Abstract
The discovery of the genetic code provided one of the basic foundations of modern molecular biology. Most organisms use the same genetic language, but there are also well-documented variations representing codon reassignments within specific groups of organisms (such as ciliates and yeast) or organelles (such as plastids and mitochondria). In addition, duality in codon function is known in the use of AUG in translation initiation and methionine insertion into internal protein positions as well as in the case of selenocysteine and pyrrolysine insertion (encoded by UGA and UAG, respectively) in competition with translation termination. Ambiguous meaning of CUG in coding for serine and leucine is also known. However, a recent study revealed that codons in any position within the open reading frame can serve a dual function and that a change in codon meaning can be achieved by availability of a specific type of RNA stem-loop structure in the 3’-untranslated region. Thus, duality of codon function is a more widely used feature of the genetic code than previously known, and this observation raises the possibility that additional recoding events and additional novel features have evolved in the genetic code.
Keywords: Selenocysteine, Pyrrolysine, Genetic Code, Codon Reassignment
Introduction
The demonstration that phenylalanine was incorporated into protein in response to poly-uracil by Nirenberg and Matthaei in 1961 (Nirenberg and Matthaei, 1961) marked the beginning of the deciphering of the genetic code. The ensuing three years represented one of the fiercest competitions known in the biological sciences that occurred between Nirenberg’s (Nirenberg et al, 1963) and Ochoa’s laboratories (Speyer et al, 1963) in using homo- and heteropolynucleotides in protein synthesis to elucidate the code. The end result of these initial studies was that codewords were most assuredly dictated by a trinucleotide and many of the 20 protein amino acids were assigned codons. The sequences of the codewords, however, could not be determined except in only a few cases that primarily involved homopolynucleotides. These studies embodied the first stage in elucidating the genetic code. The sequences of the codewords were determined in the second stage. These latter studies involved the binding of a labeled aminoacyl-tRNA to ribosomes in response to a specific trinucleoside diphosphate of known sequence (Nirenberg and Leder, 1964; Leder and Nirenberg, 1964). By 1966, the genetic code had been deciphered (Nirenberg et al, 1966). One other extremely important study during this period involved demonstrating that the genetic code was universal when Marshall, Caskey and Nirenberg found that aminoacyl-tRNAs from guinea pig liver, amphibian liver and Escherichia coli were recognized by the same codewords (Marshall et al, 1967). The deciphering of the genetic code and demonstration of its universality represent among the most important discoveries in biology that have had a most profound effect on the advancement of science.
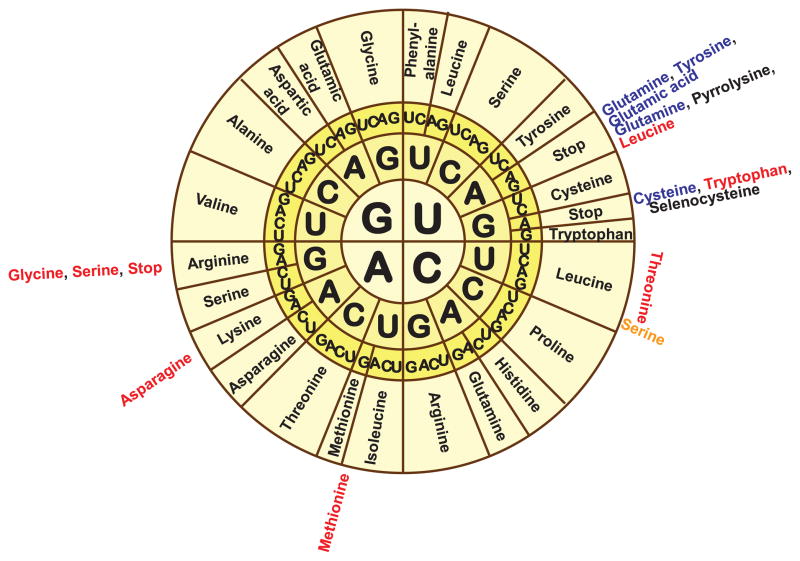
It was subsequently discovered, however, that variations in the genetic code have occurred in cellular organelles (see reviews by Osawa et al, 1992; Jukes and Osawa, 1993), and currently 23 variants of the code are known that are listed at the NCBI (Figure 1). It was also found that alternative genetic codes in nuclear and mitochondrial genomes have arisen independently more than 30 times (Knight et al, 2001; Swire et al, 2005; Soll and RajBhandary, 2006). Most often, these changes represent a simple codon reassignment, wherein a different amino acid is coded by the “universal” codon. Subsequent studies identified selenocysteine (for review, see Hatfield and Gladyshev, 2002) and pyrrolysine (Srinivasan et al, 2002) as the 21st and 22nd amino acids in the genetic code that are decoded by the termination codons, UGA and UAG, respectively. Also, it was found that the UGA termination codon in rabbit β-globin mRNA is involved in ribosomal hopping and may be suppressed by insertion of serine, tryptophan, arginine or cysteine (Chittum et al, 1998). Furthermore, a recent study reported that the same codon can have a dual function in inserting different amino acids in any position of the open reading frame and that this occurs, not only in the same organism, but even in the same gene (Turanov et al, 2009). This review discusses the best characterized cases of dual functions of codons.
