Abstract
Immunoreceptor tyrosine based activation motif (ITAM)-coupled receptors play an essential role in regulating macrophage activation and function by cross-regulating signaling from heterologous receptors. We investigated mechanisms by which ITAM-associated receptors inhibit type I interferon (IFN-α/β) signaling in primary human macrophages and tested the effects of simultaneous ligation of ITAM-associated receptors and TLR4 on TLR4-induced Jak-STAT signaling that is mediated by autocrine IFN-β. Preligation of ITAM-coupled β2 integrins and FcγRs inhibited proximal signaling by the type I IFN receptor IFNAR. Cross-inhibition of IFNAR signaling by β2 integrins resulted in decreased Jak1 activation and was mediated by partial downregulation of the IFNAR1 subunit and MAPK-dependent induction of USP18, which blocks the association of Jak1 with IFNAR2. Simultaneous engagement of ITAM-coupled β2 integrins or Dectin-1 with TLR4 did not affect TLR4-induced direct activation of inflammatory target genes such as TNF or IL6, but abrogated subsequent induction of IFN response genes that is mediated by autocrine IFN-β signaling. Type I IFNs promote macrophage death after infection by Listeria monocytogenes. Consequently, attenuation of IFN responses by β2 integrins protected primary human macrophages from Listeria monocytogenes induced apoptosis. These results provide a mechanism for cross-inhibition of type I IFN signaling by ITAM-coupledβ2 integrins and demonstrate that ITAM signaling qualitatively modulates macrophage responses to PAMPs and pathogens by selectively suppressing IFN responses.
Keywords: monocytes/macrophages, cytokines, inflammation, TLRs, signal transduction
Introduction
Numerous receptors across different immune cell subsets signal through an immunoreceptor tyrosine-based activation motif (ITAM). ITAMs are present within a receptor’s cytoplasmic tail or in ITAM-containing adaptors that associate with various cell surface receptors. In myeloid cells, the two major ITAM-containing adaptors are FcRγ and DNAX activation protein-12 (DAP12) that associate with approximately 20 different receptors (1). The ITAM contains two tyrosine residues which, upon phosphorylation, form a binding site for SH2-domain containing kinases such as Syk. Recruitment and activation of Syk leads to activation of downstream pathways and effector molecules, including MAPKs, Rho-mediated cytoskeletal rearrangement, calcium signaling, generation of reactive oxygen species, and activation of inflammosomes and NF-κB. Initially, ITAM-coupled receptors were thought to primarily initiate activating signals, but it is now established that ITAM-coupled receptors also mediate inhibitory signaling by cross-regulating various receptors including TLRs and cytokine and chemokine receptors (2). Mechanisms by which ITAMs cross-inhibit signaling include direct modulation of heterologous receptors by ITAM-associated phosphatases or kinases, or indirect crossregulation by induction of signaling inhibitors such as SOCS3 and A20 (3–6)
The ligands of many myeloid ITAM-coupled receptors are not known. The known ligands include endogenous factors expressed at sites of inflammation, and pathogen-associated molecular patterns (PAMPs) expressed by microbial pathogens. Endogenous inflammatory ligands that engage ITAM-coupled receptors include immune complexes that engage Fc receptors (FcRs) and complement degradation products and fibrin(ogen) that engage ITAM-coupled β2 integrins. Although no direct association of integrins with ITAM-containing adaptors has been reported, genetic evidence shows that ITAM-containing FcRγ and DAP12 mediate signaling byβ2 and β3 integrins (7, 8); thus, β2 integrins are functionally coupled with ITAM-associated adaptors and downstream signaling pathways. Products of necrotic cells can engage Mincle (CLEC4E) or CLEC9A (9, 10); CLEC9A contains a hemITAM, an ITAM-like motif containing only one tyrosine that utilizes similar signaling pathways as ITAMs. Various bacterial and fungal PAMPs engage ITAM-associated or hemITAM-containing C-type lectin receptors, such as Dectin-1, Dectin-2 and CLEC4E that are involved in pathogen sensing (11). Thus, ITAM-coupled receptors are engaged during innate immune responses and cooperate with other microbial sensing receptors, such as TLRs, in determining the magnitude and qualitative nature of responses to pathogens in the context of an inflammatory microenvironment. ITAM-associated receptors can either augment or suppress TLR responses (12–21), depending on context and timing of receptor ligation, and thus have been proposed to fine-tune innate and inflammatory responses.
