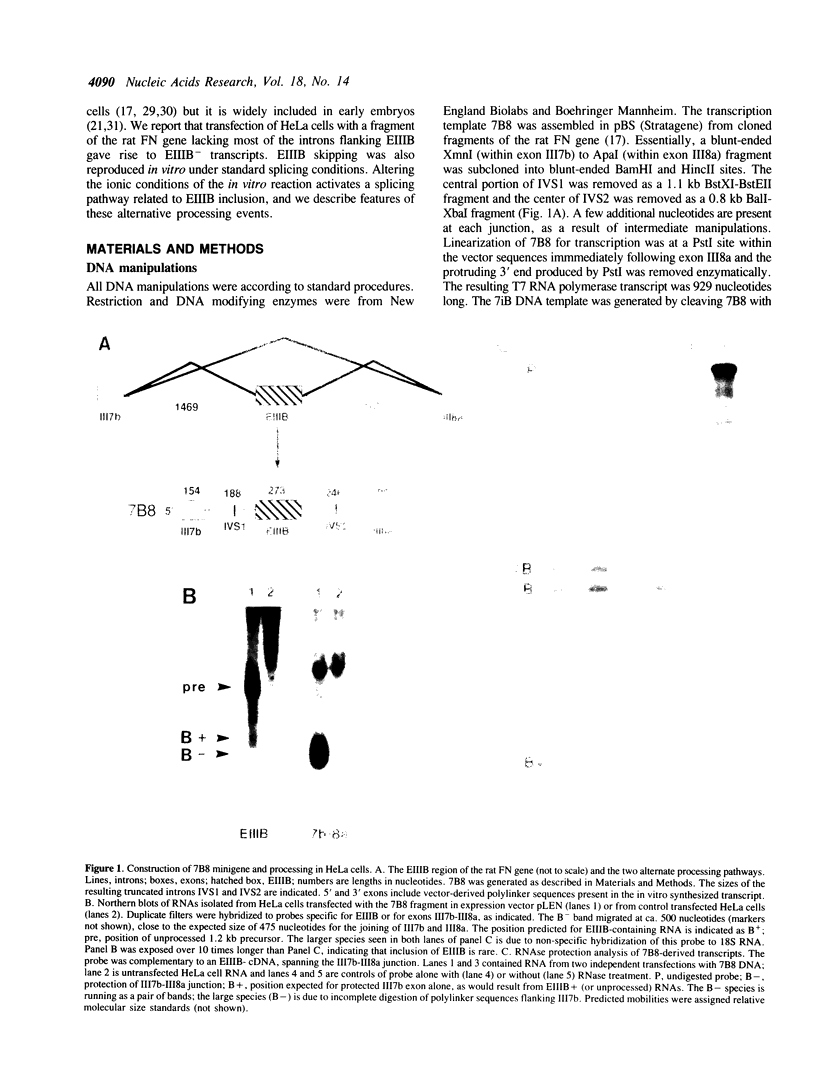
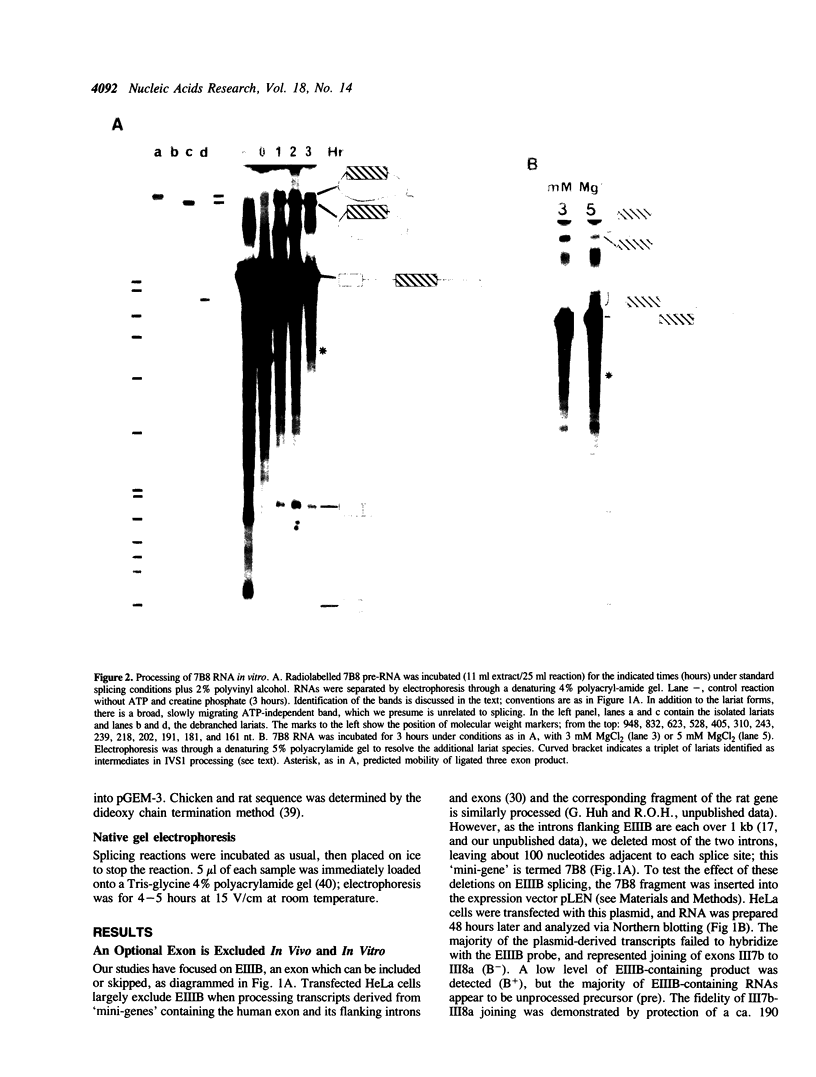
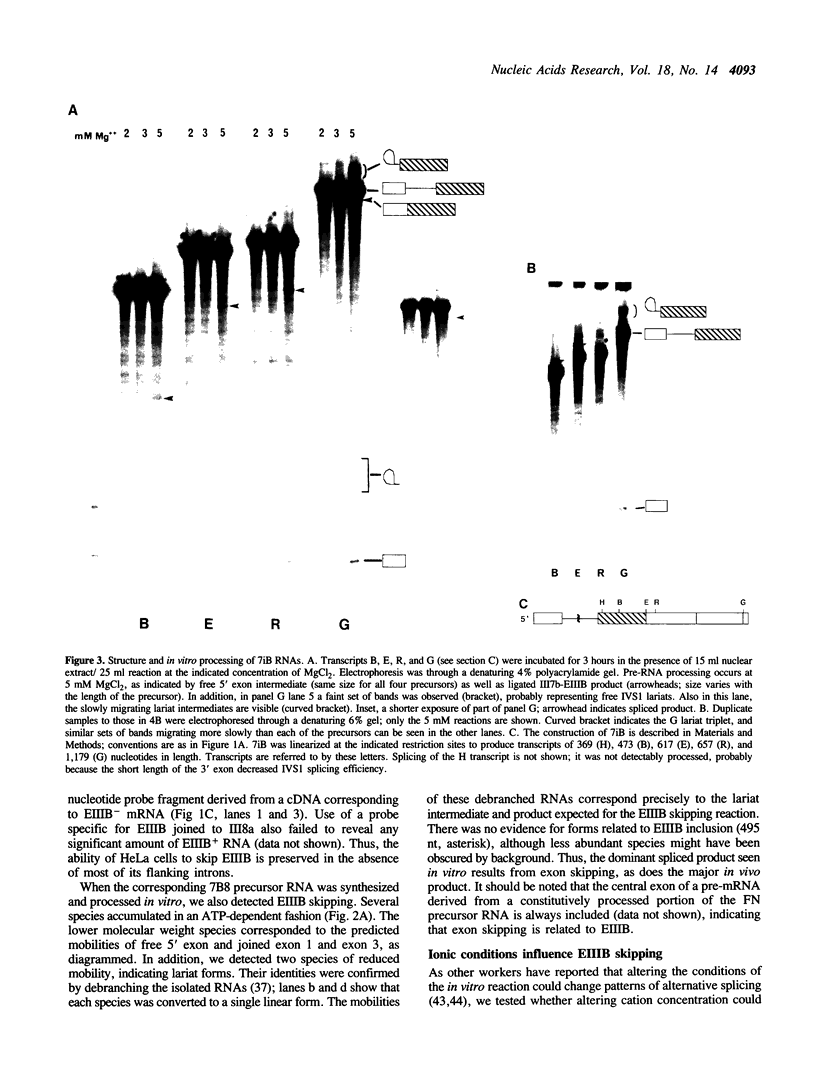
Abstract
We have investigated the alternative splicing of the EIIIB exon of the rat fibronectin gene. Mini-gene constructs containing this exon and portions of adjacent introns and exons, when transfected into HeLa cells, are transcribed and spliced, but omit the EIIIB exon. In vitro, HeLa nuclear extracts similarly splice out (skip) the EIIIB exon from similarly structured transcripts. Therefore, the HeLa splicing apparatus recognizes as atypical the EIIIB exon and its flanking intron sequences, both in vivo and in vitro. We also report that alterations in the ionic conditions of the in vitro splicing reaction can promote the initiation of EIIIB exon inclusion, as reflected by the formation of intermediate and product RNAs related to the removal of the intron upstream of EIIIB. Processing of this intron correlates with the formation of complexes resembling intermediates in spliceosome assembly. The branch sites involved in this alternative processing pathway are rather distant from the EIIIB 3' splice site, and lie within a region which is well conserved in the fibronectin genes of other species. Thus, the intron upstream of EIIIB shows singular structure and behavior which probably have a bearing on the regulated alternative splicing of this exon.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agris C. H., Nemeroff M. E., Krug R. M. A block in mammalian splicing occurring after formation of large complexes containing U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins. Mol Cell Biol. 1989 Jan;9(1):259–267. doi: 10.1128/mcb.9.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A., Gallego M. E., Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–242. doi: 10.1146/annurev.cb.03.110187.001231. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Barone M. V., Henchcliffe C., Baralle F. E., Paolella G. Cell type specific trans-acting factors are involved in alternative splicing of human fibronectin pre-mRNA. EMBO J. 1989 Apr;8(4):1079–1085. doi: 10.1002/j.1460-2075.1989.tb03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Cardone M. H., Ordahl C. P. Cis requirements for alternative splicing of the cardiac troponin T pre-mRNA. Nucleic Acids Res. 1988 Sep 12;16(17):8443–8465. doi: 10.1093/nar/16.17.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C., Hynes R. O. Alternative splicing of fibronectin is temporally and spatially regulated in the chicken embryo. Development. 1989 Jun;106(2):375–388. doi: 10.1242/dev.106.2.375. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C., Hynes R. O. Patterns of fibronectin gene expression and splicing during cell migration in chicken embryos. Development. 1988 Nov;104(3):369–382. doi: 10.1242/dev.104.3.369. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Gattoni R., Schmitt P., Stevenin J. In vitro splicing of adenovirus E1A transcripts: characterization of novel reactions and of multiple branch points abnormally far from the 3' splice site. Nucleic Acids Res. 1988 Mar 25;16(6):2389–2409. doi: 10.1093/nar/16.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Sharp P. A. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science. 1986 Sep 19;233(4770):1294–1299. doi: 10.1126/science.3638792. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Guan J. L., Trevithick J. E., Hynes R. O. Retroviral expression of alternatively spliced forms of rat fibronectin. J Cell Biol. 1990 Mar;110(3):833–847. doi: 10.1083/jcb.110.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman A., Kornblihtt A. R. Identification of a third region of cell-specific alternative splicing in human fibronectin mRNA. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7179–7182. doi: 10.1073/pnas.84.20.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., Finn L. A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988 Dec;2(12A):1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