Figure 1. The genetic code and its variations.
The figure is available in color in the online publication. The genetic code is shown in the circular form, with known alternative meanings indicated outside the circle. Differences with the standard genetic code are shown as follows: red for mitochondrial, blue for ciliate and Euplotid nuclear code, and orange for the ambiguous yeast nuclear code. Sec and Pyl are shown in black.
Methionine and translation initiation
During translation, AUG acts as an initiator of protein synthesis as well as a codon for methionine (Met) incorporation at internal protein positions in eukaryotes and an N-formylmethionine (fMet) in prokaryotes. However, the presence of AUG alone is not sufficient to start translation – initiation factors and a specific nearby sequence (or nucleotide context) are also required. Although alternative start codons in eukaryotes are rarely used (Gerashchenko et al, 2010), a recent study utilizing ribosome-profiling strategy to monitor translation in budding yeast (Ingolia et al, 2009) discovered the occurrence of pervasive initiation at specific, favorable, non-AUG sites. While further studies are needed in eukaryotes to define alternative initiation of translation, in prokaryotes the use of GUG and UUG as initiators in addition to AUG is known to be quite widespread; for example, an analysis of the E. coli genome revealed that 14% of the genes use GUG for translation initiation, and 3% utilize UUG (Blattner et al, 1997). A separate initiator tRNA is used in the initiation process, and although an alternative start codon may be used, it is still translated as Met (or fMet) by the initiator tRNAMet. During initiation, AUG in bacteria is translated as fMet; however, when the same codon is encountered in the open reading frame (ORF), Met is inserted by the internal reading tRNAMet (Sherman et al, 1985). The discriminatory mechanism ensuring the fidelity of initiation usually requires the starting AUG to occur in an optimum nucleotide context. In eukaryotes, this context is represented by the so-called "Kozak consensus sequence" (the optimal sequence is GCC(A/G)CCAUGG, but variations of it also provide the same function) (Kozak, 1991).
Thus, AUG serves as either a translation initiating codon or a Met internal codon. The dual functionality of this codon is somewhat artificial as there is no competition for amino acid insertion at either the initiator or internal reading AUG site. The initiator tRNA that binds prior to ribosome assembly only works for providing the N-terminal amino acid residue. Once translation has started, AUG acts in the same manner as any other codon and has a specific role in inserting Met in internal positions of proteins.
Selenocysteine and translation termination
Another widespread example of a codon with different functions is UGA, which is used for selenocysteine (Sec) incorporation. Sec is known as the 21st naturally occurring amino acid that is co-translationally incorporated into nascent polypeptide chains. Structurally, Sec is similar to cysteine and serine, but contains a selenium atom in place of sulfur in cysteine and oxygen in serine. Its presence in all domains of life and conservation of several components of the Sec biosynthesis and insertion machinery suggest an ancient origin of this amino acid.