The induction of type I IFNs (IFN-α/β) is an important aspect of early innate responses to viruses and bacteria. Type I IFN production is elicited by pattern recognition receptors (PRRs) that sense microbial nucleic acids, and also by TLR4 that senses bacterial lipopolysaccharides and by TLR2 sensing of viruses in a cell-type and context dependent manner. Type I IFNs are key inducers of a cellular antiviral state and thus important mediators of host defense against viruses. In contrast, type I IFNs can be either protective or detrimental in host defense against bacteria such as Listeria monocytogenes and Francisella tularensis, and their role in immunity against fungi is mostly unexplored (22). Type I IFNs signal via a heterodimeric receptor IFNAR, which is composed of IFNAR1 and IFNAR2 subunits. Ligation of IFNAR results in activation of the receptor-associated tyrosine kinases, Jak1 and Tyk2, which in turn leads to phosphorylation and activation of STAT1 and STAT2. Activated STAT1 and STAT2 associate with IRF9 to form the ISGF3 complex that activates antiviral genes via ISRE DNA elements. IFNAR signaling also activates STAT1 homodimers that induce expression of inflammatory STAT1 target genes similar to those induced by IFN-γ. A key aspect of activation of myeloid cells by nucleic acid-sensing PRRs or TLR4 is induction of an autocrine loop mediated by production of and signaling by endogenous IFN-β. Thus, PRR signaling directly and rapidly induces expression of inflammatory genes including IFNB, followed by a more delayed induction of IFN response genes by autocrine-acting IFN-β. The IFN response genes promote an antiviral cell state, and they also modulate inflammatory and antigen-presenting functions by inducing expression of chemokines such as CXCL9 and CXCL10 and costimulatory molecules such as CD80 and CD86, modulate suppressive aspects of macrophage function (23–25), and promote apoptosis and death of infected macrophages (22, 26).
Our laboratory and others have recently described several mechanisms by which preligation of ITAM-associated receptors inhibits TLR signaling (4, 5). During the course of these previous studies, we found that preligation of ITAM-associated β2 integrins also inhibited signaling by exogenously added IFN-α. In the current study, we investigated mechanisms by which ITAM-coupledβ2 integrins inhibit IFNAR signaling and tested the effects of simultaneous ligation of ITAM-associated receptors and TLR4 on the TLR4-induced IFN-β mediated autocrine loop. We found that preligation of ITAM-coupled β2 integrins by fibrinogen (Fb) inhibited proximal IFNAR signaling by modestly downregulating cell surface IFNAR1 expression but nearly completely suppressing IFN-α-induced Jak1 tyrosine phosphorylation. Cross-inhibition of IFNAR signaling developed in a time-dependent manner over several hours and was characterized by MAPK-dependent induction of inhibitors of IFNAR-Jak-STAT signaling, including SOCS3 and USP18. Consistent with a delayed inhibitory mechanism, simultaneous engagement of β2 integrins or Dectin-1 with TLR4 did not affect TLR4-induced direct signaling that activates inflammatory cytokine genes such as TNF and IL6, but instead abrogated the subsequent development of the autocrine IFN response that modulates cell function and promotes cell death in response to certain pathogens. Accordingly,β2 integrin-mediated attenuation of the IFN response to infection with Listeria monocytogenes protected primary human macrophages from IFN-induced apoptosis. Overall, our results delineate a mechanism for cross-inhibition of type I IFN signaling by ITAM-coupledβ2 integrins, and show that ITAM-associated receptors modulate the functional outcome of PAMP and pathogen sensing by macrophages by preferentially suppressing IFN responses.
