciferase activity was detected by use of Dual-Luciferase (Promega) and a luminometer (Turner Biosystems). Experiments were performed four times. Stimulation by active human TGF-β1 (0.01 and 1 nM) was a positive control; cells that lacked a threefold responsiveness were discarded.
Cell Culture
Lymphoblastoid cells.
Archived Epstein-Barr virus-immortalized cells were cultured in standard conditions by use of a THBS1 IVS8+255 G/A carrier and two wild-type controls (Coriell Cell Repository) as described, with and without puromycin D at 100 μg/ml to inhibit potential nonsense-mediated decay of misspliced RNA (8). Unstimulated cells were grown in suspension cultures in flasks until reaching a density of 3 × 105 cells/ml; then protein, RNA, and chromatin were extracted. Additional experiments were performed in the IVS8+255A mutation carrier to test for alternative splicing of THBS1 RNA in response to PAH-relevant stimuli: after reaching subconfluence, cells were placed for 4 h or 24 h in 0.1, 1, or 21% O2 with or without 10 ng/ml TNF-α and 10 nM IL-6. Experiments were done in a HERAcell 150 tri-gas incubator (Kendro).
Pulmonary artery vascular cells.
Microvascular endothelial cells (HPAEC; CC-2627) and smooth muscle cells (HPASMC; CC-2581) from human pulmonary arteries (Lonza) were expanded in growth media (EGM2-MV or SGM) in 5% CO2 at 37°C. Once subconfluent, cells were used for RNA and protein isolation or trypsinized and transferred to 96-well plates for proliferation assays at 5 × 103 cells/well in 200 μl of EGM (HPAEC; 1% gelatin-coated plates). The next morning cells were either serum starved in 0.1% FBS/RPMI for 48 h (HPASMC were overconfluent at 72 h without starvation) or not (HPAEC did not tolerate starvation). Medium was replaced with growth medium (10% FBS) containing TSP1 peptides or full-length rTSP1 (0–50 nM for peptides, 0–1.6 nM for rTSP1) or vehicle (DMSO for synthetic peptides, TBS-C2 for rTSP1). In parallel wells, platelet TSP1 was an inhibition control and medium with 20% FCS was a positive control. Higher concentrations of monomeric peptides were utilized because they are less effective than trimeric TSP1 (57). Cells were utilized in passages 4–8 (older cells lost responsiveness). All incubations were performed by with three or six well replicates per condition.
Cell Proliferation
HPAEC and HPASMC proliferation were measured at 72 h after addition of proteins by use of a tetrazolium reduction colorimetric assay (CellTiter 96, Promega) (29). Manual cell counts from plating differential cell numbers verified the accuracy of colorimetric methods for cell number assessment in both cell lines (r2 = 0.94–0.96). The average OD490 of replicates for each concentration was calculated, and the average of the vehicle wells was defined as TSP1 concentration of zero (“baseline”). Means were normalized to vehicle means; these normalized means were summed and averaged for all experiments at that concentration. Of vehicles, only DMSO (TSR peptides) had a significant effect on proliferation (small inhibitory effect); this was adjusted for in the analysis. Assays were performed at least four times for each peptide or protein.
ChIP
Chromatin was isolated from HPASMC and lymphoblasts (of an IVS8+255A carrier) by using a kit and sonication (Active Motif). Fixed protein-DNA was immunoprecipitated for 4 h with 1 μg of antibody to SP1, control rabbit IgG (both Promega), or antibody to MAZ (Aviva). After isolation of bound DNA, PCR was performed with primers flanking IVS8+255 (Table 2). The relative abundance of DNA pulled down was compared by running 10 μl of PCR products on ethidium bromide-stained agarose gels, and quantitatively by real-time SYBR green analysis of products. In SYBR green assays, DNA content was determined at midpoints in the linear amplification range as published, on an ABI 7300 platform (15). Software-determined threshold cycle values (relative to control IgG) were calculated to determine fold enrichment. Input DNA was used as a PCR control. Positive controls were performed using known strong consensus nucleotides for SP1 and MAZ binding. Negative control experiments were performed using nucleotides from the VMAT gene, proven devoid of SP1 and MAZ sites (Table 3) (2). In parallel, 1 μg of chromatin immunoprecipitation (ChIP) PCR products from input or lymphoblast DNA was subcloned into pTOPO (Invitrogen); then after transformation in E. coli 10 randomly picked colonies (blue-white selection) for each condition were miniprepped, and DNA was isolated and sequenced with M13 primer to test whether transcription factors preferentially bound to an allele. Experiments were performed at least four times.
Statistical Analysis of Data
Data were analyzed by use of Prism 3.0 software (GraphPad). Values are expressed as means ± SD. Significance was determined by an unpaired Student's t-test for two-group comparisons and by one-way ANOVA testing for comparisons of three or more groups (Kruskal-Wallis or Mann-Whitney testing if nonparametric). If ANOVA was significant, differences between any two concentrations were determined by nonparametric or parametric unpaired t-tests, as appropriate. Comparisons of categorical data were by contingency tables (Fisher exact or χ2). All experiments were performed at least three times. All analyses were two tailed, and a P < 0.05 was considered significant. Analysis of a proband from 60 families gave a power of 95% if mutations were present in 5% of the population (http://www.stat.uiowa.edu/∼rlenth/Power/).
RESULTS
Mutations in the THBS1 Type I Repeat Domains Are Present in FPAH Cohorts
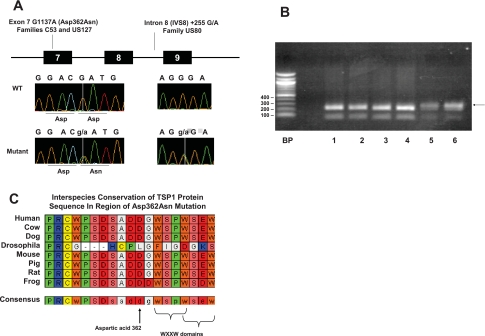
Novel mutations in three probands from 60 FPAH kindreds were found (5% of all probands). A point mutation in exon 7/TSR1 (Asp362Asn) was found in two unrelated kindreds and a point mutation in intron 8 (IVS8+255 G/A) was found in a third kindred (Fig. 2A). None of the mutations were found in large control cohorts (both healthy and chronic disease controls). Online searches of human genomic databases revealed no prior reports of these mutations, except for a single report for IVS8+255 G/A (rs55924981); however, the rare minor allele frequency (MAF) of IVS8+255A in our work confirms its classification as a mutation.
Fig. 2.
THBS1 mutations found in familial pulmonary arterial hypertension (FPAH) subjects. A: mutation location, type, and chromatograms. B: confirmation of the Asp362Asn mutation by restriction fragment length polymorphism analysis (RFLP). A BccI site is present in the wild-type allele, cutting PCR products in those subjects into fragments of 62 and 223 base pairs (bp) (lanes 1–4). Only a single strand cuts in mutation carriers (lanes 5–6; arrow denotes uncut 285-bp fragment). C: Clustal alignment in the region of the Asp362Asn mutation in TSR1. Letters are standard amino acid abbreviations. Left is NH2-terminal. Asp362 (arrow) and flanking residues are highly conserved in vertebrates. Two overlapping WXXW domains are near Asp362.