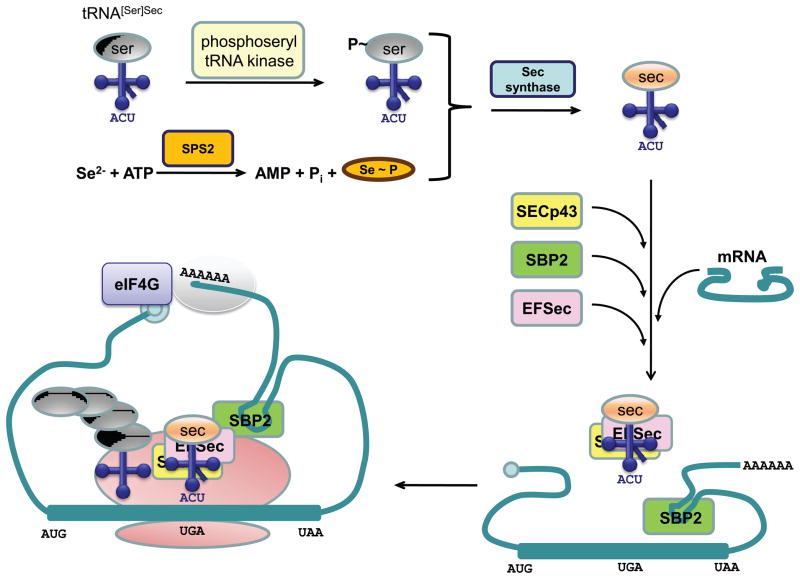
It was initially found that the gene coding for formate dehydrogenase from E. coli contains, within its open reading frame, a UGA stop codon at position 140 (Zinoni et al, 1986) that corresponded to Sec in the enzyme. In addition, a separate study (Chambers et al, 1986) found that the mouse glutathione peroxidase gene has an in-frame UGA codon, which was predicted to code for Sec at the active site of the corresponding protein. Unlike most other amino acids, Sec biosynthesis is a multistep process that occurs on its tRNA (Xu et al, 2007). First, tRNASec is aminoacylated with serine, which, after phosphorylation of the serine moiety on its tRNA in archaea and eukaryotes, is converted to the selenol group to yield Sec-tRNASec. The normal function of UGA is to terminate protein synthesis, and thus, depending on the conditions (see below), this codon may act as either a nonsense or a Sec codon. Sec incorporation at the UGA codon requires the presence of a stem-loop structure designated the Sec insertion sequence (SECIS) element and a set of Sec insertion machinery components, including SECIS binding protein 2 (SBP2), Sec specific elongation factor (EFSec), Sec tRNA, phosphoseryl-tRNA kinase (PSTK), SECp43 and Sec synthase as shown in Figure 2.
Figure 2. Selenocysteine incorporation machinery.
The figure is available in color in the online publication. The mechanism of Sec biosynthesis and incorporation into selenoproteins is shown and discussed in the text. Further discussion and identification of the various components involved in Sec biosynthesis are presented elsewhere (Xu et al, 2007).
SECIS elements have different structures in eukaryotes, bacteria and archaea (Berry et al, 1993; Rother et al, 2001; Kryukov and Gladyshev, 2004). In bacteria, the SECIS element is located immediately downstream of the UGA codon, while in eukaryotic organisms it is present in the 3'-untranslated region (3’-UTR). In archaea, SECIS elements may be located in both 5’-UTR and 3’-UTR. Several studies have been undertaken to measure the efficiency of Sec incorporation. When the fdhF SECIS element was inserted within a gst-lacZ fusion, read-through efficiency was 4–5% and overexpression of the Sec insertion machinery increased read-through to 7–10% (Suppmann et al, 1999). The efficiency of Sec incorporation into a mammalian luciferase reporter system in vitro was 5–8%, whereas read-through reached ~40% in vitro in selenoprotein P (Mehta et al, 2004). It should be noted, however, that these data were obtained from transfection experiments conducted in vitro, and thus may not reflect the true efficiency of in vivo Sec incorporation.
In prokaryotes, a single protein, elongation factor SelB, binds to selenocysteyl-tRNA and the SECIS element on the ribosome for insertion of Sec into protein (Forchhammer et al, 1989; Fourmy et al, 2002). In eukaryotes, the same process requires two proteins, the Sec-specific elongation factor, EFSec, that forms a complex with selenocysteyl-tRNA and the SECIS-binding protein, SBP2 (Copeland and Driscoll, 1999), that binds to the SECIS element in forming a complex with EFSec-selenocysteyl-tRNA for insertion of Sec into the nascent polypeptide chain. Ribosomal protein L30 (Chavatte et al, 2005) and SECp43 protein were also implicated in Sec insertion, but their precise roles are currently unknown. Recently, a crystal structure of human Sec tRNA was solved (Itoh et al, 2009). In contrast to the 7-bp acceptor stem and the 5-bp T stem in the canonicaltRNAs, human tRNASec has a 9-bp acceptor stem and a 4-bp T stem. The variable arm is comprised of a 6-bp stem and a 4-nt loop. The unusual structure of the Sec tRNA is consistent with the need for a novel elongation factor EFSec/SelB, dedicated to Sec incorporation.
In addition to the SECIS element, a Sec redefinition element (SRE) was recently described in selenoprotein N (Howard et al, 2005). SRE represents another conserved stem-loop structure that is located immediately downstream of the UGA codon. It was found that SRE increases the read-through rate two- to six-fold, depending on the presence of the SECIS element. Additional experiments supported the conclusion that SRE is a functional element regulating SelN expression, probably by promoting Sec insertion and inhibiting termination of protein synthesis.