Materials and Methods
Cell culture and reagents
Peripheral blood mononuclear cells (PBMC) obtained from blood leukocyte preparations purchased from the New York Blood Center were separated by density gradient centrifugation with Ficoll (Invitrogen, Carlsbad, CA, USA) using a protocol approved by the Hospital for Special Surgery Institutional Review Board. Human monocytes were purified from PBMCs immediately after isolation by positive selection with anti-CD14 magnetic beads, as recommended by the manufacturer (Miltenyi Biotec, Auburn, CA, USA). Monocytes were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% low IgG FBS (Invitrogen), penicillin/streptomycin (Invitrogen), L-glutamine (Invitrogen), and 15 ng/mL human macrophage colony-stimulating factor (M-CSF; Peprotech). Monocyte-derived macrophages obtained after 2 days of culture with human M-CSF were used, and purity of monocytes/macrophages was >97%, as verified by flow cytometric analysis. For stimulation with Fb, macrophages were harvested and added to pre-coated tissue culture plates; plates were coated overnight with either 10% FBS culture media as control or with Fb (Sigma Aldrich, St. Louis, MO, USA, fibrinogen #F9754;10μg/100μL in PBS, using 100–400μL depending on well size) and washed with 1x PBS before cells were added. Cells were adherent on both control and Fb-coated wells. Polymyxin B (14 μg/ml; Sigma Aldrich), which we verified did not directly stimulate macrophages but essentially completely blocked exogenous LPS up to concentrations of 10 ng/ml in our system, was used to ensure that contaminating endotoxin did not contribute to the effects observed, as previously described (4). Polymyxin B was used throughout except as noted in the text. For stimulation with IgG, plates were coated with 20 μg/mL of purified human IgG (Sigma Aldrich). For stimulation with α-CD11b (clone M1/70; Millipore, Temecula, CA, USA), plates were coated with 5 μg/mL ofα-CD11b or IgG2b isotype control (eBioscience, San Diego, CA, USA). The following ligands were used to stimulate cells: recombinant human IFN-α 2a (5–1000 U/mL; PBL Interferon Source, Piscataway, NJ, USA), LPS (10 ng/mL; Invivogen, San Deigo, CA), and zymosan (100 μg/mL; Invivogen). The following pharmacological inhibitors were used from CalBiochem (San Diego, CA, USA): SB203580 and U0126 at 15μM, GF109203X at 10μM, MG-132 at 20μM, piceatannol at 80 μM, and phosphatase inhibitor sodium stibogluconate at 10 and 100 μg/mL (27).
Immunoblot analysis
Whole-cell extracts were obtained, and protein amounts quantitated with the Bradford assay (BioRad, Hercules, CA, USA). For immunoblotting, cell lysates (10 μg) were fractionated on 7.5% or 10% polyacrylamide gels using SDS-PAGE and transferred to polyvinylidene difluoride membranes for probing with Ab. ECL was used for detection. The following polyclonal Abs were used: pY-Jak1 (Y1022/1023) (Invitrogen), pY-STAT1 (Y701) (Cell Signaling Technology, Danvers, MA, USA), SOCS3 (Cell Signaling), and p38α (C-20) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The following monoclonal Abs were used: Jak1 (BD Transduction Laboratories, Lexington, KY, USA), STAT1 (E-23) (Santa Cruz Biotechnology), and USP18 (Cell Signaling).
ELISA
Human IFN-β was measured using the Verikine ELISA kit from PBL Interferon Source following the manufacturer’s instructions.
mRNA isolation and real-time quantitative PCR (qPCR)
RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD, USA) following the manufacturer’s instructions. Quantitative real-time PCR was performed in triplicate using Fast SYBR Green Master Mix and a 7500 Fast Real-Time cycler (Applied Biosystems, Foster City, CA, USA). Expression was normalized relative to levels of GAPDH. Oligonucleotide primers for human transcripts were as follows: CXCL9: 5′-ATCAGCACCAACCAAGGGACT-3′ and 5′-GCTTTTTCTTTTGGCTGACCTG-3′; CXCL10: 5′-ATTTGCTGCCTTATCTTTCTG-3′ and 5′-TCTCACCCTTCTTTTTCATTGTAG-3′; IFIT1: 5′-TTCGGAGAAAGGCATTAGA-3′ and 5′-TCCAGGGCTTCATTCATAT-3′; MX1: 5′-AGCCACTGGACTGACGACTT-3′ and 5′-ACCACGGCTAACGGATAAG-3′; SOCS3: 5′-CACTCTTCAGCATCTCTGTCGGAAG-3′ and 5′-CATAGGAGTCCAGGTGGCCGTTGAC-3′; USP18: 5′-CGGAACTTCGGTCCCAG-3′ and 5′-TCAGGACAGCACGACTTCAC-3′ (28); TNF: 5′-AATAGGCTGTTCCCATGTAGC-3′ and 5′-AGAGGCTCAGCAATGAGTGA -3′; IL1B: 5′-TTCTTCGACACATTGGATAACG-3′ and 5′-TGGAGAACACCACTTGTTGCT-3′; IL10: 5′-TTATCTTGTCTCTGGGCTTGG-3′ and 5′-GTTGGGGAATGAGGTTAGGG-3′; IFNB: 5′-GAGCTACAACTTGCTTGGATTCC-3′ and 5′-CAAGCCTCCCATTCAATTGC-3′; INDO: 5′-TTAGAGTCAAATCCCTCAGTCC-3′ and 5′-TTTGCAGATGGTAGCTCCTC-3′; GADPH: 5′-ATCAAGAAGGTGGTGAAGCA-3′ and 5′-GTCGCTGTTGAAGTCAGAGGA-3′
RNA interference
Primary human monocytes were nucleofected immediately after isolation with On-Target plus SMARTpool short interfering RNAs (siRNA) purchased from Dharmacon Inc. (Lafayette, Colorado, USA) specific for SOCS3 or USP18. Non-targeting siRNA #5 was used as control. Cells were nucleofected using the Human Monocyte Nucleofector Kit and the AMAXA Nucleofector System according to the manufacturer’s instructions (Lonza Cologne, Cologne, Germany). Cells were harvested 3 days after nucleofection for stimulation.
