Abstract
Translocations involving the mixed-lineage leukemia gene (MLL) confer a poor prognosis in acute leukemias. In t(1;11)(q21;q23), MLL is fused reciprocally with AF1q. Here we describe a t(1;11)(q21;q23) with a secondary event involving insertion of the telomeric portion of MLL into the p arm of chromosome 11 (11p11). We show that this latter event interrupts the CUG triplet repeat binding protein-1 (CUGBP1) gene, a translational enhancer of C/EBPβ. We then showed that these cells have reduced expression of CUGBP1 and C/EBPβ when compared to other AML blasts. This is the first report to describe insertional disruption of the CUGBP1 gene and to suggest a role for the CUGBP1-C/EBPβ pathway in leukemogenesis.
1. Introduction
Translocations of chromosomal band 11q23 are among the most observed rearrangements in leukemia, occurring in 5–6% of primary acute myelogenous leukemias (AML), a majority of secondary leukemias, and nearly all infant acute lymphocytic leukemias (ALL) [1,2]. 11q23 translocations disrupt the mixed-lineage leukemia (MLL) gene and usually confer a poor prognosis [1,3]. The MLL gene is nonrandomly fused with one of several dozen MLL fusion partner genes such as AF4, AF6, ELL, and ENL [2,4–6]. Most rearrangements of the MLL gene occur within a single 8.5-kb breakpoint cluster region and can be identified by cytogenetic G-banding and fluorescent in situ hybridization (FISH) analyses [6–10]. The t(1;11)(q21;q23) fuses the MLL gene to the AF1q gene located on chromosomal band 1q21 [6,11]. To our knowledge, fewer than 20 cases of this translocation have been reported this past decade [5,6,11,12].
CCAAT/enhancer binding proteins (C/EBPs) are transcription factors known to play a critical role in myeloid differentiation. They are often dysregulated in myeloid leukemias and have been identified as targets of BCR/ABL, ETO, and FLT3 mutations. Expression of C/EBP proteins is controlled at several levels, including regulation of translation by certain RNA-binding proteins. One of these proteins, CUG triplet repeat binding protein-1 (CUGBP1), is a translational regulator of C/EBPβ that interacts with the 5′ region of C/EBPβ mRNA and increases the translation of two C/EBPβ isoforms, liver-enriched activator protein (LAP) and liver-enriched inhibitory protein (LIP) [13].
In this case report, we describe a variant t(1;11)(q21;q23) occurring in a pediatric AML patient (designated AML17) that activates MLL as described previously, but also disrupts CUGBP1, leading to decreased translation of C/EBPβ.
2. Materials and methods
2.1. Clinical history
The patient described herein (AML17) is a 7-month-old Hispanic female who presented to the local emergency room with a history of 1 week of upper respiratory symptoms, 2 days of fussiness, and fever to 40°C. The presenting white blood cell (WBC) count was 310 × 103 cells/μL with a morphology on peripheral smear suggestive of myeloblasts. Hydration reduced the WBC count to 210.6 × 103 cells/μL. A large-bore pheresis catheter was placed, and leukaphoresis was performed that reduced her WBC to 144.9 × 103 cells/μL. Peripheral blood was sent for routine hematopathologic stains, flow immunophenotyping, and cytogenetics. The smears demonstrated blasts, promonocytes, and reactive-appearing monocytes having fine granules and strong positive neuron-specific enolase (NSE) staining completely inhibited by NaF, consistent with M5 AML. Immunophenotyping revealed positive staining for cell surface markers CD4 (95%), CD11b (90%), CD13 (36%), CD15 (94%), CD33 (96%), CD(16+56) (97%), κ-light chain (86%), and λ-light chain (47%). The cytogenetics findings are described in Table 1. The patient received one dose of rasburicase and was treated according to Pediatric Oncology Group (POG) protocol 9421. The leukemia responded rapidly to therapy, and she remains in remission 1 year off therapy.
Table 1.