In summary, the UGA codon in an mRNA usually indicates translation termination. However, in some cases, wherein this codon occurs in an ORF and in the presence of the Sec insertion machinery and the SECIS element in its proper position, UGA is used for incorporation of Sec. SECIS elements display a wide range of Sec incorporation activity, with difference in efficiency reaching several thousand fold in vivo and several hundred fold in vitro (Latréche et al, 2009). This process could be highly efficient, as indicated by the existence of selenoprotein P, which may contain up to 28 Sec residues depending on the organism (Lobanov et al, 2008). Since Sec utilization may provide catalytic advantages (due to a higher catalytic efficiency of Sec-containing enzymes compared to Cys-containing ones), but does not necessarily warrant expansion of the genetic code or complete reassignment of a codon (because the number of Sec-containing proteins in an organism is limited), Nature came up with an elegant solution, enabling a dual function of UGA codon.
Pyrrolysine and translation termination
Pyrrolysine (Pyl) represents the most recently discovered addition of an amino acid to the genetic code. It is known as the 22nd naturally occurring amino acid that is inserted into protein in response to UAG codon (Hao et al, 2002). By analogy to UGA and Sec, the possibility of recoding UAG to have two meanings in the genetic code was immediately apparent. Less than 1% of all sequenced genomes contain Pyl-containing proteins, whose known distribution is currently limited to several methanogenic archaea and bacteria. Pyl incorporation requires the presence of Pyl tRNA with a CUA anticodon (encoded by the pylT gene) and an aminoacyl-tRNA synthetase (encoded by the pylS gene) responsible for aminoacylating this tRNA with Pyl (Srinivasan et al, 2002).
The utilization of Pyl by both archaea and bacteria is of special interest, since these organisms belong to different domains of life. When the utilization of Pyl by Methanosarcinaceae was first discovered, it was assumed that Pyl represents "a late archaeal invention designed to meet the specific physiological needs of a particular archaeal lineage" and "another example of genetic code evolution after the era of the last common universal ancestor" (Ambrogelly et al, 2007). But a later finding of Pyl utilization by Desulfitobacterium suggested that PylRS was already present in the last universal common ancestor, and that this system persisted only in organisms that utilized methylamines as energy sources (Nozawa et al, 2009). As a rule, a narrow phylogenetic distribution is an indicator of recent changes. The situation with Pyl is not so obvious. Phylogenetic analysis of Pyl users (Fournier, 2009) showed that pyrrolysyl-tRNA synthetase diverged from other synthetases before the split between bacteria and archaea. However, a very narrow distribution of current Pyl users poses questions regarding evolutionary pressure to either maintain or eliminate the Pyl system.
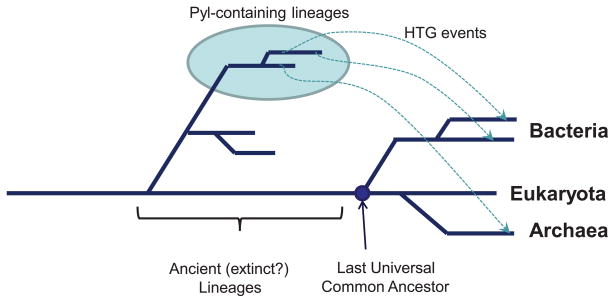
The simplest explanation of unusual Pyl distribution would be a vertical inheritance with many lineage-specific independent gene losses. It assumes that the Pyl usage machinery (together with the genes utilizing Pyl) was present in the last universal common ancestor of known life (LUCA). During evolution, these genes were then lost in all lineages except the direct ancestors of current users. The obvious problem with this hypothesis is that it requires a huge number of selective losses to explain the current distribution. Moreover, known users utilize Pyl primarily in methylamine methyltransferase enzymes (or in other proteins that exist in organisms containing methylamine methyltransferases), indicating that either Pyl usage in other systems was abandoned, or Pyl evolved for this particular set of enzymes. The former requires independent, parallel losses of entire gene families resulting in exactly the same sets of genes, and the latter would mean that Pyl utilization provides catalytic advantage, thus putting a sufficient selective pressure for the maintenance of Pyl machinery. Thus, it is likely that the system for Pyl encoding has evolved via a transfer from an ancient, currently unknown, deeply branching lineage (Fournier, 2009). Moreover, the branch lengths between the bacterial homologs of the PylS gene family, as well as between the bacterial and the archaeal homologs, indicate that if horizontal gene transfer occurred, the archaeal and bacterial genes were transferred from different donor organisms (Figure 3). It was initially assumed that Pyl evolved to be an active-site constituent. However, it was recently found that Pyl replacement with Trp in Methanosarcina acetivorans Thg1 yielded a fully active enzyme, suggesting that Pyl in this protein is a dispensable residue that appears to confer no selective advantage (Heinemann et al, 2009).