L. monocytogenes infection and CFU analysis
Purified human monocytes were isolated and cultured as described in the “Cell Culture” section except antibiotics were omitted from the media. After 2 days of culture, differentiated macrophages were plated onto Fb-coated wells or control FBS-coated wells for 18 h. Cells were plated in 12-well plates (RNA analysis) or 96-well plates (flow cytometric analysis) at a density of 2×106 per mL. Triplicate wells per condition were used for analysis of apoptosis and cell death. Cell culture media was changed prior to infection with L. monocytogenes wildtype strain 10403S, which was provided by Eric Pamer’s laboratory, and was grown to logarithmic phase in brain-heart infusion (BHI) broth. Bacteria were opsonized in RPMI media containing 50% heat inactivated human serum (Sigma Aldrich) at 37°C for 30 min, washed with PBS, and then added to cells. Macrophages were infected at MOIs of 5–20, and gentamicin sulfate (50ug/mL; Sigma Aldrich) was added to cultures 30 min later to prevent growth of extracellular bacteria. At the times indicated in figure legends, macrophages were harvested and analyzed by flow cytometry and colony forming units (CFUs) were measured by lysing cells in 0.05% Triton X-100 PBS and plating cell lysates containing viable intracellular bacteria onto BHI agar plates, as previously described (29). Bacterial colonies were counted after 24 h incubation.
Flow cytometry
To detect IFNAR expression, the following Abs were used after FcRs on the cell surface were blocked using human FcR Blocking Reagent (Miltenyi Biotec): PE-conjugated mouse α-IFNAR1 and α-IFNAR2 (PBL Interferon Source), PE-conjugated isotype controls against mouse IgG1 and IgG2a (BD Pharmingen, San Diego, CA, USA). Apoptosis and cell death were measured using the Annexin V FITC Apoptosis Detection Kit I purchased from BD Pharmingen and DAPI nuclear stain from Invitrogen.
Statistical analysis
All statistical analyses were performed with Graphpad Prism 5.0 software using the 2-tailed, paired Student t test.
Results
Ligation of ITAM-associated β2 integrins and FcγRs inhibits proximal IFNAR signaling
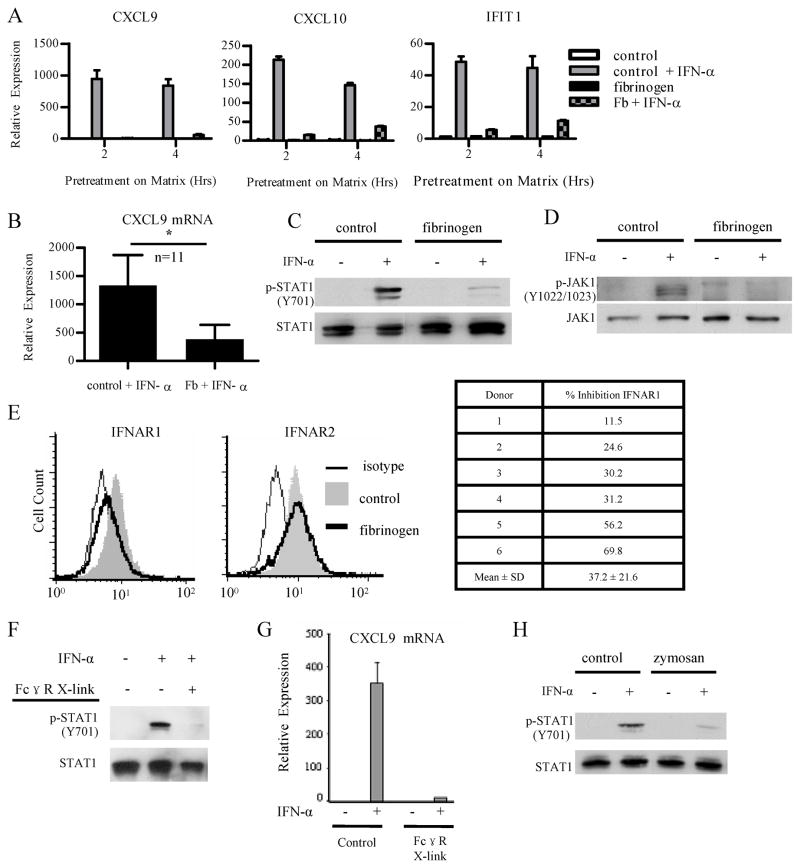
We wished to extend our previous observation that ITAM-coupled β2 integrins can inhibit type I IFN signaling by identifying the level in the IFN signaling pathway where inhibition occurs. We pre-ligated β2 integrins in primary human monocyte-derived macrophages using plate-bound fibrinogen (Fb) as previously described (4) and then added exogenous IFN-α and assessed induction of IFN response genes, Jak-STAT signaling, and IFNAR cell surface expression. Control cells plated on FBS-coated plates exhibited a clear induction of the IFN response genes CXCL9, CXCL10 and IFIT1 after addition of IFN-α, while induction of these genes in Fb-stimulated cells was strongly suppressed (Fig. 1A). Preligation of β2 integrins strongly and consistently suppressed IFN-α-induced expression of STAT1 target genes such as CXCL9 in multiple experiments with different blood donors (Fig. 1B shows cumulative data from 11 independent blood donors).
