Abstract
Identification of pathways of drug metabolism provides critical information regarding efficacy and safety of these compounds. Particularly challenging cases involve stereospecific processes. We found that broad classes of compounds containing methylsulfinyl groups are reduced to methylsulfides specifically by methionine sulfoxide reductase A, which acts on the S-stereomers of methionine sulfoxides, whereas the R-stereomers of these compounds could not be efficiently reduced by any methionine sulfoxide reductase in mammals. The findings of efficient reduction of S-methylsulfinyls and deficiency in the reduction of R-methylsulfinyls by methionine sulfoxide reductases suggest strategies for improved efficacy and decreased toxicity of drugs and natural compounds containing methylsulfinyls through targeted use of their enantiomers.
Methionine (Met) contains a methylsulfide group in its side chain, which can be easily oxidized by reactive oxygen species (ROS). Met oxidation can disrupt structure and function of proteins and contribute to the development of diseases associated with accumulation of oxidatively damaged proteins (1). To repair oxidized Met, organisms utilize methionine sulfoxide reductases (Msrs), which reduce methionine sulfoxide (Met-O) back to Met (2, 3). Msrs are regenerated by thiol oxidoreductases, such as thioredoxin (Trx) and glutaredoxin. Trx, which is powered by NADPH-dependent thioredoxin reductase (TR), is the most common and the best characterized in vivo reductant, whereas dithiothreitol (DTT) is often used for the regeneration of Msrs in in vitro assays. These reductants were also used in our current study. Met-O is composed of two diastereomers, methionine-S-sulfoxide (Met-S-SO) and methionine-R-sulfoxide (Met-R-SO), which are subject to stereospecific reduction by MsrA and MsrB, respectively (2, 4). An additional Msr type, free methionine-R-sulfoxide reductase (fRMsr), is specific for free Met-R-SO, although this protein is absent in animals (5, 6). Mammals have four Msrs: MsrA, MsrB1, MsrB2, MsrB3 (7), which account for total Msr activity in these organisms.
Like Met, many drugs and natural compounds contain methylsulfide groups, which are oxidized to methylsulfinyls upon exposure to hydrogen peroxide, hypochloric acid, and enzymes such as cytochrome p450 (Cyp450) and flavin monooxygenase (FMO) (8-10). These methylsulfinyl-containing xenobiotics are also composed of two stereoisomers, represented by R- or S-sulfoxides (10). For example, Cyp450, including its many isoforms, is responsible for oxidation of carbon or sulfur atoms in drugs and differences in these enzyme activities in ethnic populations is known to result in variations in drug responses. Numerous enzymes metabolizing drugs have been identified and much research on these proteins focused on the investigation of links between drug metabolism and drug efficacy. Our findings in the current study point to the existence of a stereospecific reductase system for any methylsulfinyl-containing compound in mammals.
We recently reported that mammals reduce free Met-S-SO, but are unable to reduce free Met-R-SO (6). This is probably because MsrA reduces both protein-based and free Met-S-SO, whereas MsrBs can only reduce protein-based Met-R-SO. Based on this finding, we hypothesized that mammals may be generally deficient in the reduction of R-stereoisomers of methylsulfinyls, but could reduce S-stereoisomers with MsrA. An indirect support for this idea was provided by the observation that the methylsulfinyl group in an anticancer compound sulindac sulfoxide could be reduced by E. coli MsrA, which acted only on the S-enantiomer of this compound (11). Our new findings of stereospecific reduction of methylsulfinyls should help improve efficacy of drugs containing this functional group.
Results and Discussion
To investigate the hypothesis that mammalian Msrs may have differences in the reduction of methylsulfinyl-containing drugs or natural chemical compounds, we used mouse MsrA (mMsrA), mouse MsrB2 (mMsrB2), and yeast fRMsr (yfRMsr) (Fig. S1), which are representative of the three Msr families, for in vitro reduction of a diverse set of methylsulfinyl-containing compounds, including an anti-schizophrenia drug mesoridazine, a cardiotonic drug sulmazole, an anti-liver fluke drug triclabendazole sulfoxide (TCBZSX), a common solvent and vehicle for application of pharmaceuticals dimethylsulfoxide (DMSO), and a natural anti-cancer compound sulforaphane (Fig. 1A). Mesoridazine, sulmazole, and TCBZSX were used as an approximately 1:1 mixture of R- and S-enantiomers. As reductants for the regeneration of Msrs, either DTT or TR/Trx/NADPH were used. Although TR/Trx/NADPH is a physiological recycling system used in vivo, DTT works efficiently as a reductant in in vitro assays (12).
Figure 1. Msrs reduce S-stereoisomer but not R-stereoisomer forms of methylsulfinyl-containing drugs and natural compounds.