- Hynes R. Molecular biology of fibronectin. Annu Rev Cell Biol. 1985;1:67–90. doi: 10.1146/annurev.cb.01.110185.000435. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rosbash M. RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5835–5839. doi: 10.1073/pnas.83.16.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Correct in vivo splicing of the mouse immunoglobulin kappa light-chain pre-mRNA is dependent on 5' splice-site position even in the absence of transcription. Genes Dev. 1988 Nov;2(11):1448–1459. doi: 10.1101/gad.2.11.1448. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Vibe-Pedersen K., Baralle F. E. Human fibronectin: cell specific alternative mRNA splicing generates polypeptide chains differing in the number of internal repeats. Nucleic Acids Res. 1984 Jul 25;12(14):5853–5868. doi: 10.1093/nar/12.14.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. Presplicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988 Sep;2(9):1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- Kubomura S., Obara M., Karasaki Y., Taniguchi H., Gotoh S., Tsuda T., Higashi K., Ohsato K., Hirano H. Genetic analysis of the cell binding domain region of the chicken fibronectin gene. Biochim Biophys Acta. 1987 Nov 20;910(2):171–181. doi: 10.1016/0167-4781(87)90070-4. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Sharp P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987 Aug;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lowery D. E., Van Ness B. G. In vitro splicing of kappa immunoglobulin precursor mRNA. Mol Cell Biol. 1987 Apr;7(4):1346–1351. doi: 10.1128/mcb.7.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Hamm J. Regulated splicing in early development and stage-specific U snRNPs. Development. 1989 Feb;105(2):183–189. doi: 10.1242/dev.105.2.183. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988 Mar;2(3):319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- Neufeld G., Mitchell R., Ponte P., Gospodarowicz D. Expression of human basic fibroblast growth factor cDNA in baby hamster kidney-derived cells results in autonomous cell growth. J Cell Biol. 1988 Apr;106(4):1385–1394. doi: 10.1083/jcb.106.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Hynes R. O. Alternative splicing of chicken fibronectin in embryos and in normal and transformed cells. Mol Cell Biol. 1987 Dec;7(12):4297–4307. doi: 10.1128/mcb.7.12.