The Asp362Asn Mutation Occurs in a Highly Conserved Region of TSR1
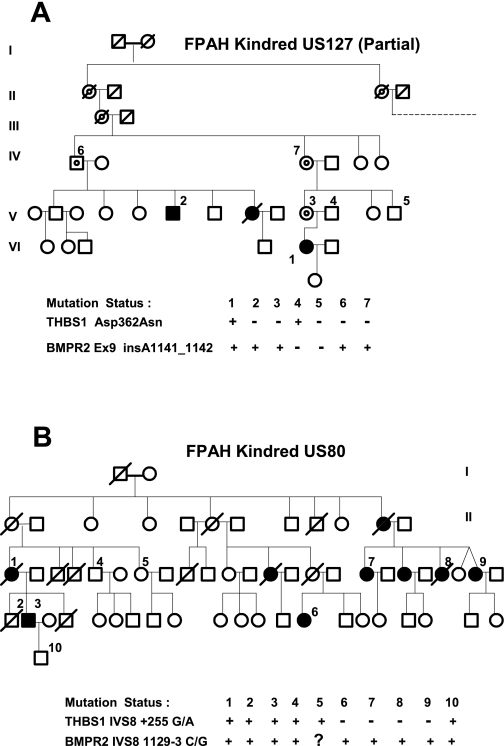
Asp362Asn occurs at nucleotide 1137 of cDNA (G1137A), causing a change of amino acid 362 from the charged aspartic acid (Asp) to an uncharged polar asparagine (Asn). The mutation was confirmed by bidirectional sequencing, RFLP (Fig. 2B), and TaqMan probe detection. Asp362Asn occurred in a highly conserved region (Fig. 2C) in probands from Vanderbilt family US127 and Colorado family C53. US127 has a known BMPR2 kinase region mutation; this region of BMPR2 was sequenced in the C53 proband but no mutation was found, proving the subjects are unrelated. Family tree data for US127 are in Fig. 3A. This mutation was not observed in 794 controls, confirming by its rarity that it fits the definition of a mutation, not a polymorphism.
Fig. 3.
Transmission of THBS1 mutations and associated BMPR2 mutations in FPAH cohorts. A: US127 family with known exon 9 BMPR2 mutation. Only a partial family tree is depicted. The parental pair on the right had 7 offspring, indicated by the dashed line; 12 descendants had PAH associated with the BMPR2 mutation, but the 7 who donated specimens were wild-type for THBS1. B: US80 family with known IVS8 BMPR2 mutation. Symbol key: 1, proband; circle, female; square, male; solid symbols, PAH diagnosis; slash through symbol, deceased; circle within symbol (target), obligate carrier of BMPR2 mutation. Generations are in Roman numerals where space allows.
A THBS1 Intron 8 Mutation (IVS8+255 G/A) Is Located Within Putative MAZ and SP1 Binding Sites
Family US80 has a point mutation in intron 8 of THBS1 (IVS8+255 G/A; Fig. 2A). In silico predictions indicated that this mutation falls within a putative zinc finger transcription factor binding site (consensus GGGAGGG, mutation site underlined) known to bind the SP1 and Myc-associated zinc finger proteins (MAZ). The mutant A allele is predicted to eliminate these sites (64). Multiple members of this family carry the mutation (Fig. 3B). This mutation was not observed in 344 controls, confirming by its rarity that it fits the definition of a mutation, not a polymorphism. Because of costs, “only” 344 controls were genotyped as IVS8+255 G/A required sequencing for genotyping (whereas a custom TaqMan probe was successfully made to detect Asp362Asn, allowing for economical genotyping in larger cohorts).
Common THBS1 Polymorphism Frequencies Are Similar in FPAH and Control Cohorts
Twelve known THBS1 SNP within resequenced domains of THBS1 were observed. The MAF of only one SNP (rs2664141) was different between groups, being higher in nonfamilial PAH (PAH 0.34, FPAH 0.26, controls 0.21; only whites analyzed; P = 0.004 adjusted for multiple hypotheses). We regard the findings as interesting, but mainly hypothesis generating.
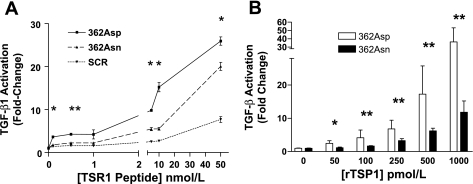
Mutant 362Asn TSP1 Displays Loss-of-Function Effects on Latent TGF-β1 Activation
Short synthetic TSR1 peptides.
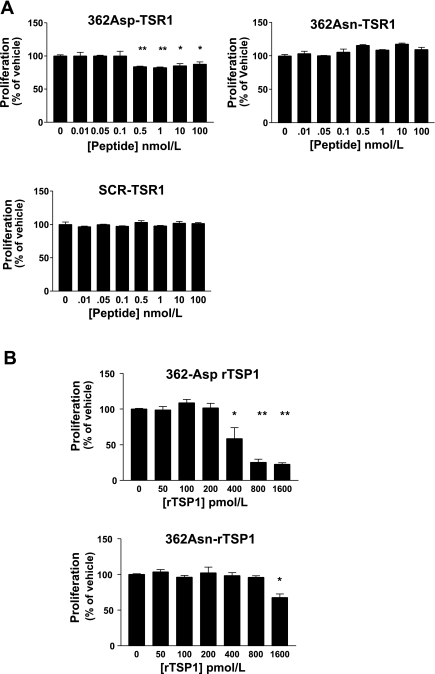
362Asn peptides displayed less activation of latent TGF-β1 than wild-type peptides over all tested concentrations, demonstrating the first evidence of loss-of-function effects from this mutation (Fig. 4A). The nanomolar concentrations of TSP1 peptides required for effects in these experiments are consistent with the known decreased potency of monomeric TSP1 proteins vs. full-length trimeric TSP1 (72).
Fig. 4.
The 362Asn TSP1 mutation displays loss-of-function characteristics on latent TGF-β1 activation in the mink lung epithelial cell bioassay. A: latent TGF-β1 activation by synthetic 13-amino acid TSR1 peptides centered on the 362Asn TSP1 locus. At all concentrations wild-type TSR1 (362Asp) is a more potent activator compared with mutant TSR1 [362Asn, which activates more than scrambled peptide (SCR) only when ≥5 nM]. 362Asp effects were always greater than SCR effects (*P < 0.05). B: trimeric recombinant TSP1 (rTSP1) is a more potent activator of TGF-β1 (pM effects). The mutant isoform is weaker at all concentrations compared with the wild-type isoform (*P < 0.01, **P < 0.001). Activation is expressed as fold change compared with vehicle (zero). Symbols indicate statistical significance by ANOVA or unpaired t-tests. Results represent summed means ± SD of ≥4 experiments.
Full-length trimeric rTSP1.
rTSP1 was of the expected size on Western blotting, was active immunologically in an ELISA, and was functionally active at physiological concentrations. Loss-of-function effects on TGF-β1 activation with 362Asn rTSP1 were impressive, demonstrating that this mutation has an important effect in the context of the native trimeric TSP1 molecule (Fig. 4B). Effects of wild-type rTSP1 on TGF-β1 activation were consistent with other reports (61).
Mutant 362Asn rTSP1 Is a Less Effective Inhibitor of HPAEC Proliferation
Most previous studies have localized the endothelial inhibitory domains of TSP1 protein to the TSR regions, particularly to TSR1 (16, 17, 48). Thus we first investigated the effects of the 362Asn mutation on proliferation of HPAEC at 72 h using short synthetic TSR1 peptides to isolate regional effects, then with full-length trimeric rTSP1.