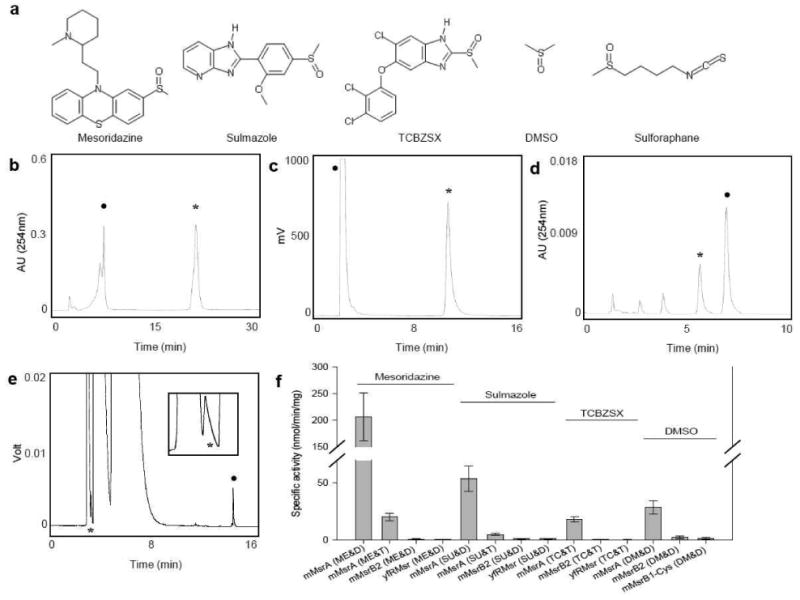
(A) Structures of representative methylsulfinyl-containing compounds. (B) HPLC analysis of mesoridazine reduction by mMsrA using DTT as a reductant. (C) HPLC analysis of sulmazole reduction by mMsrA with DTT as a reductant. Reduced and oxidized forms were measured by following fluorescence with the excitation at 330 nm and emission at 370 nm. (D) HPLC analysis of TCBZSX reduction by mMsrA with TrxR/Trx/NADPH. (E) GC analysis of DMSO reduction by mMsrA with DTT. The peak of dimethylsulfide, the reduced form of DMSO, is enlarged in the inset. In panels B-E, peaks of original substrates are marked with closed circles, and peaks of reduced forms with stars. (F) Specific activities of mMsrA, mMsrB2, and yfRMsr for reduction of mesoridazine, sulmazole, TCBZSX, and DMSO using either DTT or TrxR/Trx/NADPH as reductants. All measurements were repeated 3 times independently. Abbreviations used are: mesoridazine (ME), sulmazole (SU), TCBZSX (TC), DMSO (DM), DTT (D), TR/Trx/NADPH (T).
Reduction of mesoridazine to thioridazine was tested by an HPLC analysis, which revealed that only mMsrA could reduce this compound (Fig. 1B), whereas mMsrB2 and yfRMsr were inactive (Fig. S2). The specific activity of mMsrA for the reduction of mesoridazine was 206.8 ± 45.2 nmol/min/mg in the DTT-dependent assay and 20.2 ± 3.2 nmol/min/mg in the presence of TR/Trx/NADPH (Fig. 1F). Sulmazole was also selectively reduced by mMsrA, but we observed no activity with either mMsrB2 or yfRMsr (Fig. 1C and Fig. S3). The MsrA specific activity was 53.8 ± 4.7 nmol/min/mg in the DTT-dependent reaction and 7.1 ± 1.7 nmol/min/mg with TR/Trx/NADPH (Fig. 1F). TCBZSX underwent a partial non-enzymatic reduction in the presence of DTT, but the MsrA-dependent activity was well above the background and could be reliably assayed (in the TR/Trx/NADPH-dependent assay, it was 18 ± 2.5 nmol/min/mg) (Fig. 1D, 1F and Fig. S4), whereas other Msrs were inactive. Reduction of these two drugs by mMsrA was further confirmed by an ion-spray mass spectrometry analysis (Fig. S7, S8). Since these compounds were used in a mixed R- and S-sulfoxide form (approximately 1:1 ratio) and MsrA was specific for the S-sulfoxide form, our data suggested that only the S-sulfoxide forms of three drugs could be reduced.
We further examined DMSO and R,S-sulforaphane as substrates. A gas chromatography (GC) analysis revealed DMSO reduction by MsrA (28.6 ± 5.73 nmol/min/mg with DTT as a reductant), but not by other Msrs (Fig. 1E, F and Fig. S5). Since DMSO does not have a chiral group at the sulfur atom, this compound could be quantitatively reduced by MsrA. In the case of sulforaphane, we preincubated the substrate with excess DTT to conjugate its isothiocyanate group prior to the Msr reaction and followed generation of erucin-DTT, which is the reduced form of sulforaphane-DTT, by an HPLC analysis (Fig. S6). The R- and S-sulforaphane-DTT forms were partially converted to erucin-DTT in a non-enzymatic reaction; however, addition of MsrA to the reaction mixture increased yield of the product, whereas other Msrs had no effect. Similar results were obtained when mMsrB2 reacted with R-sulforaphane and mMsrA with S-sulforaphane (Fig. S6). Thus, we found overwhelming evidence that only the S-sulfoxide forms of methylsulfinyl-compounds are stereospecifically reduced by MsrA, while other Msrs (i.e., MsrB and yfRMsr) cannot reduce any enantiomeric forms of the drugs.
To extend these in vitro findings to the reduction of methylsulfinyls in a more biological setting, we used mouse liver to investigate selectivity of reduction of methylsulfinyl-containing drugs by Msrs. Liver lysates from wild type, MsrA knockout (KO), and MsrB1 KO mice (Fig. 2A, B) were used to examine DTT-dependent reduction of mesoridazine and sulmazole. Lysates from wild type and MsrB1 KO mice were equally effective in reducing mesoridazine and sulmazole, whereas the ability of MsrA KO liver lysates to reduce these compounds was severely compromized (Fig. 2C, D) (p<0.05).