Figure 3. Horizontal gene transfer of the pyrrolysine trait.
The figure is available in color in the online publication. The model for a possible horizontal gene transfer of the Pyl trait to archaea and bacteria is shown. Ancient (probably extinct) Pyl users are indicated by a light blue circle, and the last universal common ancestor (LUCA) of eukaryotes, bacteria and archaea is shown in dark blue. Dotted lines show a possible Pyl trait transfer.
There are some differences in bacterial and archaeal Pyl systems. For example, PylS in bacteria is encoded by two adjacent genes, while in archaea it is present as a single gene product. Of interest is the fact that in methanogenic archaea there is no proof of specific UAG usage as a stop codon, while in D. hafniense the UAG codon is used as a specific translation termination signal in many proteins (Zhang et al, 2005). Moreover, because the Pyl incorporation machinery is clustered as a “cassette”, it is relatively easy to transfer the Pyl trait to other genomes. It has been demonstrated that the presence of the PylRS-tRNA(Pyl) system allows site-specific incorporation of the range of amino acids (such as different L-lysine derivatives) in mammalian cells (Mukai et al, 2008) and E. coli (Namy et al, 2007).
Thus, there are several important differences between the Sec and Pyl protein insertion machineries. First, Sec is synthesized on Sec tRNA, which is first aminoacylated with Ser, while Pyl is synthesized prior to its aminoacylation to Pyl tRNA. Sec incorporation requires the Sec-specific elongation factor, EFSec, while Pyl is served by the canonical elongation factor EF-Tu. Also, in order for Sec to be included into protein, the presence of a cis-SECIS element in mRNA is required; for Pyl, no similar element has been found. Trans-acting elements needed for Sec insertion include EFSec/SBP2 (or SelB in prokaryotes), while trans-elements for Pyl are currently unknown. Known enzymes for Sec biosynthesis include SelA and SelD, and for Pyl biosynthesis PylS, PylB, PylC and PylD (Srinivasan et al, 2002). Each amino acid is served by non-canonical tRNAs (i.e., Sec tRNA and Pyl tRNA) and is inserted into a polypeptide in response to UGA (for Sec) or UAG (for Pyl) codons.
It is interesting to note that two independent studies, a search for tRNA genes for novel amino acids (Lobanov et al, 2006) and a protein-level search for stop-codon-encoded amino acids (Fujita et al, 2007), were successful in identifying Sec and Pyl tRNAs, and Sec-containing and Pyl-containing proteins; however, no new amino acid candidates were found by either approach, suggesting that unknown amino acids encoded by stop codons either do not exist, or their phylogenetic distribution is very limited.
Ambiguous serine and leucine insertion
In 1989, it was found that asporogenic yeast, Candida cylindracea, utilizes CUG (a universal leucine codon) for serine insertion (Kawaguchi et al, 1989). A series of in vitro experiments demonstrated that in six out of fourteen species examined, CUG was used as a serine codon, while in the remaining eight species it coded for leucine. A specific tRNA responsible for the translation of CUG as serine was detected in all six species, in which CUG was translated as serine. When this tRNA was analyzed in more detail, it was discovered that it evolved from a serine tRNA via insertion of a single adenosine in the second position of the anticodon, changing it to CAG (Suzuki et al, 1994). Further analysis showed that CUG was decoded as both serine and leucine in vivo and that C. albicans tolerates up to 28.1% of leucine mis-incorporation at CUG positions (Gomes et al, 2007). Unlike Sec or Pyl insertion, the choice of leucine or serine is completely random. In other words, each protein in the cell is represented by a unique combination of proteins containing leucine or serine at positions encoded by CUG codons.
Because of the difference in properties of these two amino acids (serine is polar and leucine is hydrophobic), random selection has a potential to drastically affect properties of proteins. An important biological outcome is that this mechanism provides an extensive and unanticipated phenotypic variability (Yokogawa et al, 1992). The possibility to insert both amino acids indicates dual functionality of this CUG codon. However, the fact that the choice of an amino acid to be incorporated into a polypeptide chain is made at random does not qualify this system as being a true dual function codon and reduces the usefulness of this model for practical applications. Still, the case of Candida, where one amino acid could be partially replaced by another, demonstrates that the genetic code is not rigid.