Patient characteristics of primary AML samples used in quantitative RT-PCR study
| Sample | Age at diagnosis | FAB | Reported clinical karyotype |
|---|---|---|---|
| AML10 | 6 years | M5 | 45,X,add(X)(q?25),t(2;7)(p21;q36),t(2;11)(q23;p13),del(3)(q21),del(5)(q15q31),del(14)(q?24),-17,add(22)(q11.2)[18]46,XX2] |
| AML13 | 11 years | M2 | 46,XX,t(4;20)(q31;q11)[19]/46,XX[1] |
| AML17 | 7 months | M5 | 46,XX,t(1;11)(q21;q23),inv(11)(q13q23)[20] |
| AML19 | 8 years | M1, MDS-related | 47,XX,t(1;19)(p22;q13),del(2)(q33),del(3)(q21),+i(5)(p10),add(6)(p2?3)[14] |
| AML21 | 12 years | M1/M2 | 46,XY[20] |
Leukocytes were obtained by leukopheresis from patients with AML at initial diagnosis. Hematopathologic subtypes and cytogenetic abnormalities were determined by the clinical laboratory.
2.2. Patient samples and cell lines
Leukemic myeloblasts were obtained by leukapheresis of newly diagnosed pediatric patients with very high peripheral blood blast counts (Table 1) under an institutional review board (IRB)-approved collection and banking protocol. Mononuclear cells were isolated by density gradient centrifugation over Lymphoprep (Axis-Shield PoC AS, Oslo, Norway). Cells were viably frozen in freezing media [RPMI 1640, 20% fetal bovine serum (FBS), 10% DMSO] and stored in liquid nitrogen. BS LCL is an EBV-transformed lymphoblastoid cell line (LCL) from a normal healthy donor. NIH 3T3 and HeLa cell lines were obtained from American Type Culture Collection (Manassas, VA, USA).
2.3. Conventional cytogenetic analysis
Cytogenetic evaluations were conducted on all AML patient samples as clinically indicated. Routine G-banding karyotype analyses were performed by the clinical laboratory to identify chromosomal translocations and other abnormalities as indicated clinically. Fluorescence in situ hybridization (FISH) was performed on metaphase cells of AML17 using LSI MLL dual color break apart rearrangement probe and TelVysion telomere region-specific probes for chromosomes 1 and 11 (Abbott Molecular/Vysis, Des Plaines, IL, USA).
2.4. Array comparative genomic hybridization
Genomic DNA was isolated by QIAamp DNA Mini Kit (Qiagen, Inc., Valencia, CA, USA). Array comparative genomic hybridization (aCGH) was performed using the SpectralChip 2600, a bacterial artificial chromosome (BAC) array with an average resolution of 1 MB across the genome. Briefly, genomic DNA was fragmented and labeled by a poplymerase reaction using fluorophore labeled dNTPs and random hexamer primers. DNA was then hybridized to the chip according to the manufacturer’s protocol. The chips were scanned using a GenePix 3000B scanner running GenePix Pro 6.0.1 and analyzed using SpectralWare v2.
2.5. Long-distance inverse polymerase chain reaction (LDI-PCR)
The DNA sample was treated and analyzed as described [14]. Briefly, 1 μg of genomic DNA was digested with restriction enzymes and re-ligated to form DNA circles before LDI-PCR using MLL-specific primers. Restriction polymorphic PCR amplimers were isolated from the gel and subjected to DNA sequence analyses to obtain the patient-specific fusion sequences [5,14].
2.6. Quantitative reverse-transcription polymerase chain reaction (RT-PCR)
RNA was isolated using the SV Total RNA Isolation Kit (Promega Corp., Madison, WI, USA). Quantitative RT-PCR was performed in triplicate on RNA isolated from each sample (Table 1) using the One-Step RT-PCR Kit (Qiagen) with Taqman probes (Applied Biosystems, Foster City, CA, USA) for CUGBP1 (Hs00198069_m1), HOXA9 (Hs00365956_m1), and 18s rRNA (Hs99999901_s1), following the manufacturer’s protocol. After 30 minutes of reverse transcription at 48°C and 10 minutes of enzyme activation at 95°C, 40 cycles of amplification were performed (15 seconds of denaturation at 95°C followed by 1 minute of annealing and elongation at 60°C). Fluorescence was measured after each cycle. Quantititative RT-PCR was performed using an Opticon thermocycler (MJ Research, Hercules, CA, USA). The expression levels were normalized to 18s rRNA.