Figure 2. Roles of Msrs in the reduction of methylsulfinyl-containing drugs by liver lysates.
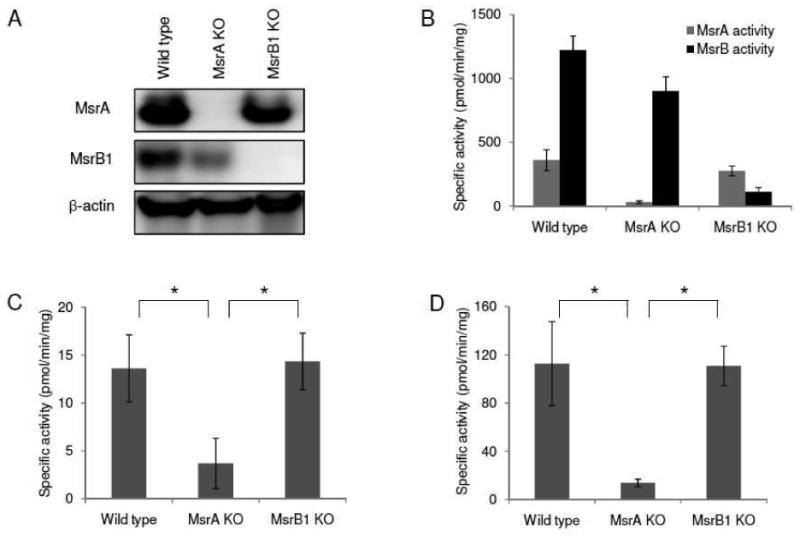
(A) Western blot analysis of MsrA, MsrB1, and β-actin in liver lysates from wild type, MsrA KO, and MsrB1 KO mice. (B) Specific MsrA (gray bars) and MsrB (black bars) activities in liver lysates of wild type, MsrA KO, and MsrB1 KO mice. Dabsylated Met-S-SO and dabsylated Met-R-SO were used as substrates for measurement of MsrA and MsrB activities, respectively. (C) Specific activity for the reduction of sulmazole in liver lysates from wild type, MsrA KO, and MsrB1 KO mice. (D) Specific activity for the reduction of mesoridazine in liver lysates from wild type, MsrA KO and MsrB1 KO mice. All measurements were repeated 3 times independently. Data was analyzed with a Student's t-test (*: p<0.05).
Cyp450 and FMO are microsomal proteins capable of oxidizing sulfur atoms in diverse compounds; thus, reoxidation of methylsulfides to methylsulfinyls by these and other enzymes might influence the reduction rate of methylsulfinyls by Msrs. To examine this possibility, we subjected liver cytosolic and microsomal fractions from wild type, MsrA KO, and MsrB1 KO mice to the reduction and oxidation assays using sulmazole, mesoridazine, and thioridazine as substrates. The cytosolic fraction lacking MsrA showed a significant decrease in the reduction of mesoridazine (Fig. S9A,B, S10B) and sulmazole (Fig. S10A), whereas this activity was little affected by MsrB1 KO (Fig. S9C, S10A,B). Using microsomal fractions, we found that generation of mesoridazine was proportional to protein concentration (Fig. S10C), suggesting that microsomes can oxidize thioridazine; however, the specific oxidase activity was 100 fold lower compared to the specific activity for the reduction of mesoridazine by the cytosol. In addition, no difference in the oxidation activity was detected between wild type, MsrA KO, and MsrB1 KO mice (Fig. S10D). Thus, although microsomes do oxidize thioridazine to mesoridazine, this activity has little influence on the overall reduction of mesoridazine by mouse liver.
Next, we investigated if the stereospecific reduction of methylsulfinyls influences properties of drugs and natural compounds in vivo. To test this possibility, HEK 293 cells, which were transfected with mMsrA or mMsrB2 constructs, were treated with various concentrations of mesoridazine. mMsrA-expressing cells showed decreased viability in the presence of mesoridazine compared with control and mMsrB2-expressing cells (25 μM mesoridazine, p<0.05; 50 μM, p<0.05; 100 μM, p<0.01) (Fig. 3A), suggesting that reduction of this compound by mMsrA enhanced toxicity of this drug. Consistent with this idea, the reduction product of mesoridazine, thioridazine, was found to be more toxic than mesoridazine (Fig. 3B).
Figure 3. Selective reduction of mesoridazine affects cell viability.