Figure 1. Ligation of ITAM-associated β2 integrins and FcγRs inhibits proximal IFNAR signaling.
Primary human monocyte-derived macrophages were plated onto control FBS-coated wells, fibrinogen (Fb) coated wells, or IgG coated wells for the indicated times, and then IFN-α (1000 U/mL) was added for an additional 2 h to assess induction of IFN-α response genes (A,B,G) or 15 min to assess phosphorylation of Jak1 and STAT1 (C,D,F).
A, Macrophages were plated for 2 and 4 h prior to addition of IFN-α. mRNA expression was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA. Data are representative of at least 3 independent experiments.
B, Cumulative data from macrophages plated for 2 h prior to addition of IFN-α. mRNA expression was measured by qPCR and normalized relative to GAPDH mRNA. Results are presented as mean ± SEM of 11 independent donors. Statistical analysis was performed using the 2-tailed, paired Student t test, p = 0.0203.
C and D, Macrophages were plated for 6 h prior to addition of IFN-α. Whole cell lysates were immunoblotted with Abs against phospho-STAT1 (Y701) and STAT1 (C) and Abs against phospho-JAK1 (Y1022/1023) and JAK1 (D). Data are representative of at least 3 independent experiments.
E, Cell surface expression of both subunits of the type I IFN receptor was measured by flow cytometry. Macrophages were plated for 3 or 4 h. Flow cytometry data from an experiment showing strong downregulation of IFNAR1 are shown (top panels) and cumulative data from 6 independent experiments are shown in the table (bottom). Percent inhibition was calculated by subtracting isotype control staining and comparing IFNAR1 MFI of control and Fb-stimulated cells.
F, Macrophages were plated for 1.5 h on control or IgG coated wells prior to addition of IFN-α. Whole cell lysates were immunoblotted with Abs against phospho-STAT1 (Y701) and STAT1. Data are representative of at least 3 independent experiments.
G, Macrophages were plated for 1.5 h on control or IgG coated wells prior to addition of IFN-α. mRNA expression was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA. Data are representative of at least 3 independent experiments.
H, Macrophages were stimulated for 4 h with zymosan (10 ug/mL) prior to addition of IFN-α. Whole cell lysates were immunoblotted with Abs against phospho-STAT1 (Y701) and STAT1. Data are representative of 3 independent experiments.