2.7. Western blot analysis
Proteins were isolated from freshly thawed cells and analyzed by Western blotting as described [13,15]. Briefly, cells were homogenized in buffer A containing 20 mmol/L Tris-HCl, pH 7.5, 30 mmol/L NaCl, and 2 mmol/L MgCl2. Nuclei were pelleted by centrifugation at 12,000 rpm for 10 minutes. The supernatant (cytoplasm) was used for the examination of CUGBP1. A nuclear extract was obtained by treatment of nuclei with buffer B (0.42 mol/L NaCl, 20 mmol/L Tris-HCL, pH 7.5, 5 mmol/L DTT, 2 mmol/L MgCl2, and 25% sucrose). Inhibitors of proteases and phosphatases were included in all buffers used for the isolation of proteins. Cytoplasmic and nuclear proteins were separated by denaturing SDS 6–18% gradient gel (BioRad, Hercules, CA, USA) and transferred onto the membrane. Proteins were detected with antibodies against C/EBPβ and CUGBP1 (clones C19 and 3B1, respectively; Santa Cruz Biotechnologies, Inc., Santa Cruz, CA, USA). The membranes were re-probed with antibodies to β-actin and stained with Coomassie to assess for equal protein loading.
3. Results
3.1. A variant t(1;11)(q21;q23) identified by cytogenetics and FISH
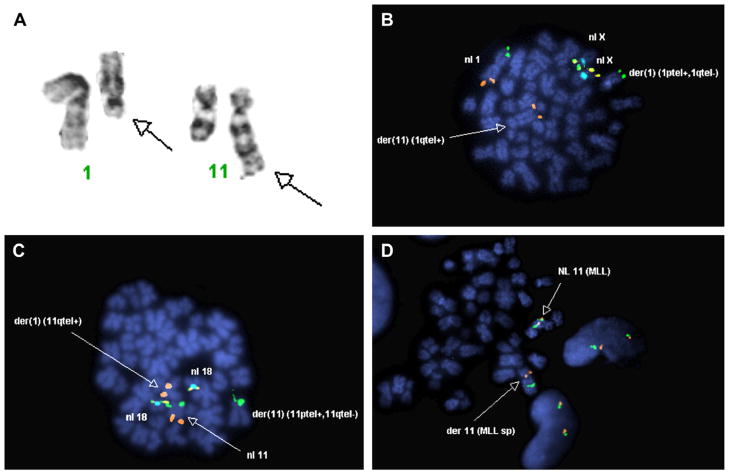
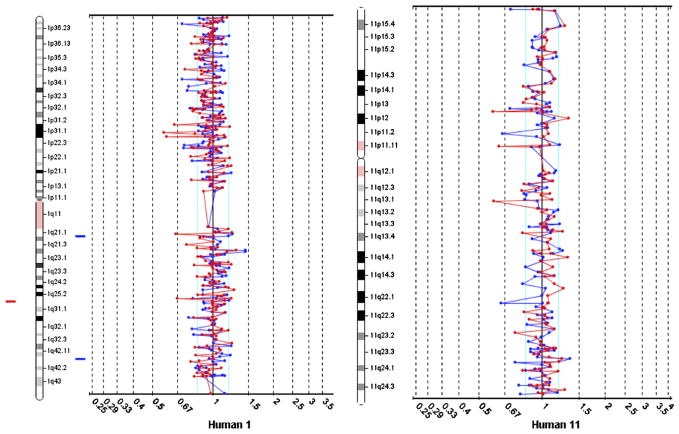
The rearrangement of chromosomal bands 1q21 and 11q23 was identified in the routine G-banding analysis (Fig. 1a). Metaphase FISH using region-specific probes for chromosome 1 shows normal 1p and 1q telomeric hybridization signals on one copy of chromosome 1 but only a 1p signal on the homologous chromosome. The remaining 1q signal is located on the derivative chromosome 11 (Fig. 1b). Similarly, metaphase FISH using region-specific probes for chromosome 11 shows normal 11p and 11q telomeric hybridization signals on one copy of chromosome 11, but an 11p only on the homologous pair and the remaining 11q on the derivative chromosome 1 (Fig. 1c), suggesting a balanced translocation. However, FISH analysis for the MLL gene using a break-apart rearrangement probe shows the telomeric hybridization signal of the MLL probe (orange) is located in the p-arm of chromosome 11 (Fig. 1d) rather than on chromosome 1, where it is expected. Array comparative genomic hybridization (aCGH) reveals no genomic losses or gains within the detection limits of this assay (Fig. 2).