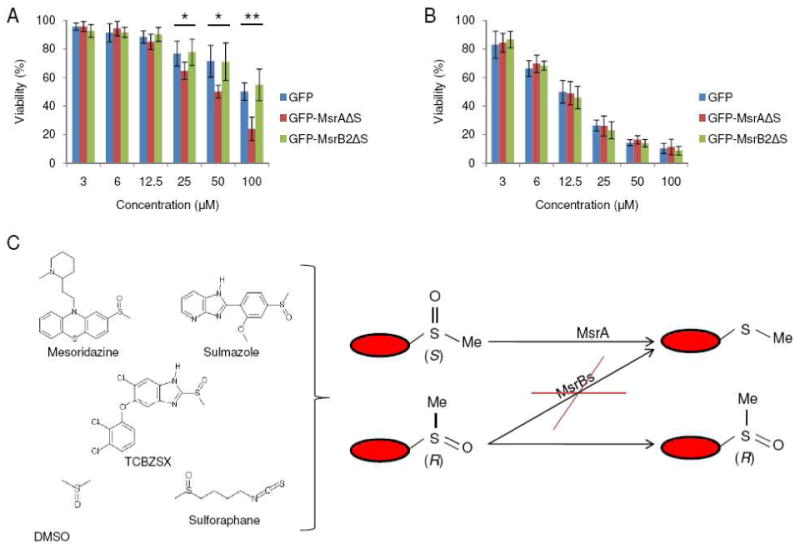
(A) Cell viability analysis of HEK 293 cells expressing GFP (control), GFP-fused mMsrAΔS, or GFP-fused mMsrB2ΔS following treatment of cells with mesoridazine (3, 6, 12.5, 25, 50, 100 μM) for 24 h. Constructs were designed to express proteins in the cytosol (where indicated by removing the signal peptide marked as ΔS). (B) Cell viability analysis of HEK 293 cells transfected with GFP (control), GFP-fused mMsrAΔS, or GFP-fused mMsrB2ΔS following treatment with thioridazine (3, 6, 12.5, 25, 50, 100 μM) for 24 h. Constructs were designed to express proteins in the cytosol (ΔS indicates that the signal peptide was removed). All measurements were repeated 6 times independently, and the data analyzed with a Student's t-test (**: p<0.01 and *: p<0.05). (C) An overall scheme of stereospecific reduction of methylsulfinyl-containing drugs.
Thus, the idea that the two Msr families in mammals, MsrA and MsrB, have completely different catalytic preferences for the reduction of methylsulfinyl-containing compounds is fully supported by our study. Previous research provided some clues. For example, E. coli MsrA could efficiently reduce free Met-S-SO, whereas E. coli MsrB possessed a 1000-fold lower activity with free Met-R-SO (13, 14). Also, we previously reported that human SK-Hep1 cells could not use free Met-R-SO as a source of Met for its growth, because this compound was not reduced by MsrBs (6). In addition, it was found that mouse plasma had only a single type of free Met sulfoxide, Met-R-SO, whereas Met-S-SO could only be detected in the plasma of MsrA KO mice (it was observed together with Met-R-SO) (6). One study examined the reduction of an anti-cancer compound sulindac by E. coli Msrs and partially purified cellular fractions (11). In this experiment, purified MsrA and a membranous fraction reduced sulindac and it was suggested that MsrA was specific for the reduction of the S-sulfoxide form, while a putative membrane-bound Msr, mem-R,S-Msr, reduced the R-sulfoxide form. Mammals have four Msrs: MsrA, MsrB1, MsrB2, and MsrB3, which account for the reduction of methionine sulfoxides in these organisms. Three MsrBs share a similar function by acting in different cellular compartments, whereas a single MsrA is targeted to cellular compartments by virtue of its signal peptide and alternative first exon splicing (15).
In support of our hypothesis that mammalian MsrBs cannot reduce R-methylsulfinyls, we found that only mMsrA reduced methylsulfinyls in a broad set of compounds, whereas neither mMsrB nor yfRMsr were active. This selective reduction was further confirmed by examining reduction of DMSO and sulforaphane. DMSO does not have a chiral group, but only MsrA showed activity towards this compound. Also, only S-sulforaphane was reduced by mMsrA, though a non-enzymatic reduction in the presence of DTT was detected with both stereomers. Moreover, liver lysates from MsrA KO mice could only reduce mesoridazine and sulmazole with low efficiency, whereas liver lysates from MsrB1 KO mice were as efficient as those from wild type mice. Accordingly, stereospecificity for S-sulfoxides and a broad substrate specificity of MsrA, i.e., its reduction of any S-methylsulfinyl, should lead to the stereospecific reduction of mixtures of S-sulfoxide and R-sulfoxide forms of drugs (Figure 3C). This specific MsrA-based reduction of methylsulfinyls is apparently due to the fact that MsrA recognizes methylsulfinyl as a substrate, whereas MsrB and fRMsr require larger functional groups (16-19).
Aside from the selective reduction of compounds by mMsrA, MsrA KO mice exhibited a low activity towards methylsulfinyls (Figure 2C and D). This activity is consistent with the occurrence of an unknown low efficiency reductase. A recent study suggested that the R-enantiomer of sulindac may be reduced by mammalian MsrB1 (20). Although purified MsrB2, MsrB3, and the Cys mutant of MsrB1 did not reduce this R-enantiomer in vitro, the function of wild type MsrB1 could not be tested (20). In the case of sulmazole, we found that these enzymes also could not reduce this compound, and the MsrB1 KO liver lysate had the same activity towards sulmazole and mesoridazine as the wild type lysate. Nevertheless, it is possible that an unknown reductase is involved in the low activity reduction of methylsulfinyls in sulmazole and mesoridazine. It is unclear if this activity is higher in the case of sulindac (20) or if the sulindac structure (e.g., being less charged and less bulkier) is different enough from the compounds tested in our study, so that it is more efficiently reduced by MsrB1. Regardless of the possible low activity of MsrB1 towards some methylsulfinyl-containing xenobiotics, our data strongly support the idea that MsrA is the major reductase for the S-stereoisomer reduction and that MsrBs lack or exhibit low efficiency in the R-stereoisomer reduction.