Effects of TSR1 peptides.
Mutant 362Asn TSP1 and scrambled peptides had no inhibitory effects. Wild-type peptides inhibited proliferation at all tested concentrations ≥0.1 nM, with a peak effect of 14.9 ± 8.3% inhibition of growth at 0.5 nM (Fig. 5A).
Fig. 5.
THBS1 Asp362Asn mutation effects on inhibition of endothelial cell (HPAEC) proliferation at 72 h. A: effects of 13-amino acid TSR1 peptides. Wild-type 362Asp peptides inhibited proliferation at concentrations ≥0.5 nM; although mutant 362Asn and SCR peptides had no effect. B: effects of rTSP1 on proliferation. Wild-type rTSP1 had more potent effects than mutant rTSP1. Results expressed as means ± SD, summed results of 4 experiments. *P < 0.05 **P < 0.01 vs. vehicle effects.
Effects of full-length trimeric rTSP1.
Loss-of-function effects on inhibition of endothelial proliferation with the 362Asn mutation were marked in the context of rTSP1 (Fig. 5B). 362Asn rTSP1 displayed an IC50 of >1.6 nM, whereas wild-type rTSP1 displayed an IC50 of 0.64 nM (mutant rTSP1 inhibited growth only at the highest concentration). Microscopy revealed that wild-type rTSP1 actually caused cell death, consistent with prior reports, as cell numbers with higher doses were far below the numbers initially plated (17).
Mutant 362Asn rTSP1 Is a Less Effective Inhibitor of Proliferation of HPASMC
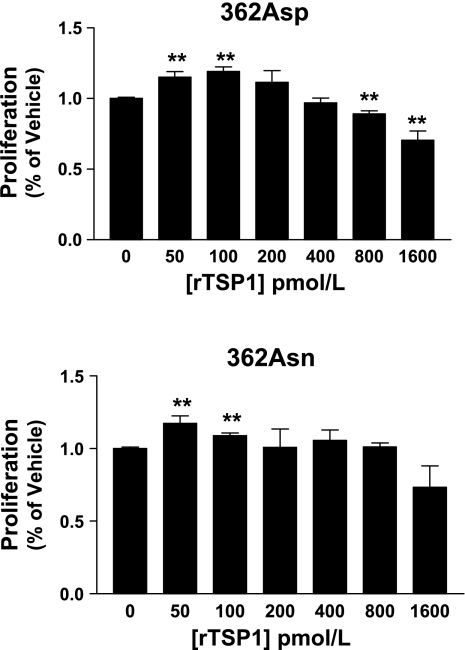
Given that smooth muscle cell proliferation is also a hallmark of PAH, we next evaluated the effects of the Asp362Asn mutation on HPASMC proliferation at 72 h. Since multiple TSP1 regions have been implicated for its effects on SMC (not just TSR regions), these experiments were performed only with full-length rTSP1. Wild-type rTSP1 decreased proliferation at concentrations of 0.8 and 1.6 nM, with a peak inhibition of 25.8% at 1.6 nM (Fig. 6); although inhibitory effects were lost with mutant rTSP1, it did not inhibit HPASMC growth at any concentration (P = 0.34 for 1.6 nM). Notably, effects of rTSP1 on HPASMC were less prominent than seen with endothelial cells, which are known to be exquisitely sensitive to TSP1 (from inhibition to actual apoptosis). Small increases in proliferation were seen with both rTSP1 isoforms at low concentrations, but these did not differ between genotypes (see discussion).
Fig. 6.
362Asn mutation displays loss-of-function properties on TSP1 inhibition of human pulmonary artery smooth muscle cell (HPASMC) proliferation at 72 h. Full-length rTSP1 was used for all studies. Only wild-type 362Asp rTSP1 had significant inhibitory effects. Stimulatory effects on proliferation were seen with both isoforms at low concentrations. Results are expressed as means ± SD, summed results of 4 experiments. *P < 0.05 **P < 0.01 vs. vehicle (zero) values.
THBS1 Mutation IVS8+255 G/A Alters Local Binding of Transcription Factors MAZ and SP1
To examine effects of the THBS1 IVS8+255 G/A mutation on local transcription factor binding, EMSA and ChIP experiments were performed.
EMSA results.
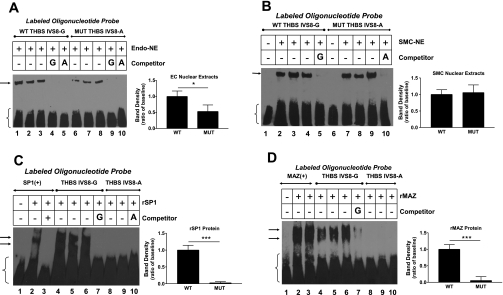
Both wild-type (WT; IVS8+255G) and mutant (MUT; IVS8+255A) oligonucleotides (oligos) avidly bound NE from both HPAEC and HPASMC. However, MUT oligos bound HPAEC NE at only 53 ± 20% of WT binding (Fig. 7A). Binding to HPASMC NE was not significantly different between oligo genotypes (P = 0.39; Fig. 7B). We then tested whether IVS8 region oligos specifically bound recombinant human SP1 and MAZ proteins (rSP1, rMAZ) and whether the mutation changed binding. MUT oligos bound rSP1 and rMAZ poorly (Fig. 7, C and D). Moreover, excess unlabeled WT oligos were an efficient competitor for rSP1 binding to a positive control oligo, confirming that this region of THBS1 avidly binds SP1 (WT at 50× inhibited SP1 binding to 22.5 ± 10.8% of baseline, WT at 100× inhibited to 21.9 ± 9.2 of baseline; P < 0.001 for both, means ± SD). However, excess unlabeled MUT oligo did not compete for rSP1 binding to an SP1-consensus oligo, even at a 100× greater concentration (0% inhibition at 100×). Potent inhibition of rMAZ binding to a MAZ strong consensus oligo was also seen with unlabeled IVS8 WT oligos (50× WT inhibited rMAZ binding to 68.8 ± 5.2% of baseline, 100× WT inhibited binding to 2.5 ± 0.5% of baseline; P < 0.01 and P < 0.001, respectively; means ± SD). EMSA experiments were performed with appropriate positive and negative controls for SP1 and MAZ binding, demonstrating that PASMC-NE contained proteins that specifically bound to both SP1 and MAZ (+) control oligos, and that rMAZ and rSP1 bound to their respective (+) control oligos but not to their respective (−) control oligos.
Fig. 7.