Fig. 1.
Cytogenetics of AML17. (A) G-banding analysis shows derivative chromosomes 1 and 11 (arrows). Metaphase FISH using telomere region–specific probes for chromosomes 1 (B) and 11 (C) shows a pattern consistent with a balanced reciprocal translocation. (D) FISH using a break-apart rearrangement probe for the MLL gene shows both portions of MLL on the derivative chromosome 11.
Fig. 2.
Array CGH of AML17. DNA was purified from banked samples of AML17 and hybridized to the BAC-based human genome array as described, showing no apparent losses or gains of genetic material. Chromosomes 1 (left) and 11 (right) are shown.
3.2. Genomic breakpoints determined by LDI-PCR
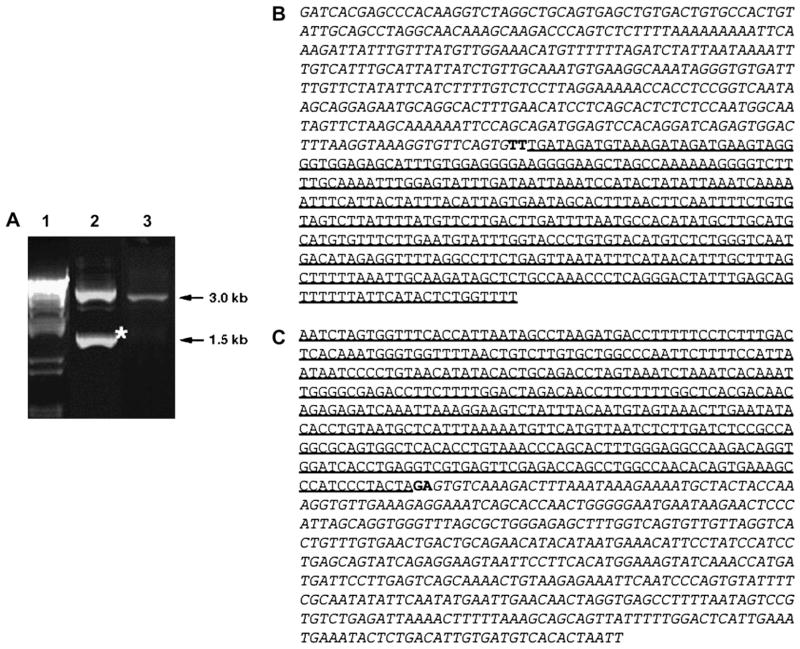
To determine the genomic breakpoints of the t(1;11) in AML17, the fusion region was amplified by LDI-PCR for the 5′ MLL fusion (Fig. 3a, lane 2) and the 3′ MLL fusion (Fig. 3a, lane 3). The upper band in lane 2 is consistent with the wild-type MLL. The sequence of the lower band in lane 2 reveals a fusion of 5′ MLL at intron 10/11 with AF1q at intron 1/2, linked by two filler nucleotides (Fig. 3b). Sequencing of the band in lane 3 reveals a fusion of 3′ MLL at intron 10/11 with CUGBP1 at intron 9/10, linked by two filler nucleotides (Fig. 3c). This sequence indicates that CUGBP1 and MLL are fused in opposing transcriptional directions.