Potential differences in the metabolism and efficacy of R- or S-sulfoxide forms of drugs and natural compounds previously received little attention. However, our findings suggest that the differences in metabolizing these forms are an important factor that can be used in development of drugs with improved efficacy and decreased toxicity. It was previously reported that the affinity of mesoridazine for dopaminergic and α-adrenergic receptors was higher than that of thioridazine, while mesoridazine had a lower affinity for the muscarinic receptor. This differential binding upon oxidation was further supported by clinical data on the side effects that correlated with the use of these drug forms (21). For TCBZ, it was suggested that the sulfoxidation status of the methylsulfide group may be associated with the increased resistance of F. hepatica to drug treatment (22, 23). In the case of sulforaphane, studies have shown that the sulfoxide form of this compound was approximately 10 times more effective in inducing NAD(P)H:quinone oxidoreductase (NQO1) activity than the sulfide form (24). Finally, our toxicity assay data showed that the R-sulfoxide form of mesoridazine had lower toxicity than the mixture of two enantiomers, whereas the S-sulfoxide form was more toxic due to its conversion to the sulfide form by MsrA. Consequently, if the oxidized forms of methylsulfinyl-containing drugs or natural compounds, such as mesoridazine and sulforaphane, are less toxic or show higher efficacy than the reduced forms, the R-enantiomers of such compounds can be used for improved drug efficacy. In contrast, if the reduced forms are more active or less toxic, the S-enantiomers of these pro-drugs should be utilized. Although our study examined only a limited number of methylsulfinyl-containing compounds, the overall findings are clear and should apply to most other methylsulfinyl/methylsulfide-containing drugs, such as enoximone, nifuratel, albendazole, pergolide, lincomycin, triethylperazine, fensulfothion, clindamycin, captodiame, flosequinan, and thiocolchicoside. Some newly developed drugs will surely also include this functional group.
Methods
Cloning, expression, and purification of mouse MsrA, mouse MsrB2, yeast fRMsr, and mouse MsrB1-Cys
Previously established expression constructs for mouse MsrB1 (mutant in which Sec is replaced with Cys for increased protein expression) (25), mouse MsrB2 (25), mouse MsrA (26), and yeast fRMsr (27) were transformed into BL21 (DE3) E. coli. Cells with the plasmid in 500 ml LB medium containing 50 μg/ml ampicillin were grown until OD600 reached 0.6-0.8, followed by addition of IPTG to 0.3 mM. Protein expression was induced at 30 °C for 4 h, followed by harvesting cells by centrifugation at 4,000 rpm for 5 min. Cells were washed with PBS and stored at -70 °C until use.
To purify proteins, cell pellet was dissolved in resuspension buffer (Tris-HCl, pH 7.5, 15 mM imidazole, 300 mM NaCl), and PMSF was added to a final concentration of 0.5 mM. After sonication, supernatant was collected by centrifugation at 8,000 rpm for 30 min. The supernatant was loaded onto a cobalt Talon resin (Clontech) pre-equilibrated with resuspension buffer. Following washing with the same buffer, the protein was eluted with elution buffer (Tris-HCl, pH 7.5, 300 mM imidazole, 300 mM NaCl). Fractions containing the expressed protein were pooled together and dialyzed overnight against PBS in a dialysis cassette (Pierce).
Preparation of mesoridazine, triclabendazole sulfoxide, sulmazole, and R- and S-sulforaphane-DTT
Solutions of mesoridazine (Sigma), triclabendazole sulfoxide (AmXpress), sulmazole (Sigma) were prepared in either ethanol or distilled water prior to enzyme assays. R- and S-sulforaphane (LKT Laboratories, Inc.) were incubated with DTT at a 1 to 10 ratio for 1 h at 37 °C and then used in assay mixtures to test enzyme activity.
Activity assays of mouse MsrA, mouse MsrB2, and yeast fRMsr using DTT
Mouse MsrA, mouse MsrB2, and yeast fRMsr were utilized in the reduction assays in the presence of DTT. Dabsylated Met-R-SO, dabsylated Met-S-SO, and free Met-R-SO were prepared as previously described (6). In the DTT-dependent reaction, the reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 20 mM DTT, 500 μM Met-O (dabsylated Met-R-SO, dabsylated Met-S-SO, or free Met-R-SO), and purified proteins. Reactions were carried out at 37 °C for 30 min and then were stopped by adding 200 μl of acetonitrile. The reduced product was analyzed by HPLC as described previously (6, 25).