We next examined the effects of β2 integrin preligation on IFN-induced Jak-STAT signaling. To check activation of STAT1, we measured p-STAT1 (Y701) after IFN-α stimulation of control or Fb-treated cells. β2 integrin ligation inhibited activation of STAT1 in response to subsequent IFN-α stimulation (Fig 1C); comparable results were obtained in more than 15 independent experiments with different blood donors. Diminished STAT1 tyrosine phosphorylation could be explained by decreased upstream signaling by Jaks or by increased dephosphorylation of STAT1. We addressed this issue by assessing IFN-α-induced activation of Jak1 by measuring p-Jak1 (Y1022/1023) levels. Preligation ofβ2 integrins consistently and nearly completely inhibited IFN-α-induced Jak1 activation (Fig 1D), indicating that Fb-mediated inhibition occurs upstream of STAT1. We next tested if diminished Jak1 activation could be explained by a decrease in cell surface IFNAR expression. Preligation of β2 integrins did not affect cell surface IFNAR2 expression, but did induce a partial downregulation of cell surface IFNAR1 (Fig. 1E, left). However, the extent of IFNAR1 downregulation varied among donors (Fig. 1E, chart) and did not strictly correlate with the extent of signaling inhibition. Thus, although downregulation of cell surface IFNAR1 expression may contribute to suppression of IFN-α-induced signaling, such downregulation is insufficient to fully explain the observed block in IFN responses. Our data show that stimulation with fibrinogen suppresses IFNAR signaling by inhibiting Jak activation.
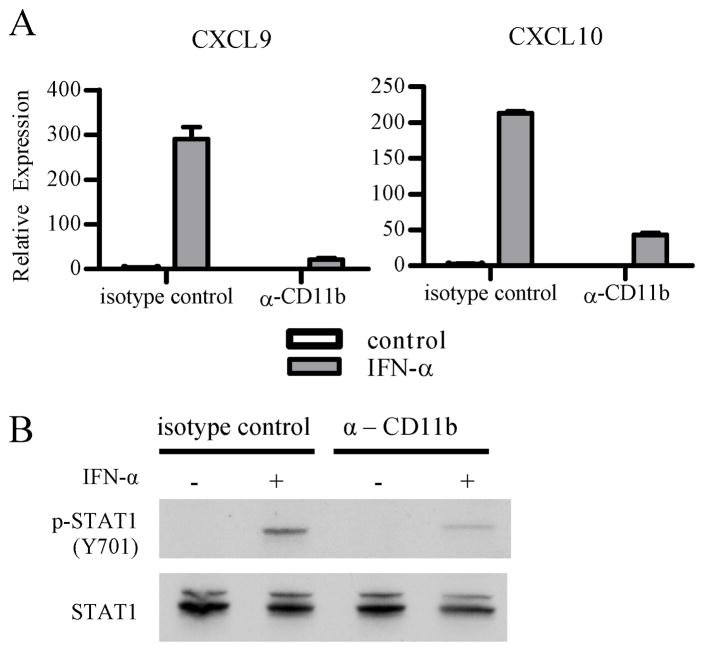
In addition to DAP12-mediated signaling, β2 integrins can activate non-ITAM dependent signaling pathways. Thus, to provide additional support for inhibition of IFNAR responses by ITAM signaling, we used immune complexes that signal via FcγRs and the associated ITAM-containing FcRγ adaptor protein. Preligation of FcγRs effectively suppressed IFN-α-induced STAT1 activation and downstream gene expression (Fig. 1F and 1G). In addition, zymosan, which activates hemi-TAM signaling by ligating Dectin-1, suppressed IFN-α-induced STAT1 activation (Fig 1H). As specificity controls, the TLR2 ligand Pam3Cys had modest and inconsistent effects on IFNAR signaling (data not shown) and the TLR4 ligand LPS actually induced IFNAR signaling (see below). To further support a role for ITAM signaling in inhibition of IFN responses, we tested the effects of inhibiting Syk, a key mediator of ITAM signaling. Indeed, inhibition of Syk substantially attenuated the ability of Fb to inhibit IFN-induced gene expression (Supp. Fig. 1). To further implicate a role for β2 integrins in inhibition of IFN signaling, we specifically ligated αMβ2 integrins, comprised of CD11b (αM) and CD18 (β2) subunits, using CD11b antibodies. In contrast to the isotype control, preligation of CD11b suppressed IFN-α-induced gene expression and STAT1 activation (Fig. 2A and 2B). Overall the data show that myeloid FcγRs and β2 integrins ligated by endogenous factors present at sites of inflammation cross-inhibit IFNAR signaling.
Figure 2. Ligation of β2 integrins by CD11b antibodies inhibits IFN-α signaling.