Fig. 3.
Genomic breakpoints within MLL. (A) LDI-PCR analysis of AML17 genomic DNA. Lane 1, marker; lane 2, LDI-PCR analysis of MLL-translocation partner (TP) fusion showing the wild-type band and the additional MLL-TP band (asterisk); lane 3, LDI-PCR analysis of TP-MLL fusion (in this case, only the TP-MLL amplimer appeared). (B) The sequence of the MLL-TP amplimer reveals a fusion between intron 10/11 of MLL (italic) and intron 1/2 of AF1q (underline) linked by two filler nucleotides (bold). (C) The sequence TP-MLL reveals a fusion between intron 9/10 of CUGBP1 (underline) and intron 10/11 of MLL (italic) linked by two filler nucleotides (bold).
3.3. Expression of CUGBP1 is reduced and HOXA9 is elevated
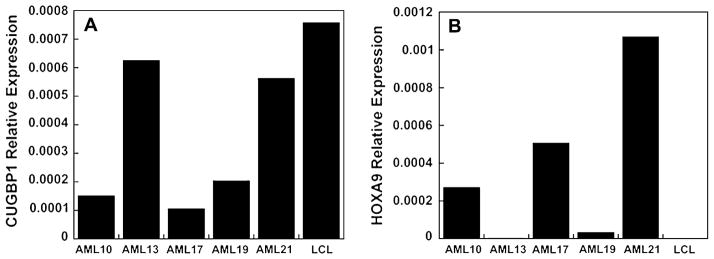
Since the CUGBP1-MLL fusion did not result in a transcribable fusion but, rather, disrupted the CUGBP1 gene, we evaluated whether transcription of CUGBP1 was affected. In addition, to confirm that MLL-AF1q was an activating fusion product, we assessed transcriptional levels of HOXA9 as a known target of MLL. The mRNA levels of CUGBP1 and HOXA9 in AML17 were compared to four other AML samples (Table 1) and one LCL by quantitative RT-PCR using Taqman probes. AML17 has decreased CUGBP1 (Fig. 4a) and increased HOXA9 expression (Fig. 4b) compared to the other cells tested.
Fig. 4.
Quantitative RT-PCR analysis. Quantitative RT-PCR for CUGBP1 (A) and HOXA9 (B) was performed on RNA from representative primary AML blasts and a control LCL line. Expression levels are normalized to 18S rRNA.
3.4. Expression of C/EBPβ isoforms are reduced
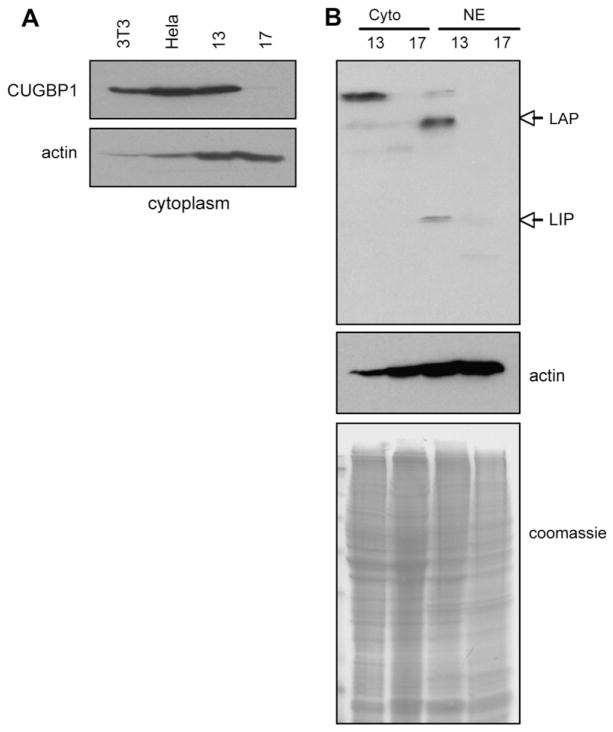
CUGBP1 binds to the 5′ region of C/EBPβ mRNA and increases the translation of the C/EBPβ-LIP and C/EBPβ-LAP isoforms [16]. The decrease in CUGBP1 expression suggests that its leukemogenic role in this patient might be through a loss of post-transcriptional regulation of C/EBPβ. Therefore we assessed the protein expression of these genes in AML17 and compared them to AML13, a sample with high mRNA levels of CUGBP1. The reduced protein level of CUGBP1 as determined by Western blot probe (Figure 5a) is consistent with the reduced mRNA levels shown by quantitative RT-PCR (Figure 4a). Moreover, Western blot for C/EBPβ showed that the protein levels of both the LIP and LAP isoforms of C/EBPβ are markedly decreased in AML17 compared to the levels of these isoforms in AML13 (Figure 5b). This inhibition of C/EBPβ translation is consistent with the hypothesis that the interrupted CUGBP1 gene leads to a low level of CUGBP1 and thus to reduced translation of C/EBPβ.
Fig. 5.