Reduction assays for analyses by ion-spray mass spectrometry, HPLC, and GC
Mouse MsrA, mouse MsrB2, mouse MsrB1-Cys and yeast fRMsr were used in the reduction assays in the presence of DTT or Trx/TR/NADPH. Mesoridazine, triclabendazole sulfoxide, sulmazole, DMSO, R-sulforaphane-DTT, and S-sulforaphane-DTT were used as substrates. In the DTT-dependent reaction, a reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 20 mM DTT, substrates (500 μM mesoridazine, 500 μM sulmazole, 500 μM triclabendazole sulfoxide, 500 μM DMSO, 5 mM R-sulforaphane-DTT or 5 mM S-sulforaphane-DTT) and purified proteins. In the Trx-dependent reaction, a reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 6.8 μM E. coli Trx (Sigma), 0.2 mM NADPH, 0.4 μM E. coli TR, substrates (500 μM mesoridazine, 500 μM sulmazole, or 500 μM triclabendazole sulfoxide) and purified proteins. The reactions were carried out at 37 °C for 30 min and then were stopped by adding 300 μl acetonitrile (ACN) in the case of mesoridazine reaction, 900 μl ethanol in the case of DMSO reaction, and 10 μl trichloroacetic acid (50%) in the case of sulmazole, triclabendazole sulfoxide, R- or S-sulforaphane-DTT reactions. After 5 min incubation at 4 °C, the reaction mixtures were centrifuged at 13,000 rpm for 20 min. Then, supernatants were taken for further ion-spray mass spectrometry, HPLC, or GC analyses. Additionally, each peak from HPLC analyses was collected and subjected to ion-spray mass spectrometry analyses to confirm identity of compounds together with internal standards.
Ion-spray mass spectrometry analysis
MS and MS/MS spectra were obtained on a Sciex API-III triple-quadrupole mass spectrometer equipped with an atmospheric pressure ionization source. The mass spectrometer was operated in the negative mode. The samples (20 μl) were introduced in distilled water/acetonitrile/formic acid (50:50:0.3 by volume) (180 μl). Scanning was done with m/z from 100 to 1000.
Animal studies
Care and treatment of experimental animals were approved by the Animal Care and Use Committee at the University of Nebraska-Lincoln (UNL). 6 month old C57BL/6 wild type, MsrA knockout, and MsrB1 knockout mice were used. MsrA and MsrB1 knockout mice were previously described (28).
Sample preparation
Animals were sacrificed, and their livers were dissected and subjected to protein expression and activity assays. Tissues were homogenized in PBS containing protease inhibitors (Roche), and the homogenates were normalized with regard to protein concentration. Western blotting analyses and MsrA and MsrB activity assays were then carried out with these samples.
Protein expression analysis
Tissue homogenates were separated on SDS-PAGE gels (40 μg of protein was loaded). Then, the proteins were transferred onto PVDF membranes and probed with the antibodies indicated (MsrA, MsrB1, and β-actin). Secondary HRP-linked anti-rabbit or anti-mouse antibodies, and ECL substrate detection were from GE HealthCare.
Isolation of cytosolic and microsomal liver fractions from wild type, MsrA KO, and MsrB1 KO mice and oxidation of thioridazine with the microsomal fractions
Fresh mouse liver was homogenized in PBS (pH 7.5) containing protease inhibitor cocktail and centrifuged at 13,000 g for 30 min. The supernatant was collected and centrifuged again at 100,000 g for 2 h. The resulting supernatant was collected as the cytosolic fraction, and the pellet was washed with PBS, recentrifuged for 1 h at 100,000 g and collected as the microsomal fraction. These fractions were resuspended in 2 ml of PBS and subjected to the assays as described below. Microsomal fractions were used to assay thioridazine oxidation (20). Briefly, 500 μM thioridazine was incubated with 40, 80, 120, 160 μg microsomal fractions in PBS (pH 7.5) in the presence of 1.5 mM NADPH, 5 mM glucose-6-phosphate, 300 ng glucose-6-phosphate dehydrogenase, and 5 mM MgCl2 for 60 min at 37 °C. A total volume of the reaction was 100 μl. After stopping the reaction by adding 300 μl acenotritrile and centrifuging the mixture for 15 min at 13,000 g, 50 μl of the supernatant was injected onto HPLC and analyzed as described below.
Reduction assays of mesoridazine and sulmazole using liver lysates and liver cytosolic fractions
80-200 μg liver lysates or their cytosolic and microsomal fractions from wild type, MsrA KO, and MsrB1 KO mice were used in the reduction assays in the presence of DTT. Mesoridazine and sulmazole were used as substrates. In the DTT-dependent reaction, a reaction mixture (100 μl) contained 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 20 mM DTT, substrates (500 μM mesoridazine or sulmazole) and liver lysate. Products were also analyzed by using HPLC as described below.
Transfection of mammalian cells with mouse MsrA and mouse MsrB2 and drug toxicity assays
Constructs expressing GFP fused with mouse MsrA (26) or GFP fused with mouse MsrB2 lacking the signal peptide (29) were transfected into HEK 293 cells using Lipofectamin 2000 (Invitrogen). After 36 h, indicated concentrations of mesoridazine, thioridazine, sulmazole, DMSO, or triclabendazole sulfoxide were added to cells and cells were analyzed for 24 h. Cell viability was measured with 0.4 % trypan blue staining by using Countess® Automated Cell Counter (Invitrogen).
HPLC analysis of mesoridazine and its reduced form
50 μl supernatant from the mesoridazine reduction assay was injected onto ZORBAX RX-C18 column (4.6×150 mm) (Agilent). In this assay, solvent A (20 mM sodium acetate, pH 4.0) and solvent B (ACN) were used. Mesoridazine and thioridazine were separated from other compounds at room temperature at a flow rate of 1 ml/min using a linear gradient of 30-50 % solvent B from 0 to 30 min, and 50-100 % solvent B from 30 to 33 min. Detection was at 254 nm using a Waters 996 PDA detector.