Primary human macrophages were plated onto control IgG2b coated wells or anti-CD11b coated wells.
A, Macrophages were plated for 2 h and then IFN-α (50 U/mL) was added for an additional 2 h to assess induction of IFN-α response genes. mRNA expression was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA. Data are representative of 3 independent experiments.
B, Macrophages were plated for 4 hr and then IFN-α (500 U/mL) was added for an additional 15 min to assess phosphorylation of STAT1. Whole cell lysates were immunoblotted with Abs against phospho-STAT1 (Y701) and STAT1. Data are representative of 3 independent experiments.
β2integrin ligation induces negative regulators of IFN-α signaling in a MAPK-dependent manner
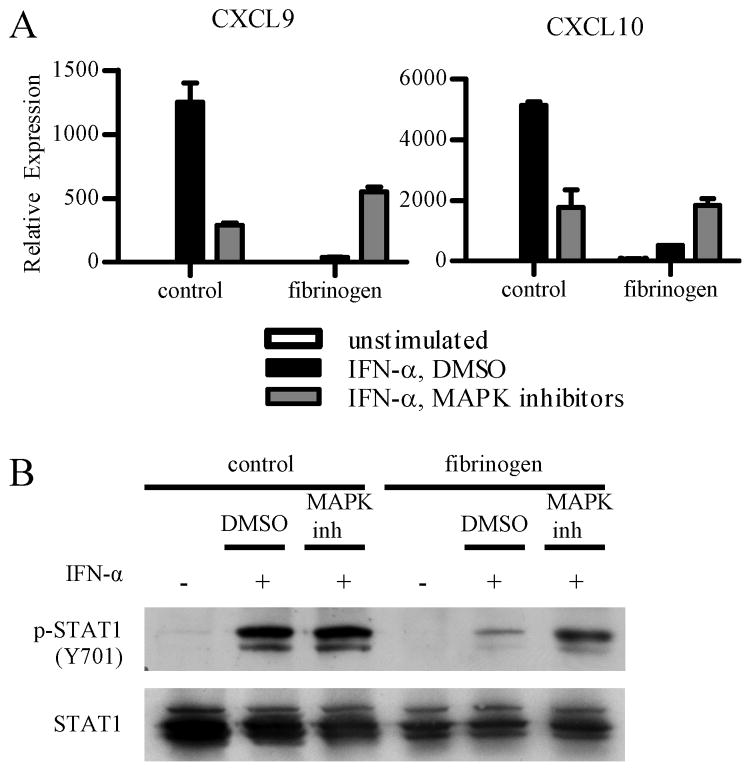
We wished to identify the β2 integrin-induced signaling pathway that inhibits IFNAR responses. ITAM signaling leads to downstream activation of MAPKs, which have been implicated in modulation of cytokine signaling in other systems (21). Thus, we tested whether inhibition of MAPKs would reverse Fb-induced suppression of IFNAR responses. Inhibition of ERK or p38 activation alone had minimal effects (data not shown), and we next tested the combination of SB203580 and U0126, which respectively inhibit p38 and MEK (thus preventing activation of ERKs). Primary human macrophages were treated with both inhibitors and plated onto control or Fb coated wells. IFN-α was added and gene induction and activation of STAT1 were measured. As expected, combined inhibition of p38 and ERK partially attenuated the robust induction of CXCL9 and CXCL10 by IFN-α on control plates (Fig. 3A), while Fb alone strongly suppressed gene expression (Fig. 3A, bar 5). Inhibition of p38 and ERK in Fb-stimulated cells restored CXCL9 and CXCL10 expression (Fig. 3A, bar 6). Although gene expression was not fully restored to the maximal levels of induction by IFN-α in control cells (Fig. 3A, bar 2), it was restored to the level observed in IFN-α-stimulated control cells in which MAPKs were inhibited (Fig. 3A, bar 3), which corresponds to a ceiling on the level of gene induction when MAPKs are inhibited. In parallel with restoration of gene expression, inhibition of p38 and ERK restored IFN-α-induced STAT1 tyrosine phosphorylation in Fb-inhibited macrophages (Fig. 3B, lanes 5 and 6). Collectively, our results show that β2 integrins inhibit type I IFN signaling in a MAPK-dependent manner.
Figure 3. β2 integrin inhibition of IFN-α signaling is mediated by a p38 and ERK-dependent pathway.