Expression of CUGBP1 and C/EBPβ proteins. (A) Western blot analysis of CUGBP1 in protein lysates from AML13 and AML17 with actin-loading controls. Protein lysates from 3T3-L1 and HeLa cells were used as controls. (B) Western blot analysis of C/EBPβ in cytoplasmic (Cyto) and nuclear extract (NE) proteins from AML13 and AML17. Positions of LAP and LIP isoforms are shown by arrows. Coomassie staining and actin show equal protein loading of all lanes.
4. Discussion
The germline MLL gene encodes a protein of 3,972 amino acids (431 kD), which is a common target for chromosomal translocations in acute leukemias [11,17]. The human MLL gene is homologous to Drosophila trithorax, which is a homeotic transcriptional regulator and contains PHD finger motifs along with DNA-binding motifs such as AT hooks and a DNA methyltransferase homology region [11,18–20]. Translocations involving the MLL gene form various in-frame fusion transcripts with many different partner genes throughout the genome [5,17]. The N-terminal portion of the MLL gene that contains the AT hook DNA-binding motifs and the methyltransferase homology is necessary for the oncogenic effect when fused with the C-terminal portion of the partner gene [11,18,20,21]. In AML17, the MLL-AF1q fusion contains this oncogenic region of MLL, as expected. The second rearrangement in AML17 fuses the telomeric end of the MLL gene with the 5′ segment of the CUG triplet repeat binding protein-1 gene (CUGBP1) in opposing transcriptional directions.
CUGBP1 is a downstream target of the epidermal growth factor receptor (EGFR) signaling pathway and a translational regulator that interacts with the CUG repeats in the 5′ region of C/EBPβ mRNA [13,22]. The binding of CUGBP1 to the mRNA of C/EBPβ increases translation of the C/EBPβ-LIP and C/EBPβ-LAP isoforms [13,16,22]. C/EBPβ, also known as nuclear factor of interleukin-6 (NF-IL6), regulates the transcriptional activities of many processes, including the production of several cytokine genes and myeloid differentiation. Although C/EBPβ is not required for steady-state granulopoiesis [23,24], it is required for normal and emergency myeloid differentiation [25,26]. In AML17, disruption of CUGBP1 by the C-terminal portion of the MLL gene and subsequent loss of C/EBPβ translation suggests a role for CUGBP1 mutations in leukemogenesis.
These data suggest that the complex rearrangement in AML17 caused both oncogenic conversion of MLL by fusion to AF1q and decreased C/EBPβ expression by the loss of translational enhancement mediated by CUGBP1. This rearrangement could have resulted from (1) two sequential events (independent translocation and deletion/insertion events) or (2) a single cross-over event involving four recombination sites. The sequences obtained by LDI-PCR showed complete conservation of the genomic MLL sequence, suggesting the latter mechanism. This is the first report showing a possible role for the CUGBP1-C/EBPβ pathway in leukemogenesis.