HPLC analysis of triclabendazole sulfoxide and its reduced form
25 μl supernatant from the reduction of triclabendazole sulfoxide was mixed with 25 μl distilled water (0.05 % trifluoroacetic acid (TFA)) prior to injection onto a ZORBAX RX-C18 column (4.6×150 mm) (Agilent). For this analysis, solvent A (distilled water containing 0.1 % TFA) and solvent B (ACN containing 0.1 % TFA) were used. Triclabendazole and triclabendazole sulfoxide were separated from other compounds at room temperature at a flow rate of 1.5 ml/min using an isocratic of 45 % solvent B until 8 min and then a linear gradient of 45-100 % solvent B from 8 to 10 min. Detection was at 254 nm using a Waters 996 PDA detector.
HPLC analysis of sulmazole and its reduced form
2.5 μl supernatant sample from the reduction reaction of sulmazole was mixed with 47.5 μl of 20 mM sodium phosphate (pH 7.4)/ACN/distilled water (2:1:3, v/v) prior to injection onto a ZORBAX RX-C18 column (4.6×150 mm) (Agilent). For this analysis, solvent A (20 mM sodium phosphate, pH 7.4/ACN, 2:1 ratio in v/v) and solvent B (ACN) were used. Sulmazole and its reduced form were separated from other compounds at room temperature at a flow rate of 1.5 ml/min using an isocratic of 100 % solvent A until 15 min. Detection was by fluorescence using a Waters 474 scanning fluorescence detector with excitation at 330 nm and emission at 370 nm.
HPLC analysis of sulforaphane-DTT and its reduced form
A 10 μl supernatant sample from the reaction of R- or S-sulforaphane-DTT was mixed with 40 μl distilled water (0.06 % TFA) prior to injection onto a ZORBAX RX-C18 column (4.6×150 mm) (Agilent). For this analysis, solvent A (0.06 % TFA) and solvent B (ACN containing 0.06 % TFA) were used. R- or S-sulforaphane-DTT conjugate and its reduced form were separated from other compounds at room temperature at a flow rate of 1.0 ml/min using a linear gradient of 0 – 40 % solvent B from 0 to 40 min, and 40 – 100 % from 40 to 45 min. Detection was at 214 nm using a Waters 996 PDA detector.
GC analysis of DMSO and its reduced form
20 μl of supernatant from the reaction with DMSO was mixed with 980 μl ethanol and then this mixture was analyzed by GC. For this analysis, 6890N gas chromatograph (Agilent Technologies). Hydrogen gas was used as a carrier. Detection was done in the range from 40-250 °C by using a FID detector.
Supplementary Material
Acknowledgments
This study was supported by NIH grant AG021518 to VNG. The authors thank H. Park and M. Gerashchenko for help with GC analyses, X. Liang for Msr proteins, and A. Raza (all at University of Nebraska-Lincoln) for mass-spectrometry analyses.
Footnotes
Supporting Information: Supporting information includes Figures S1-S10. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stadtman ER, Moskovitz J, Levine RL. Oxidation of Methionine Residues of Proteins: Biological Consequences. Antioxid Redox Signal. 2003;5:577–582. doi: 10.1089/152308603770310239. [DOI] [PubMed] [Google Scholar]
- 2.Weissbach H, Resnick L, Brot N. Methionine Sulfoxide Reductases: History and Cellular Role in Protecting Against Oxidative Damage. Biochim Biophys Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Imlay JA. Cellular Defenses Against Superoxide and Hydrogen Peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadtman ER. Cyclic Oxidation and Reduction of Methionine Residues of Proteins in Antioxidant Defense and Cellular Regulation. Arch Biochem Biophys. 2004;423:2–5. doi: 10.1016/j.abb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, Lowther WT. Free Methionine-(R)-Sulfoxide Reductase from Escherichia Coli Reveals a New GAF Domain Function. Proc Natl Acad Sci U S A. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BC, Le DT, Gladyshev VN. Mammals Reduce Methionine-S-Sulfoxide with MsrA and are Unable to Reduce Methionine-R-Sulfoxide, and this Function can be Restored with a Yeast Reductase. J Biol Chem. 2008;283:28361–28369. doi: 10.1074/jbc.M805059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HY, Gladyshev VN. Methionine Sulfoxide Reductases: Selenoprotein Forms and Roles in Antioxidant Protein Repair in Mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 8.Lynch T, Price A. The Effect of Cytochrome P450 Metabolism on Drug Response, Interactions, and Adverse Effects. Am Fam Physician. 2007;76:391–396. [PubMed] [Google Scholar]
- 9.Beedham C. The Role of Non-P450 Enzymes in Drug Oxidation. Pharm World Sci. 1997;19:255–263. doi: 10.1023/a:1008668913093. [DOI] [PubMed] [Google Scholar]
- 10.Bentley R. Role of Sulfur Chirality in the Chemical Processes of Biology. Chem Soc Rev. 2005;34:609–624. doi: 10.1039/b418284g. [DOI] [PubMed] [Google Scholar]