EMSAs evaluating binding of proteins from vascular cell nuclear extracts (NE) and recombinant SP1 and MAZ proteins (rSP1, rMAZ) to biotin-labeled oligonucleotides (oligos) of the THBS1 IVS8+255A mutation region. Labeled oligos are depicted at the top of figures, including positive controls. NE are from HPASMC (SMC) or HPAEC (Endo). Unlabeled competitors were added at 100-fold excess to assess specificity or competition. A: mutant IVS8-A oligos (MUT; lanes 6–8) bind Endo-NE less than wild-type oligos (WT; lanes 1–3). Excess unlabeled IVS8-A oligos remain an effective inhibitor of labeled-IVS8-G oligo binding to Endo-NE, consistent with some protein binding being maintained by the mutant sequence (lane 5). B: IVS8 oligo binding to SMC-NE was not different (P = 0.39). Excess of either unlabeled oligo abrogates binding (lanes 5 and 10). C: IVS8-A oligos bind rSP1 poorly (lanes 8–10). rSP1 binds avidly to a positive control SP1 oligo (lanes 1–3) and to IVS8-G oligos (lanes 4–6) in a specific manner (lane 7). D: IVS8-A oligos bind rMAZ poorly (lanes 8 and 9) compared with avid binding to IVS8-G oligos (lanes 4–6; specificity shown in lane 7) and to rMAZ positive control oligos (lanes 1–3). Arrows indicate shifted bands; brackets indicate free labeled probe. Analysis of band densities from all experiments is depicted by graphs to the right. Results are expressed as means ± SD as the sum of 3 experiments. Radiographs are representative. *P < 0.05; **P < 0.01, or ***P < 0.001 vs. baseline values. The presence of 2 shifted bands with rMAZ and rSP1 (vs. 1 band with NE) in EMSA is related to the tendency of these purified proteins to form both monomers and dimers.
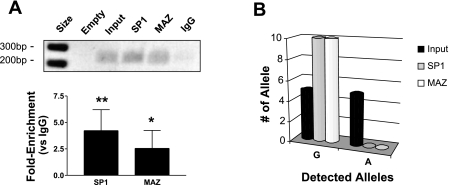
ChIP results.
ChIP experiments using HPASMC revealed that DNA of the THBS1 IVS8 region bound endogenous SP1 and MAZ. This area of chromatin was enriched 4.2 ± 2.0-fold with SP1 antibody and 2.6 ± 1.7-fold with MAZ antibody (Fig. 8A). Experiments using lymphoblasts heterozygous for the IVS8+255A mutation demonstrated that only the wild-type chromatin strand bound endogenous SP1 and MAZ because only the wild-type strand was pulled down by these antibodies, again consistent with a loss of SP1 and MAZ binding due to the mutant A allele (Fig. 8B). Accuracy of ChIP experiments was confirmed with controls: SP1 and MAZ binding were both over 10-fold enriched in the regions of known positive control gene sequences (PCR primers localized to sites in COL1A and c-myc genes, respectively; P = 0.03 for comparison of SP1 and MAZ values to control IgG), and no enrichment of SP1 or MAZ was seen at negative control sites (PCR primers localized to site in VMAT gene, P < 0.05).
Fig. 8.
A: data from chromatin immunoprecipitation (ChIP) experiments using chromatin from normal HPASMC grown under standard conditions. Chromatin was fixed and sheared, and then specific antibodies to SP1 or MAZ were used to pull down chromatin that endogenously bound SP1 or MAZ proteins (rabbit IgG was control antibody). Immunoprecipitated chromatin was amplified with primers flanking the THBS1 IVS8+255 mutation; then equal aliquots of PCR products were run on an agarose gel, shown at the top of the representative figure (ethidium-stained gel; empty, empty lane; input, amplification of input chromatin, never treated with antibody; DNA bp size marker is in leftmost lane). The fold enrichment of immunoprecipitated chromatin (relative to control IgG antibody) for specific proteins is shown in graph at bottom as determined by quantitative SYBR-green PCR (sum of 6 experiments, means ± SD; * P < 0.05, ** P < 0.01). B: analysis of THBS1 IVS8+255 G/A site occupancy in mutant IVS8+255A carrier lymphoblasts after ChIP as in A. After PCR amplification of the THBS1 IVS8 region, PCR products were subcloned and 10 randomly selected colonies (single clones) were isolated and sequenced for IVS8+255 genotype. Input DNA contained both alleles, whereas immunoprecipitation for SP1 or MAZ revealed only the presence of the G allele, demonstrating that that the A allele disrupted binding of SP1 and MAZ to THBS1 DNA at this locus in intact cells (P < 0.006; 2-tailed χ2).
The THBS1 Mutation IVS8+255 G/A Does Not Affect RNA Splicing
Presence of the IVS8+255A allele did not change alternative splicing of THBS1 mRNA in carrier lymphoblasts in resting or stimulated conditions, or in whole blood RNA from a carrier. This was assessed by absence of change in RT-PCR product band sizes (not shown) and by sequencing PCR products to rule out minor splicing events. Serum from an IVS8+255A carrier showed TSP1 protein similar in size to controls, without any evidence of truncated TSP1 protein (not shown).
DISCUSSION
We found novel and functional missense and intronic mutations in THBS1 in FPAH kindreds. These mutations were not observed in large multiracial healthy cohorts or in subjects with nonfamilial PAH. A missense mutation (Asp362Asn) in the first type I repeat (TSR1) occurred in two unrelated families. A mutation of intron 8 (IVS8+255 G/A) occurred in a third family. The finding of Asp362Asn in unrelated families is reminiscent of the descriptions of shared BMPR2 mutations among unrelated FPAH kindreds (42). FPAH is a rare disease, but clinical PH referral centers see many of these patients so our data may have implications in the genetic screening of subjects at such centers. However, the major import of these mutations and their functional effects may be to highlight the important of TSP1 in pulmonary vascular biology and disease.
The 362Asn THBS1 mutation is of particular interest because it is a missense change in a highly conserved area: TSR1. TSR have been identified as activators of latent TGF-β1, as inhibitors of smooth muscle cell growth (52), and as potent natural inhibitors of endothelial cell proliferation (30). WXXW domains in these repeats also tightly bind and sequester the potent endothelial mitogens bFGF and VEGF, inhibiting their binding to receptors (20, 45, 66), thereby inhibiting cell proliferation (11, 55). This is relevant to PAH because both exuberant TGF-β and endothelial mitogen activity exist in PAH lungs (25, 49). Mutant 362Asn TSP1 was much less effective at activating latent TGF-β1 and at inhibiting endothelial cell growth; these effects were apparent both in a regional context (TSR1 peptides) and with trimeric rTSP1. The short domain encompassed by the TSR1 peptides we employed includes two overlapping WXXW sequences, which have been described as important for docking of TSP1 with discrete LAP regions. In the latent LAP-TGF-β complex, these interactions stabilize the downstream KRFK interaction with LAP that is the most potent effector of latent TGF-β activation. Interestingly, we found that the overlapping WXXW region of TSR1 is itself an activator of latent TGF-β1 in the context of the 13-amino acid peptides we employed; others have not found such activity in shorter TSR peptides (GGWSHW) in different assays (60). This may be related to the inclusion of two WXXW sequences in our peptides compared with other studies and needs to be distinguished from the in vitro inhibitory actions of WXXW peptides on trimeric TSP1 activation of latent TGF-β due to mimicry/blocking effects on native WXXW domains (60). The Asp362Asn mutation is near the NH2-terminal end of TSR1, 51 amino acids away from the KRFK domain at the COOH-terminal end of TSR1. Since TSR 1–3 have been shown to have important interactions with each other (41), it seems likely that the 362Asn mutation affects latent TGF-β activation both through a loss of ancillary docking strength to LAP and from effects on the tertiary structure of TSR1 due to the substitution of an uncharged polar residue (Asn) for a nonpolar acidic residue (Asp). This is a nonconservative substitution that in other proteins has been associated with prominent functional and structural effects (24, 39). The even more prominent loss-of-function effects on latent TGF-β that we observed with mutant trimeric rTSP1 suggests a structural change that sterically hinders access to the KRFK sequence. Further work is required to determine the structural basis of the functional effects we observed with the 362Asn mutation.