Acknowledgments
We thank Dr. Deb Shardy for her technical help. This work was supported by research funding to W.T.C. from the American Society of Hematology (Research Trainee Award); to D.A.L. from the American Society for Clinical Oncology (Young Investigator Award), the For Julie Foundation, and the National Institutes of Health (R21 CA114251 and K12 CA90433); to R.M. from the Wilhelm-Sander-Foundation (grant 2001.061.2); and to N.A.T. from the National Institutes of Health (R01 CA100070).
W.T.C. performed the RT-PCR experiments and was the primary author of the manuscript. M.F. performed the array CGH, and M.A. perfomed the G-banding analysis and metaphase FISH. R.N. coordinated and interpreted the clinical cytogenetic data. C.M. and E.K. performed the LDI-PCR and interpreted the data with the assistance of R.M. N.T. performed and interpreted the Western blot. D.A.L. coordinated the research direction of the project, collected and banked the patient samples under an IRB-approved research protocol, and assisted in writing the manuscript.
References
- 1.Thirman MJ, Gill HJ, Burnett RC, Mbangkollo D, McCabe NR, Kobayashi H, Zieminvan der Poel S, Kaneko Y, Morgan R, Sandberg AA, Chaganti RSK, Larson RA, Le Beau MM, Diaz MO, Rowley JD. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329:909–14. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Behm FG, Downing JR. 11q23 rearrangements in acute leukemia. Leukemia. 1996;10:74–82. [PubMed] [Google Scholar]
- 3.Mrozek K, Heinonen K, Lawrence D, Carroll AJ, Koduru PR, Rao KW, Strout MP, Hutchison RE, Moore JO, Mayer RJ, Schiffer CA, Bloomfield CD. Adult patients with de novo acute myeloid leukemia and t(9; 11)(p22; q23) have a superior outcome to patients with other translocations involving band 11q23: a cancer and leukemia group B study. Blood. 1997;90:4532–8. [PubMed] [Google Scholar]
- 4.Raffini LJ, Slater DJ, Rappaport EF, Lo Nigro L, Cheung NK, Biegel JA, Nowell PC, Lange BJ, Felix CA. Panhandle and reverse-panhandle PCR enable cloning of der(11) and der(other) genomic breakpoint junctions of MLL translocations and identify complex translocation of MLL, AF-4, and CDK6. Proc Natl Acad Sci USA. 2002;99:4568–73. doi: 10.1073/pnas.062066799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer C, Schneider B, Jakob S, Strehl S, Attarbaschi A, Schnittger S, Schoch C, Jansen MW, van Dongen JJ, den Boer ML, Pieters R, Ennas MG, Angelucci E, Koehl U, Greil J, Griesinger F, Zur Stadt U, Eckert C, Szczepanski T, Niggli FK, Schafer BW, Kempski H, Brady HJ, Zuna J, Trka J, Nigro LL, Biondi A, Delabesse E, Macintyre E, Stanulla M, Schrappe M, Haas OA, Burmeister T, Dingermann T, Klingebiel T, Marschalek R. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–84. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 6.Busson-Le Coniat M, Salomon-Nguyen F, Hillion J, Bernard OA, Berger R. MLL-AF1q fusion resulting from t(1;11) in acute leukemia. Leukemia. 1999;13:302–6. doi: 10.1038/sj.leu.2401299. [DOI] [PubMed] [Google Scholar]
- 7.Bernard OA, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 8.Young BD, Saha V. Chromosome abnormalities in leukaemia: the 11q23 paradigm. Cancer Surv. 1996;28:225–45. [PubMed] [Google Scholar]
- 9.Waring PM, Cleary ML. Disruption of a homolog of trithorax by 11q23 translocations: leukemogenic and transcriptional implications. Curr Top Microbiol Immunol. 1997;220:1–23. doi: 10.1007/978-3-642-60479-9_1. [DOI] [PubMed] [Google Scholar]