Stop codons and read-through events
While translation is usually terminated when ribosomes encounters an in-frame stop codon, it has been demonstrated that in many viruses and some other organisms only a portion of ribosomes terminate translation, while other ribosomes incorporate an amino acid in place of stop codon and continue to synthesize a read-through protein. The efficiency of this process depends greatly on nucleotide sequence context, and can be significantly changed in response to both cis- and trans-acting factors, such as nearby pseudoknots (Wills et al, 1991; Feng et al, 1992) and distant 3’ sequences (Brown et al, 1996). Other factors may affect the probability of read-through to an extent that competition for UAA, UAG and UGA between a release factor and near-cognate tRNAs would favor the latter.
Studies of tobacco mosaic virus (TMV), one of the most intensively investigated read-through cases, have demonstrated that 25 nucleotides around the targeted stop codon are sufficient for a successful read-through event to occur (Skuzeski et al, 1990). In addition, the six consensus nucleotides conforming to CARYYA pattern and located immediately downstream of the UAG codon were found to be essential for read-through (Skuzeski et al, 1991; Stahl et al. 1995). In vivo efficiency of read-through in TMV is approximately 10%, and the ratio of full-length and non-read-through proteins was shown to be crucial for the viral life-cycle (Ishikawa et al, 1991).
The examples of read-through proteins are not limited to viruses; for example, 149 read-through candidates were found in the Drosophila genome (Lin et al, 2007). The study of several developmental mutants, such as headcase (Steneberg et al, 1998), showed that two types of proteins are translated from a single mRNA: a shorter form resulting from termination at the first stop codon (UAA in this case), and a longer read-through form. The ratio of these forms depends on several factors, such as tissue and developmental stage, and the rate of read-through can reach 20%. Thus, in the presence of read-through factors, UAA, UAG and UGA codons can act as stop signals or be used to incorporate amino acids and therefore these codons play a dual function.
Targeted cysteine and selenocysteine insertion
Although UGA and UAG stop codons serve dual functions, they support the insertion of single amino acids (i.e., Sec and Pyl), in competition with termination. There are, however, many more variations in the genetic code (Fig.1). For example, the function of UGA as a stop codon was changed in mitochondria of vertebrates to code for tryptophan, whereas this codeword dictates Cys in the Euplotid nuclear code. The latter case posed an interesting question: if UGA is a Cys codon in Euplotes, and if, like many eukaryotes, this organism contains selenoproteins, then which codon is assigned to Sec? An examination of selenoprotein genes in E. crassus revealed the presence of 8 selenoprotein genes in which UGA codes for Sec (Turanov et al, 2009). An analysis of the Euplotes genome also revealed three tRNA genes with the UCA anticodon. Phylogenetic analysis of these tRNA genes showed that one of them was a mitochondrial Trp tRNA, another evolved from Cys tRNA, and the third was a typical Sec tRNA containing a long variable arm, a characteristic feature of all Sec tRNAs. Thus, in E. crassus, the same codon, UGA, codes for three amino acids, with two of them, Sec and Cys, represented in the nuclear code.
Interestingly, four selenoprotein genes containing multiple UGA codons were found, and further experimental analysis revealed that Sec was only incorporated in the active site of thioredoxin reductase eTR1, while the remaining six slots in the protein were filled with Cys. This situation differs dramatically from the yeast alternative genetic code, where serine and leucine are randomly inserted. There is also a difference with Sec incorporation in mammals, where UGA codes for Sec when SECIS element is present in the mRNA but acts as termination codon when the SECIS is missing. It is clear that there must be a mechanism that prevents Sec insertion at some positions in the mRNA even when SECIS is present in the 3’-UTR. The authors hypothesized that the eTR1 mRNA undergoes conformational changes to make the SECIS element available for Sec insertion only at the Sec position. As the ribosome moves along the mRNA during translation, the mRNA structure changes to form the SECIS element or to expose this element for interaction with the translational machinery. Overall, this study found that the genetic code can be extended to recode a specific subset of codons in an organism for insertion of a different amino acid at internal decoding positions of proteins, and this occurs even within the same gene.
Conclusion
Shortly after the genetic code was deciphered, the concept of a "frozen accident" was proposed by Crick to explain the origin of the code (Crick, 1968). The frozen accident hypothesis assumed that all present day organisms use the same universal and invariant genetic code. The discoveries of alternative genetic languages used in cellular organelles of various organisms and in certain taxonomic groups led to a revision of this theory, but it still indicated that the core of the code was established prior to the divergence of the three domains of life (Knight et al, 2001).