The profound effects on endothelial cells seen with wild-type trimeric rTSP1 were not seen with wild-type TSR1 peptides, consistent with the potent apoptotic effects previously described for native TSP1 but not its derived monomeric peptides (17). Moreover, although the TSR1 monomeric peptides we tested can bind latent TGF-β and some growth factors, they cannot engage most of the nine known TSP1 receptors in vascular cells, several of which are effectors of TSP1-mediated apoptosis (22, 32). This may be related to a loss of inhibitory sequestration of bFGF and VEGF attributed to the WXXW regions of TSR in wild-type TSP1, but assessing such specific effects was outside the scope of our project (20, 66). Inhibitory effects of wild-type rTSP1 on HPASMC proliferation were clear (though modest compared with effects on HPAEC) but were lost with mutant 362Asn rTSP1. Interestingly, low concentrations of rTSP1 in the 50–100 pM range caused proproliferative effects on HPASMC for both rTSP1 isoforms, which may be consistent with the known inhibition of cAMP synthesis that occurs in smooth muscle cells via TSP1 binding to CD47 (at the COOH-terminus of TSP1), a distant motif unlikely to be affected by the 362Asn mutation (70). At higher concentrations such proproliferative effects were absent, consistent with a dominant effect of TSP1 receptors such as CD36 (which binds TSP1 in the TSR 1–3 region) in this concentration range (23). CD36 activation typically requires high picomolar concentrations of TSP1, whereas CD47 is ligated at low picomolar TSP1 concentrations. Such biphasic effects of TSP1 may occur in vivo as well, where substantial variation in TSP1 levels occurs based on location or presence of injury. During inflammation and clotting TSP1 is released from platelets and leukocytes; at this time adjoining cells may be exposed to TSP1 concentration spikes as high as 1–50 nM (12, 13). In contrast, typical plasma levels of TSP1 in humans are 0.2 nM, representative of the quiescent state. TSP1 levels of 10 nM appear to be at or above the dissociation constant (Kd) for known TSP1 receptors, with the Kd of CD47 being the lowest at 12 pM (7, 21). Thus the picomolar to low nanomolar concentrations of trimeric TSP1 that we tested are likely present in the unperturbed lung, relevant to early phases of PAH pathogenesis.
Since the IVS8+255 G/A mutation fell within a putative MAZ/SP1 zinc finger transcription factor site based on an unbiased in silico analysis (64), we tested its effects on binding of nuclear extracts from PASMC and HPAEC, on binding of SP1 and MAZ specifically, and on RNA splicing (in case these transcription factors had such a role at the mutation site). IVS8+255A nearly eliminated MAZ and SP1 binding based on EMSA and ChIP analyses, but it had no apparent effects on splicing. Interestingly, MAZ appears to have an important effect on DNA strand bending outside of regulatory regions, influencing transcriptional initiation and termination (3). SP1 and MAZ are also important regulators of other genes involved in PAH pathogenesis such as endoglin, endothelial nitric oxide synthase (eNOS), serotonin 1A receptor, c-myc, TGF-β type I receptor, α-smooth muscle actin, and phosphodiesterase type 5 (5, 26, 28, 31, 40, 56, 65). The THBS1 promoter is also constitutively activated by SP1 (36). SP1 and MAZ typically costimulate a promoter at their overlapping binding sites, but in some cases they may antagonize each other's effects (31). In most of these genes SP1 has both constitutive and inducible stimulatory effects. In the human endoglin promoter, SP1 cooperatively increases the stimulatory effects of TGF-β and smad proteins (5). SP1 is known to have effects on gene transcription even when it binds at intronic locations thousands of base pairs from a gene promoter, such effects are postulated to involve increased delivery of SP1 to the promoter via trapping and subsequent release of nuclear SP1 from such intronic sites (5, 9, 31). We postulate that the IVS8+255A mutation has effects that decrease the constitutive stimulatory effects of SP1 on the THBS1 promoter via decreased SP1 trapping. Such effects would be expected to decrease THBS1 promoter activity and RNA expression. Proof of this will require the future procurement of blood lymphoblasts from enough IVS8+255A carriers in the US80 kindred to test for genotype-specific differences in THBS1 expression. Since only one subject with this mutation had archived lymphoblasts, we did not have enough samples to perform a meaningful analysis of the effects of this mutation on THBS1 expression. Likewise, there were no archived explanted lung specimens, lung biopsies, or bronchoalveolar lavage fluid to test for lung TSP1 expression effects of mutations in these kindreds. Since only a fraction of the US80 kindred donated DNA specimens the data were inadequate to perform a trait-transmission analysis to confirm phenotype effects of IVS8+255 G/A. This ascertainment bias toward symptomatic cases is characteristic of inherited disease registries, where asymptomatic relatives often avoid genetic testing owing to personal concerns. However, subjects with PAH in the US80 and US127 families who had both BMPR2 and THBS1 mutations had a median age of disease onset at age 38, vs. median onset of PAH at age 42 for those with only BMPR2 mutations. Moreover, of the 17 subjects in both families who could be genotyped, the second and third youngest members with PAH (ages 28 and 38 at diagnosis) had both BMPR2 and THBS1 mutations. This suggests that PAH may occur earlier in the presence of both mutations. Analysis of the 18 genotyped members between the three FPAH cohorts examined did not reveal any obvious pattern of extrapulmonary disease in THBS1 mutation carriers. The expression of TSP1 is variable but widespread in mammalian tissues, so why the phenotype effects of the THBS1 mutations we observed are lung limited is unclear but likely relates to the same reasons why the TSP1-null mouse has a mostly lung-limited phenotype. TSP1 appears to have a key role in lung homeostasis.
These mutations position THBS1 as a novel modifier gene in FPAH pathogenesis. Functional THBS1 mutations are likely to be synergistic toward FPAH pathogenesis by causing further disruptions in TGF-β or cell growth regulation pathways. A protective effect of THBS1 mutations in FPAH via abrogation of TGF-β activation seems unlikely; the loss of the normal inhibitory effects on vascular cell proliferation that we demonstrate for the 362Asn mutation predicts an increased risk of developing PAH (19, 25, 58). Moreover, although exuberant TGF-β signaling activity is well described in end-stage PAH lungs (49), TGF-β dysregulation (too little TGF-β as well as too much) may be important in early PAH pathogenesis; for instance, TGF-β-null mice have early and marked pulmonary vascular smooth muscle proliferation (10). It may be that effects of TSP1 mutations are important during early FPAH pathogenesis (before the increase in TGF-β signaling described in end-stage FPAH lungs), or mainly in endothelial cells, where TSP1 had the most profound effects in our work. Relevant to these observations are reports that endothelial-selective BMPR2-null mice develop PH even though BMPR2 expression is normal in vascular smooth muscle (19) and apoptosis-resistant endothelial cells are prevalent in the pulmonary vasculature of PAH subjects (46). Thus current evidence supports the assertion that the functional aberrations we observed with mutant 362Asn TSP1 increase the risk of PAH in BMPR2 mutation carriers and/or hasten its appearance at an earlier age. Proving such a role will require work in several areas. A resequencing of the entire THBS1 gene in FPAH kindreds is needed to assess for other functional mutations that would further support such a role, since we only resequenced 5% of the entire THBS1 gene in a domain-focused look at regions thought most relevant to PAH pathogenesis. Ascertainment of genotypes from all or most unaffected members in at least a few kindreds is needed to perform a trait-transmission analysis capable of determining the effect of THBS1 genotype on penetrance, age of onset, and severity of PAH when superimposed on BMPR2 mutations. Lastly, knowledge of the role that TSP1 plays in the homeostasis of the normal pulmonary vasculature and in PH pathogenesis needs to be enhanced, particularly in other animal models of PH.