- 11.Etienne F, Resnick L, Sagher D, Brot N, Weissbach H. Reduction of Sulindac to its Active Metabolite, Sulindac Sulfide: Assay and Role of the Methionine Sulfoxide Reductase System. Biochem Biophys Res Commun. 2003;312:1005–1010. doi: 10.1016/j.bbrc.2003.10.203. [DOI] [PubMed] [Google Scholar]
- 12.Sagher D, Brunell D, Hejtmancik JF, Kantorow M, Brot N, Weissbach H. Thionein can Serve as a Reducing Agent for the Methionine Sulfoxide Reductases. Proc Natl Acad Sci U S A. 2006;103:8656–8661. doi: 10.1073/pnas.0602826103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of Oxidized Proteins. Identification of a New Methionine Sulfoxide Reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 14.Lee BC, Gladyshev VN. The Biological Significance of Methionine Sulfoxide Stereochemistry. Free Radic Biol Med. 2011;50:221–227. doi: 10.1016/j.freeradbiomed.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and Evolution of Selenoprotein Methionine Sulfoxide Reductases. Biochim Biophys Acta. 2009;1790:1471–1477. doi: 10.1016/j.bbagen.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruez A, Libiad M, Boschi-Muller S, Branlant G. Structural and Biochemical Characterization of Free Methionine-R-Sulfoxide Reductase from Neisseria Meningitidis. J Biol Chem. 2010;285:25033–25043. doi: 10.1074/jbc.M110.134528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boschi-Muller S, Gand A, Branlant G. The Methionine Sulfoxide Reductases: Catalysis and Substrate Specificities. Arch Biochem Biophys. 2008;474:266–273. doi: 10.1016/j.abb.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Neiers F, Sonkaria S, Olry A, Boschi-Muller S, Branlant G. Characterization of the Amino Acids from Neisseria Meningitidis Methionine Sulfoxide Reductase B Involved in the Chemical Catalysis and Substrate Specificity of the Reductase Step. J Biol Chem. 2007;282:32397–32405. doi: 10.1074/jbc.M704730200. [DOI] [PubMed] [Google Scholar]
- 19.Ranaivoson FM, Antoine M, Kauffmann B, Boschi-Muller S, Aubry A, Branlant G, Favier F. A Structural Analysis of the Catalytic Mechanism of Methionine Sulfoxide Reductase A from Neisseria Meningitidis. J Mol Biol. 2008;377:268–280. doi: 10.1016/j.jmb.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Brunell D, Sagher D, Kesaraju S, Brot N, Weissbach H. Studies on the Metabolism and Biological Activity of the Epimers of Sulindac. Drug Metab Dispos. 2011;39:1014–1021. doi: 10.1124/dmd.110.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bylund DB. Interactions of Neuroleptic Metabolites with Dopaminergic, Alpha Adrenergic and Muscarinic Cholinergic Receptors. J Pharmacol Exp Ther. 1981;217:81–86. [PubMed] [Google Scholar]
- 22.Alvarez LI, Solana HD, Mottier ML, Virkel GL, Fairweather I, Lanusse CE. Altered Drug influx/efflux and Enhanced Metabolic Activity in Triclabendazole-Resistant Liver Flukes. Parasitology. 2005;131:501–510. doi: 10.1017/S0031182005007997. [DOI] [PubMed] [Google Scholar]
- 23.Brennan GP, Fairweather I, Trudgett A, Hoey E, McCoy, McConville M, Meaney M, Robinson M, McFerran N, Ryan L, Lanusse C, Mottier L, Alvarez L, Solana H, Virkel G, Brophy PM. Understanding Triclabendazole Resistance. Exp Mol Pathol. 2007;82:104–109. doi: 10.1016/j.yexmp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Ahn YH, Hwang Y, Liu H, Wang XJ, Zhang Y, Stephenson KK, Boronina TN, Cole RN, Dinkova-Kostova AT, Talalay P, Cole PA. Electrophilic Tuning of the Chemoprotective Natural Product Sulforaphane. Proc Natl Acad Sci U S A. 2010;107:9590–9595. doi: 10.1073/pnas.1004104107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HY, Gladyshev VN. Different Catalytic Mechanisms in Mammalian Selenocysteine- and Cysteine-Containing Methionine-R-Sulfoxide Reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HY, Gladyshev VN. Role of Structural and Functional Elements of Mouse Methionine-S-Sulfoxide Reductase in its Subcellular Distribution. Biochemistry. 2005;44:8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- 27.Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, Kaya A, Hacioglu E, Kwak GH, Koc A, Kim HY, Gladyshev VN. Functional Analysis of Free Methionine-R-Sulfoxide Reductase from Saccharomyces Cerevisiae. J Biol Chem. 2009;284:4354–4364. doi: 10.1074/jbc.M805891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novoselov SV, Kim HY, Hua D, Lee BC, Astle CM, Harrison DE, Friguet B, Moustafa ME, Carlson BA, Hatfield DL, Gladyshev VN. Regulation of Selenoproteins and Methionine Sulfoxide Reductases A and B1 by Age, Calorie Restriction, and Dietary Selenium in Mice. Antioxid Redox Signal. 2010;12:829–838. doi: 10.1089/ars.2009.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY, Gladyshev VN. Methionine Sulfoxide Reduction in Mammals: Characterization of Methionine-R-Sulfoxide Reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