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. J., Kornblihtt A. R., Baralle F. E. Fibronectin, the generation of multiple polypeptides from a single gene. Oxf Surv Eukaryot Genes. 1986;3:141–160. [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Paolella G., Henchcliffe C., Sebastio G., Baralle F. E. Sequence analysis and in vivo expression show that alternative splicing of ED-B and ED-A regions of the human fibronectin gene are independent events. Nucleic Acids Res. 1988 Apr 25;16(8):3545–3557. doi: 10.1093/nar/16.8.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. S., Odermatt E., Schwarzbauer J. E., Hynes R. O. Organization of the fibronectin gene provides evidence for exon shuffling during evolution. EMBO J. 1987 Sep;6(9):2565–2572. doi: 10.1002/j.1460-2075.1987.tb02545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P., Gattoni R., Keohavong P., Stévenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987 Jul 3;50(1):31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Mulligan R. C., Hynes R. O. Efficient and stable expression of recombinant fibronectin polypeptides. Proc Natl Acad Sci U S A. 1987 Feb;84(3):754–758. doi: 10.1073/pnas.84.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Patel R. S., Fonda D., Hynes R. O. Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J. 1987 Sep;6(9):2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi K., Klos A. M., Kurachi K., Yoshitake S., Hakomori S. Human liver fibronectin complementary DNAs: identification of two different messenger RNAs possibly encoding the alpha and beta subunits of plasma fibronectin. Biochemistry. 1986 Aug 26;25(17):4936–4941. doi: 10.1021/bi00365a032. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibe-Pedersen K., Kornblihtt A. R., Baralle F. E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984 Nov;3(11):2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M., Zapp M. L., Berget S. M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988 Feb;8(2):814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]