It remains unclear whether these THBS1 mutations have effects in the lung via dysfunctional TSP1 protein (e.g., TSP1 Asn362) or via a global decrease in TSP1 levels (as we postulate occurs for IVS8+255A). In a hypoxic rodent model of PAH, Ochoa and colleagues (54) found that TSP1-null mice still developed PH; but the PH and pulmonary vascular remodeling were actually less than seen in wild-type mice. A blunted hypoxic pressor response was also seen in these mice. This suggests that a global decrease in TSP1 levels may not be a likely event in PAH pathogenesis, unlike the well-described deficiencies of the nitric oxide synthase and prostacyclin synthase in PAH lungs. In fact, human PAH lungs appear to have preserved TSP1 expression (6). It seems more likely that an imbalance of TSP1 domain signaling through specific receptors is the important determinant for effects of these mutations in human PAH. The hypoxic rodent model of PH is a useful tool, but its relevance to the factors that result in human PAH is limited outside of the subset of patients with PH associated with chronic hypoxemia. Moreover, the TSP1-null mouse loses all TSP1 signaling effects, a situation that apparently does not occur in humans.
Such data for partial protection against PH, in the absence of all TSP1-related functions, likely relates to effects of CD47 ligation by TSP1. In rodents TSP1-CD47 ligation causes tonic eNOS inhibition (21). Thus removal of all TSP1-CD47 binding (null mouse) could ameliorate PAH by derepressing lung NO production. Since neither deletion of the entire THBS1 gene nor mutations of the CD47 binding region have been reported, the implications of this data are unclear. Since we did not find THBS1 mutations in nonfamilial PAH subjects (BMPR2 status unknown), it may be that THBS1 mutations predispose to PAH only in subjects with BMPR2 mutations; confirmation of this will require THBS1 resequencing in a cohort of idiopathic PAH subjects with BMPR2 mutations. If THBS1 is an important modifier gene in FPAH pathogenesis, it seems rational that functional mutations in its receptors (such as CD36 and CD47) could also influence FPAH pathogenesis via impaired TSP1 signaling. However, although an inherited mutant CD36 null phenotype exists in humans, these subjects have a principal phenotype of hyperlipidemia; PH has not been described (69). As a large multidomain protein, TSP1 signaling through its receptors is complex, and which of these receptors influence pulmonary vascular homeostasis remains unclear.
In conclusion, this research adds TSP1 to a small list of genes known to have functional mutations in inherited forms of PH. Perhaps most importantly, these data highlight the importance of TSP1 in pulmonary vascular biology. An evaluation for mutations in other THBS1 regions and further work on the role of TSP1 in pulmonary vascular homeostasis are warranted to better understand the genetics and pathophysiology of PH.
GRANTS
This research was funded in part by National Institutes of Health Grants HL071618 (J. P. Maloney) and PO1-072058 (J. E. Loyd) and by Vanderbilt institutional research grant M01-RR-00095.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank the patients and family members who enrolled in the familial PH study at Vanderbilt University and in research protocols at the University of Colorado. Joanne Murphy-Ullrich, Deane Mosher, and Doug Annis provided helpful comments.
REFERENCES
- 1. Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem 216: 276–284, 1994 [DOI] [PubMed] [Google Scholar]
- 2. Almeida-Vega S, Catlow K, Kenny S, Dimaline R, Varro A. Gastrin activates paracrine networks leading to induction of PAI-2 via MAZ and ASC-1. Am J Physiol Gastrointest Liver Physiol 296: G414–G423, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashfield R, Patel AJ, Bossone SA, Brown H, Campbell RD, Marcu KB, Proudfoot NJ. MAZ-dependent termination between closely spaced human complement genes. EMBO J 13: 5656–5667, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bossone SA, Asselin C, Patel AJ, Marcu KB. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc Natl Acad Sci USA 89: 7452–7456, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Botella LM, Sanchez-Elsner T, Rius C, Corbi A, Bernabeu C. Identification of a critical Sp1 site within the endoglin promoter and its involvement in the transforming growth factor-beta stimulation. J Biol Chem 276: 34486–34494, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Botney MD, Kaiser LR, Cooper JD, Mecham RP, Parghi D, Roby J, Parks WC. Extracellular matrix protein gene expression in atherosclerotic hypertensive pulmonary arteries. Am J Pathol 140: 357–364, 1992 [PMC free article] [PubMed] [Google Scholar]
- 7. Boukerche H, Berthier-Vergnes O, Tabone E, Bailly M, Dore JF, McGregor JL. Thrombospondin modulates melanoma–platelet interactions and melanoma tumour cell growth in vivo. Br J Cancer 72: 108–116, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cogan JD, Pauciulo MW, Batchman AP, Prince MA, Robbins IM, Hedges LK, Stanton KC, Wheeler LA, Phillips JA, 3rd, Loyd JE, Nichols WC. High frequency of BMPR2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med 174: 590–598, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Courey AJ, Holtzman DA, Jackson SP, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59: 827–836, 1989 [DOI] [PubMed] [Google Scholar]
- 10. Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, Bouck N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell 93: 1159–1170, 1998 [DOI] [PubMed] [Google Scholar]
- 11. Dawson DW, Pearce SF, Zhong R, Silverstein RL, Frazier WA, Bouck NP. CD36 mediates the in vitro inhibitory effects of thrombospondin-1 on endothelial cells. J Cell Biol 138: 707–717, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dudek AZ, Mahaseth H. Circulating angiogenic cytokines in patients with advanced non-small cell lung cancer: correlation with treatment response and survival. Cancer Invest 23: 193–200, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Fike CD, Slaughter JC, Kaplowitz MR, Zhang Y, Aschner JL. Reactive oxygen species from NADPH oxidase contribute to altered pulmonary vascular responses in piglets with chronic hypoxia-induced pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol 295: L881–L888, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freishtat RJ, Benton AS, Watson AM, Wang Z, Rose MC, Hoffman EP. Delineation of a gene network underlying the pulmonary response to oxidative stress in asthma. J Investig Med 57: 756–764, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, Nelson SF, Horvath S, Berliner JA, Kirchgessner TG, Lusis AJ. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA 103: 12741–12746, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Good DJ, Polverini PJ, Rastinejad F, Le Beau MM, Lemons RS, Frazier WA, Bouck NP. A tumor suppressor-dependent inhibitor of angiogenesis is immunologically and functionally indistinguishable from a fragment of thrombospondin. Proc Natl Acad Sci USA 87: 6624–6628, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo N, Krutzsch HC, Inman JK, Roberts DD. Thrombospondin 1 and type I repeat peptides of thrombospondin 1 specifically induce apoptosis of endothelial cells. Cancer Res 57: 1735–1742, 1997 [PubMed] [Google Scholar]