Most often, variations in the genome code represent a codon reassignment, wherein a codon disappears from the coding sequence and then reappears in a new role. Frequently, this occurs when stop codons are assigned a new function, such as coding for tryptophan in mitochondria or for glutamine in ciliates. However, two cases stand apart: insertion of Sec and Pyl at UGA and UAG codons, respectively, in competition with translational termination. Their insertion into nascent polypeptides in most cases is influenced by nucleotide context and potentially some other factors, wherein the same codon could serve as the codon for amino acid insertion in some genes, and signal termination in others.
However, a recent study reported the dual function of a codon at internal positions of proteins in E. crassus. In that study, instead of complete codon reassignment or competition with translation termination, the same codeword encoded two amino acids at internal positions of proteins. UGA was found to code for both Cys and Sec, and the dual function of UGA may occur even within the same gene (Turanov et al, 2009). Current data indicate that incorporation of these amino acids is specific, i.e., Sec can be incorporated only at certain UGA positions, and Cys only at other UGA positions. This finding provided novel insights in the evolution of the genetic code. It is now possible to identify coding events where a subset of codons in an organism is recoded to insert a different amino acid. With the availability of complete sequences of over 1000 genomes, it would be of interest to scrutinize these sequences for multiple coding functions of all 64 codons in the genetic code.
Acknowledgments
This work was supported by National Institutes of Health grants to VNG and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to DLH.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Söll D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci USA. 2007;104:3141–6. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–22. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, et al. The Complete Genome Sequence of Escherichia coli K-12. Science. 1997;277:1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Brown CM, Dinesh-Kumar SP, Miller WA. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J Virol. 1996;70:5884–5892. doi: 10.1128/jvi.70.9.5884-5892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the 'termination' codon, TGA. EMBO J. 1986;5:1221–7. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit β-Globin Is Extended Beyond Its UGA Stop Codon by Multiple Suppressions and Translational Reading Gaps. Biochemistry. 1998;37:10866–70. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–54. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. General nature of the genetic code for proteins. Nature. 1961;192:1227–32. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- Crick FH. On the origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Feng YX, Yuan H, Rein A, Levin JG. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- Fourmy D, Guittet E, Yoshizawa S. Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J Mol Biol. 2002;324:137–150. doi: 10.1016/s0022-2836(02)01030-6. [DOI] [PubMed] [Google Scholar]
- Fournier G. Horizontal gene transfer and the evolution of methanogenic pathways. Methods Mol Biol. 2009;532:163–79. doi: 10.1007/978-1-60327-853-9_9. [DOI] [PubMed] [Google Scholar]
- Fujita M, Mihara H, Goto S, Esaki N, Kanehisa M. Mining prokaryotic genomes for unknown amino acids: a stop-codon-based approach. BMC Bioinformatics. 2007;8:225. doi: 10.1186/1471-2105-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Su D, Gladyshev VN. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J Biol Chem. 2010 doi: 10.1074/jbc.M109.070532. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AC, Miranda I, Silva RM, Moura GR, Thomas B, Akoulitchev A, Santos MA. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007;8:R206. doi: 10.1186/gb-2007-8-10-r206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A New UAG-Encoded Residue in the Structure of a Methanogen Methyltransferase. Science. 2002;296:1462–66. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, O'Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Söll D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci USA. 2009;106:21103–8. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005;24:1596–1607. doi: 10.1038/sj.emboj.7600642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Meshi T, Ohno T, Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Chiba S, Sekine S, Yokoyama S. Crystal structure of human selenocysteine tRNA. Nucl Acids Res. 2009;37:6259–68. doi: 10.1093/nar/gkp648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Osawa S. Evolutionary changes in the genetic code. Comp Biochem Physiol B. 1993;106:489–94. doi: 10.1016/0305-0491(93)90122-l. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Honda H, Taniguchi-Morimura J, Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989;341:164–6. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Knight RD, Landweber LF, Yarus M. How mitochondria redefine the code. J Mol Evol. 2001;53:299–313. doi: 10.1007/s002390010220. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–70. [PubMed] [Google Scholar]
- Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–43. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrèche L, Jean-Jean O, Driscoll DM, Chavatte L. Novel structural determinants in human SECIS elements modulate the translational recoding of UGA as selenocysteine. Nucleic Acids Res. 2009;37:5868–80. doi: 10.1093/nar/gkp635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder ZP, Nirenberg M. RNA codewords and protein synthesis. II. Nucleotide sequence of a valine RNA codeword. Proc Natl Acad Sci USA. 1964;52:420–7. doi: 10.1073/pnas.52.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder ZP, Nirenberg MW. RNA Codewords and Protein Synthesis, III. On the Nucleotide Sequence of a Cysteine and a Leucine RNA Codeword. Biochemistry. 1964;52:1521–29. doi: 10.1073/pnas.52.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Carlson JW, Crosby MA, Mathews BB, Yu C, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Kryukov GV, Hatfield DL, Gladyshev VN. Is there a twenty third amino acid in the genetic code? Trends Genet. 2006;22:357–360. doi: 10.1016/j.tig.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RE, Caskey CT, Nirenberg M. Fine Structure of RNA Codewords Recognized by Bacterial, Amphibian, and Mammalian Transfer RNA. Science. 1967;155:820–826. doi: 10.1126/science.155.3764.820. [DOI] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–9. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–22. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–68. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Namy O, Zhou Y, Gundllapalli S, Polycarpo CR, Denise A, Rousset JP, Söll D, Ambrogelly A. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581:5282–8. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei HJ. The Dependence Of Cell- Free Protein Synthesis In E. coli Upon Naturally Occurring Or Synthetic Polyribonucleotides. Proc Natl Acad Sci USA. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MW, Jones OW, Leder P, Clark BFC, Sly WC, Pestka S. On the Coding of Genetic Information. Cold Spring Harb Symp Quant Biol. 1963;28:549–57. [Google Scholar]
- Nirenberg MW, Leder P. RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes. Science. 1964;145:1399–407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- Nirenberg M, Caskey T, Marshall R, Brimacombe R, Kellogg D, Doctor B, Hatfield D, Levin J, Rottman F, Pestka S, Wilcox M, Anderson F. The RNA code and protein synthesis. Cold Spring Harb Symp Quant Biol. 1966;31:11–24. doi: 10.1101/sqb.1966.031.01.008. [DOI] [PubMed] [Google Scholar]
- Nozawa K, O’Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Soll D, Nureki O. Pyrrolysyl-tRNA synthetase:tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–67. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–64. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M, Resch A, Wilting R, Böck A. Selenoprotein synthesis in archaea. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- Sherman F, Stewart JW, Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985;3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF. Analysis of leaky viral translation termination codons in vivo by transient expression of improved β-glucuronidase vectors. Plant Mol Biol. 1990;15:65–79. doi: 10.1007/BF00017725. [DOI] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- Steneberg P, Englund C, Kronhamn J, Weaver TA, Samakovlis C. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the Drosophila trachea. Genes Dev. 1998;12:956–967. doi: 10.1101/gad.12.7.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D, RajBhandary UL. The genetic code — thawing the ‘frozen accident’. J Biosci. 2006;31:459–63. doi: 10.1007/BF02705185. [DOI] [PubMed] [Google Scholar]
- Speyer FJ, Lengyel P, Basilio C, Wahba AJ, Gardner RS, Ochoa S. Synthetic polynucleotides and the amino acid code. Cold Spring Harb Symp Quant Biol. 1963;28:559–67. [Google Scholar]
- Srinivasan G, James CM, Krzycki JA. Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-Decoding Specialized tRNA. Science. 2002;296:1459–62. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- Suppmann S, Persson BC, Böck A. Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 1999;18:2284–93. doi: 10.1093/emboj/18.8.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda T, Yokogawa T, Nishikawa K, Watanabe K. Characterization of serine and leucine tRNAs in an asporogenic yeast Candida cylindracea and evolutionary implications of genes for tRNA(Ser)CAG responsible for translation of a non-universal genetic code. Nucleic Acids Res. 1994;22:115–23. doi: 10.1093/nar/22.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueda T, Watanabe K. The 'polysemous' codon--a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–34. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swire J, Judson OP, Burt A. Mitochondrial genetic codes evolve to match amino acid requirements of proteins. J Mol Evol. 2005;60:128–39. doi: 10.1007/s00239-004-0077-9. [DOI] [PubMed] [Google Scholar]
- Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, Klobutcher LA, Hatfield DL, Gladyshev VN. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323:259–61. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational readthrough of the Moloney muribe leukemia virus gag stop codon. Proc Natl Acad Sci USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T, Suzuki T, Ueda T, Mori M, Ohama T, Kuchino Y, Yoshinari S, Motoki I, Nishikawa K, Osawa S. Serine tRNA complementary to the nonuniversal serine codon CUG in Candida cylindracea: evolutionary implications. Proc Natl Acad Sci USA. 1992;89:7408–11. doi: 10.1073/pnas.89.16.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280:20740–51. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- Zinoni F, Birkmann A, Stadtman TC, Böck A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4650–54. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]