- 10.Meloni-Balliet AM, Morgan R, Piatt J, Sandberg AA. Translocation t(1;11)(q21;q23), a new subgroup within M4 acute nonlymphocytic leukemia. Cancer Genet Cytogenet. 1989;37:269–71. doi: 10.1016/0165-4608(89)90058-7. [DOI] [PubMed] [Google Scholar]
- 11.Tse W, Zhu W, Chen HS, Cohen A. A novel gene, AF1q, fused to MLL in t(1;11)(q21;q23), is specifically expressed in leukemic and immature hematopoietic cells. Blood. 1995;85:650–6. [PubMed] [Google Scholar]
- 12.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15(417–74) doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 13.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–25. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer C, Schneider B, Reichel M, Angermueller S, Strehl S, Schnittger S, Schoch C, Jansen MW, van Dongen JJ, Pieters R, Haas OA, Dingermann T, Klingebiel T, Marschalek R. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci USA. 2005;102:449–54. doi: 10.1073/pnas.0406994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timchenko LT, Iakova P, Welm AL, Cai ZJ, Timchenko NA. Calreticulin interacts with C/EBPalpha and C/EBPbeta mRNAs and represses translation of C/EBP proteins. Mol Cell Biol. 2002;22:7242–57. doi: 10.1128/MCB.22.20.7242-7257.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karagiannides I, Thomou T, Tchkonia T, Pirtskhalava T, Kypreos KE, Cartwright A, Dalagiorgou G, Lash TL, Farmer SR, Timchenko NA, Kirkland JL. Increased CUG triplet repeat binding protein-1 predisposes to impaired adipogenesis with aging. J Biol Chem. 2006;281:23025–33. doi: 10.1074/jbc.M513187200. [DOI] [PubMed] [Google Scholar]
- 17.Kohlmann A, Schoch C, Dugas M, Schnittger S, Hiddemann W, Kern W, Haferlach T. New insights into MLL gene rearranged acute leukemias using gene expression profiling: shared pathways, lineage commitment, and partner genes. Leukemia. 2005;19:953–64. doi: 10.1038/sj.leu.2403746. [DOI] [PubMed] [Google Scholar]
- 18.Harris BN, Davis EM, Le Beau MM, Bitter MA, Kaminer LS, Morgan E, Rowley JD. Variant translocations (9;11): identification of the critical genetic rearrangement. Cancer Genet Cytogenet. 1988;30:171–5. doi: 10.1016/0165-4608(88)90108-2. [DOI] [PubMed] [Google Scholar]
- 19.Ma Q, Alder H, Nelson KK, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce CM, Siracusa LD, Buchberg AM. Analysis of the murine All-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferases. Proc Natl Acad Sci USA. 1993;90:6350–4. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst P, Wang J, Korsmeyer SJ. The role of MLL in hematopoiesis and leukemia. Curr Opin Hematol. 2002;9:282–7. doi: 10.1097/00062752-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Raimondi SC, Peiper SC, Kitchingman GR, Behm FG, Williams DL, Hancock ML, Mirro J., Jr Childhood acute lymphoblastic leukemia with chromosomal breakpoints at 11q23. Blood. 1989;73:1627–34. [PubMed] [Google Scholar]
- 22.Baldwin BR, Timchenko NA, Zahnow CA. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPbeta-LIP in mammary epithelial cells. Mol Cell Biol. 2004;24:3682–91. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, Bistoni F, Frati L, Cortese R, Gulino A, Ciliberto G, Constantini F, Poli V. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. Embo J. 1995;14:1932–41. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–61. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 25.Mueller BU, Pabst T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol. 2006;13:7–14. doi: 10.1097/01.moh.0000190110.08156.96. [DOI] [PubMed] [Google Scholar]
- 26.Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–9. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]