- 18. He B, Weber GF. Synergistic activation of the CMV promoter by NF-kappaB P50 and PKG. Biochem Biophys Res Commun 321: 13–20, 2004 [DOI] [PubMed] [Google Scholar]
- 19. Hong KH, Lee YJ, Lee E, Park SO, Han C, Beppu H, Li E, Raizada MK, Bloch KD, Oh SP. Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension. Circulation 118: 722–730, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iruela-Arispe ML, Lombardo M, Krutzsch HC, Lawler J, Roberts DD. Inhibition of angiogenesis by thrombospondin-1 is mediated by 2 independent regions within the type 1 repeats. Circulation 100: 1423–1431, 1999 [DOI] [PubMed] [Google Scholar]
- 21. Isenberg JS, Annis DS, Pendrak ML, Ptaszynska M, Frazier WA, Mosher DF, Roberts DD. Differential interactions of thrombospondin-1, -2, and -4 with CD47 and effects on cGMP signaling and ischemic injury responses. J Biol Chem 284: 1116–1125, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Isenberg JS, Hyodo F, Matsumoto K, Romeo MJ, Abu-Asab M, Tsokos M, Kuppusamy P, Wink DA, Krishna MC, Roberts DD. Thrombospondin-1 limits ischemic tissue survival by inhibiting nitric oxide-mediated vascular smooth muscle relaxation. Blood 109: 1945–1952, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Isenberg JS, Martin-Manso G, Maxhimer JB, Roberts DD. Regulation of nitric oxide signalling by thrombospondin 1: implications for anti-angiogenic therapies. Nat Rev Cancer 9: 182–194, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishimura-Oka K, Semenkovich CF, Faustinella F, Goldberg IJ, Shachter N, Smith LC, Coleman T, Hide WA, Brown WV, Oka K, Chan L. A missense (Asp250→Asn) mutation in the lipoprotein lipase gene in two unrelated families with familial lipoprotein lipase deficiency. J Lipid Res 33: 745–754, 1992 [PubMed] [Google Scholar]
- 25. Izikki M, Guignabert C, Fadel E, Humbert M, Tu L, Zadigue P, Dartevelle P, Simonneau G, Adnot S, Maitre B, Raffestin B, Eddahibi S. Endothelial-derived FGF2 contributes to the progression of pulmonary hypertension in humans and rodents. J Clin Invest 119: 512–523, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Izzo MW, Strachan GD, Stubbs MC, Hall DJ. Transcriptional repression from the c-myc P2 promoter by the zinc finger protein ZF87/MAZ. J Biol Chem 274: 19498–19506, 1999 [DOI] [PubMed] [Google Scholar]
- 27. Jaffe E, Bornstein P, Disteche CM. Mapping of the thrombospondin gene to human chromosome 15 and mouse chromosome 2 by in situ hybridization. Genomics 7: 123–126, 1990 [DOI] [PubMed] [Google Scholar]
- 28. Ji C, Casinghino S, McCarthy TL, Centrella M. Multiple and essential Sp1 binding sites in the promoter for transforming growth factor-beta type I receptor. J Biol Chem 272: 21260–21267, 1997 [DOI] [PubMed] [Google Scholar]
- 29. Jones LJ, Gray M, Yue ST, Haugland RP, Singer VL. Sensitive determination of cell number using the CyQUANT cell proliferation assay. J Immunol Methods 254: 85–98, 2001 [DOI] [PubMed] [Google Scholar]
- 30. Karagiannis ED, Popel AS. A systematic methodology for proteome-wide identification of peptides inhibiting the proliferation and migration of endothelial cells. Proc Natl Acad Sci USA 105: 13775–13780, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karantzoulis-Fegaras F, Antoniou H, Lai SL, Kulkarni G, D'Abreo C, Wong GK, Miller TL, Chan Y, Atkins J, Wang Y, Marsden PA. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem 274: 3076–3093, 1999 [DOI] [PubMed] [Google Scholar]
- 32. Kaur S, Martin-Manso G, Pendrak ML, Garfield SH, Isenberg JS, Roberts DD. Thrombospondin-1 inhibits VEGF receptor-2 signaling by disrupting its association with CD47. J Biol Chem 285: 38923–38932, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koch AE, Friedman J, Burrows JC, Haines GK, Bouck NP. Localization of the angiogenesis inhibitor thrombospondin in human synovial tissues. Pathobiology 61: 1–6, 1993 [DOI] [PubMed] [Google Scholar]
- 34. Koehler R, Grunig E, Pauciulo MW, Hoeper MM, Olschewski H, Wilkens H, Halank M, Winkler J, Ewert R, Bremer H, Kreuscher S, Janssen B, Nichols WC. Low frequency of BMPR2 mutations in a German cohort of patients with sporadic idiopathic pulmonary arterial hypertension. J Med Genet 41: e127, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuhn C, Mason RJ. Immunolocalization of SPARC, tenascin, and thrombospondin in pulmonary fibrosis. Am J Pathol 147: 1759–1769, 1995 [PMC free article] [PubMed] [Google Scholar]
- 36. Laherty CD, Gierman TM, Dixit VM. Characterization of the promoter region of the human thrombospondin gene. DNA sequences within the first intron increase transcription. J Biol Chem 264: 11222–11227, 1989 [PubMed] [Google Scholar]
- 37. Lawler J, Hynes RO. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J Cell Biol 103: 1635–1648, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawler J, Sunday M, Thibert V, Duquette M, George EL, Rayburn H, Hynes RO. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J Clin Invest 101: 982–992, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lefkowitz JB, Nuss R, Haver T, Jacobson L, Thompson AR, Manco-Johnson M. Factor IX Denver, ASN 346–>ASP mutation resulting in a dysfunctional protein with defective factor VIIIa interaction. Thromb Haemost 86: 862–870, 2001 [PubMed] [Google Scholar]
- 40. Lin CS, Chow S, Lau A, Tu R, Lue TF. Human PDE5A gene encodes three PDE5 isoforms from two alternate promoters. Int J Impot Res 14: 15–24, 2002 [DOI] [PubMed] [Google Scholar]
- 41. Liu Y, Mosher DF. Interactions among stalk modules of thrombospondin-1. J Biol Chem 284: 28563–28570, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Machado RD, Aldred MA, James V, Harrison RE, Patel B, Schwalbe EC, Gruenig E, Janssen B, Koehler R, Seeger W, Eickelberg O, Olschewski H, Elliott CG, Glissmeyer E, Carlquist J, Kim M, Torbicki A, Fijalkowska A, Szewczyk G, Parma J, Abramowicz MJ, Galie N, Morisaki H, Kyotani S, Nakanishi N, Morisaki T, Humbert M, Simonneau G, Sitbon O, Soubrier F, Coulet F, Morrell NW, Trembath RC. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum Mutat 27: 121–132, 2006 [DOI] [PubMed] [Google Scholar]
- 43. Machado RD, Pauciulo MW, Thomson JR, Lane KB, Morgan NV, Wheeler L, Phillips JA, 3rd, Newman J, Williams D, Galie N, Manes A, McNeil K, Yacoub M, Mikhail G, Rogers P, Corris P, Humbert M, Donnai D, Martensson G, Tranebjaerg L, Loyd JE, Trembath RC, Nichols WC. BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet 68: 92–102, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest 107: 899–907, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Margosio B, Marchetti D, Vergani V, Giavazzi R, Rusnati M, Presta M, Taraboletti G. Thrombospondin 1 as a scavenger for matrix-associated fibroblast growth factor 2. Blood 102: 4399–4406, 2003 [DOI] [PubMed] [Google Scholar]
- 46. Masri FA, Xu W, Comhair SA, Asosingh K, Koo M, Vasanji A, Drazba J, Anand-Apte B, Erzurum SC. Hyperproliferative apoptosis-resistant endothelial cells in idiopathic pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol 293: L548–L554, 2007 [DOI] [PubMed] [Google Scholar]
- 47. McLaughlin VV, Shillington A, Rich S. Survival in primary pulmonary hypertension: the impact of epoprostenol therapy. Circulation 106: 1477–1482, 2002 [DOI] [PubMed] [Google Scholar]
- 48. Miao WM, Seng WL, Duquette M, Lawler P, Laus C, Lawler J. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-beta-dependent and -independent mechanisms. Cancer Res 61: 7830–7839, 2001 [PubMed] [Google Scholar]
- 49. Morrell NW, Yang X, Upton PD, Jourdan KB, Morgan N, Sheares KK, Trembath RC. Altered growth responses of pulmonary artery smooth muscle cells from patients with primary pulmonary hypertension to transforming growth factor-beta(1) and bone morphogenetic proteins. Circulation 104: 790–795, 2001 [DOI] [PubMed] [Google Scholar]
- 50. Morse J, Barst R, Horn E, Cuervo N, Deng Z, Knowles J. Pulmonary hypertension in scleroderma spectrum of disease: lack of bone morphogenetic protein receptor 2 mutations. J Rheumatol 29: 2379–2381, 2002 [PubMed] [Google Scholar]
- 51. Newman JH, Phillips JA, 3rd, Loyd JE. Narrative review: the enigma of pulmonary arterial hypertension: new insights from genetic studies. Ann Intern Med 148: 278–283, 2008 [DOI] [PubMed] [Google Scholar]
- 52. Ochoa CD, Baker H, Hasak S, Matyal R, Salam A, Hales CA, Hancock W, Quinn DA. Cyclic stretch affects pulmonary endothelial cell control of pulmonary smooth muscle cell growth. Am J Respir Cell Mol Biol 39: 105–112, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ochoa CD, Hasak SL, Al-Ansari E, Yu L, Hales CA, Quinn DA. Thrombospondin-1 in hypoxic pulmonary vascular remodeling. FASEB J 21: A1434–A1435, 2007 [Google Scholar]
- 54. Ochoa CD, Yu L, Al-Ansari E, Hales CA, Quinn DA. Thrombospondin-1 null mice are resistant to hypoxia-induced pulmonary hypertension. J Cardiothorac Surg 5: 32, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oganesian A, Armstrong LC, Migliorini MM, Strickland DK, Bornstein P. Thrombospondins use the VLDL receptor and a nonapoptotic pathway to inhibit cell division in microvascular endothelial cells. Mol Biol Cell 19: 563–571, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parks CL, Shenk T. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J Biol Chem 271: 4417–4430, 1996 [DOI] [PubMed] [Google Scholar]
- 57. Qabar AN, Bullock J, Matej L, Polverini P. Expression and characterization of novel thrombospondin 1 type I repeat fusion proteins. Biochem J 346: 147–153, 2000 [PMC free article] [PubMed] [Google Scholar]
- 58. Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 118: 2372–2379, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sage E, Mercier O, Van den Eyden F, de Perrot M, Barlier-Mur AM, Dartevelle P, Eddahibi S, Herve P, Fadel E. Endothelial cell apoptosis in chronically obstructed and reperfused pulmonary artery. Respir Res 9: 19, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schultz-Cherry S, Chen H, Mosher DF, Misenheimer TM, Krutzsch HC, Roberts DD, Murphy-Ullrich JE. Regulation of transforming growth factor-beta activation by discrete sequences of thrombospondin 1. J Biol Chem 270: 7304–7310, 1995 [DOI] [PubMed] [Google Scholar]
- 61. Schultz-Cherry S, Ribeiro S, Gentry L, Murphy-Ullrich JE. Thrombospondin binds and activates the small and large forms of latent transforming growth factor-beta in a chemically defined system. J Biol Chem 269: 26775–26782, 1994 [PubMed] [Google Scholar]
- 62. Shamis Y, Hasson E, Soroker A, Bassat E, Shimoni Y, Ziv T, Sionov RV, Mitrani E. Organ-specific scaffolds for in vitro expansion, differentiation, and organization of primary lung cells. Tissue Eng Part C Methods 17: 861–870 [DOI] [PubMed] [Google Scholar]
- 63. Short SM, Derrien A, Narsimhan RP, Lawler J, Ingber DE, Zetter BR. Inhibition of endothelial cell migration by thrombospondin-1 type-1 repeats is mediated by beta1 integrins. J Cell Biol 168: 643–653, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Song J, Ugai H, Ogawa K, Wang Y, Sarai A, Obata Y, Kanazawa I, Sun K, Itakura K, Yokoyama KK. Two consecutive zinc fingers in Sp1 and in MAZ are essential for interactions with cis-elements. J Biol Chem 276: 30429–30434, 2001 [DOI] [PubMed] [Google Scholar]
- 65. Subramanian SV, Polikandriotis JA, Kelm RJ, Jr, David JJ, Orosz CG, Strauch AR. Induction of vascular smooth muscle alpha-actin gene transcription in transforming growth factor beta1-activated myofibroblasts mediated by dynamic interplay between the Pur repressor proteins and Sp1/Smad coactivators. Mol Biol Cell 15: 4532–4543, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Taraboletti G, Belotti D, Borsotti P, Vergani V, Rusnati M, Presta M, Giavazzi R. The 140-kilodalton antiangiogenic fragment of thrombospondin-1 binds to basic fibroblast growth factor. Cell Growth Differ 8: 471–479, 1997 [PubMed] [Google Scholar]
- 67. Wang IM, Stepaniants S, Boie Y, Mortimer JR, Kennedy B, Elliott M, Hayashi S, Loy L, Coulter S, Cervino S, Harris J, Thornton M, Raubertas R, Roberts C, Hogg JC, Crackower M, O'Neill G, Pare PD. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med 177: 402–411, 2008 [DOI] [PubMed] [Google Scholar]
- 68. Wight TN, Raugi GJ, Mumby SM, Bornstein P. Light microscopic immunolocation of thrombospondin in human tissues. J Histochem Cytochem 33: 295–302, 1985 [DOI] [PubMed] [Google Scholar]
- 69. Yanai H, Chiba H, Morimoto M, Abe K, Fujiwara H, Fuda H, Hui SP, Takahashi Y, Akita H, Jamieson GA, Kobayashi K, Matsuno K. Human CD36 deficiency is associated with elevation in low-density lipoprotein-cholesterol. Am J Med Genet 93: 299–304, 2000 [DOI] [PubMed] [Google Scholar]
- 70. Yao M, Roberts DD, Isenberg JS. Thrombospondin-1 inhibition of vascular smooth muscle cell responses occurs via modulation of both cAMP and cGMP. Pharmacol Res 63: 13–22, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, Brower RG, Barnes KC, Garcia JG. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 171: 361–370, 2005 [DOI] [PubMed] [Google Scholar]
- 72. Young GD, Murphy-Ullrich JE. The tryptophan-rich motifs of the thrombospondin type 1 repeats bind VLAL motifs in the latent transforming growth factor-beta complex. J Biol Chem 279: 47633–47642, 2004 [DOI] [PubMed] [Google Scholar]