Primary human macrophages were pretreated with either the vehicle control DMSO or two MAPK inhibitors: p38 inhibitor SB203580 (15 μM) and MEK inhibitor U0126 (15 μM) for 30 min. Macrophages were then plated onto control wells or fibrinogen (Fb) coated wells, and IFN-α (1000 U/mL) was added for an additional 2 h to assess induction of IFN-α response genes (A) or 15 min to assess phosphorylation of STAT1 (B).
A, Macrophages were plated for 2 h prior to addition of IFN-α. mRNA expression was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA. Data are representative of at least 3 independent experiments.
B, Macrophages were plated for 4 h prior to addition of IFN-α. Whole cell lysates were immunoblotted with Abs against phospho-STAT1 (Y701) and STAT1. Data are representative of 2 independent experiments.
Previous work from our lab showed that pharmacological activation of PKC by PMA rapidly (within minutes) inhibited IFNAR signaling by a mechanism that was independent of MAPKs but dependent on PKC-mediated recruitment of SHP-2 to IFNAR (30). The more physiological cross-inhibition of IFNAR signaling by β2 integrins observed in the current study showed very different kinetics, as inhibition increased over several hours after Fb stimulation (Supp Fig. 2). Although the difference in kinetics suggested a distinct inhibitory mechanism, we tested the role of PKC and SHP-2 in Fb-induced inhibition of IFN responses. Consistent with our prediction, inhibition of PKC and SHP-2 (and SHP-1) had no effect on Fb-mediated inhibition of IFNAR responses (Supp. Fig. 3A and 3B). Thus, the mechanism of delayed MAPK-dependent inhibition of IFNAR signaling by Fb differs from the rapid and direct PKC- and SHP-2-dependent inhibitory mechanism activated by PMA.
Given the delayed kinetics of Fb-mediated inhibition, we reasoned that Fb induced expression of inhibitors of signaling, and that this induction occurs in a MAPK-dependent manner. Our attempts to directly demonstrate a requirement for induction of inhibitory proteins by Fb were not successful, as use of cycloheximide to inhibit de novo synthesis of inhibitors resulted in a rapid diminution of IFNAR signaling in control cells (likely secondary to diminished IFNAR protein expression) (data not shown). This precluded our ability to measure reversal of Fb-mediated inhibition when protein synthesis was blocked. Instead, we used microarray analysis in an attempt to identify Fb-induced inhibitors of IFNAR signaling. Fb stimulation of primary human macrophages induced expression of several molecules that can inhibit Jak-STAT signaling, including SOCS3, SOCS2, SOCS1, PTPN1, PTPN2, and USP18. As SOCS proteins are the best established inhibitors of Jak-STAT signaling (31), SOCS3 has been implicated in inhibition of IFNAR signaling, and induction of SOCS3 is dependent on MAPKs in other systems, we first tested the role of SOCS3 in Fb-induced inhibition of IFNAR signaling. We confirmed that induction of SOCS3 was MAPK-dependent in our system (Supp. Fig. 4A), but found that RNAi-mediated knockdown of SOCS3 had minimal effects on Fb-mediated suppression of IFN-α-induced gene expression (Supp. Fig. 4B). These results suggest that SOCS3 does not play a nonredundant role in Fb-mediated suppression of IFNAR signaling, possibly because of very low expression of SOCS3 protein in human macrophages (4). In addition, IL-10, which contributes to induction of SOCS3 expression, did not play a detectable role in mediating Fb-induced suppression of IFN-α signaling, as IL-10 neutralizing and IL-10 receptor blocking antibodies had no effect (4).
We next tested the role of USP18, another negative regulator of IFN signaling. USP18 was originally identified as a protease which cleaves ISG15, a ubiquitin-like molecule, from conjugated proteins (32). Cells from USP18 null mice exhibit enhanced Jak-STAT signaling and are hyperresponsive to IFN-β (33, 34). Inhibition of IFN responses by USP18 is independent of cleavage of ISG15; instead, USP18 binds to IFNAR2 and prevents its association with Jak1 (35, 36). Fb induced expression of USP18 mRNA and protein, and this induction was dependent on MAPKs (Fig. 4A and 4B). Fb-mediated induction of USP18 was abrogated when Syk was inhibited (Fig. 4C), further supporting a role for ITAM signaling in downregulation of IFN responses. We then used USP18-specific siRNA to knock down USP18 expression in primary human macrophages. Efficient knockdown of USP18 expression (Fig. 4D) resulted in partial restoration of IFN-α-induced gene expression (Fig. 4E). For statistical analysis of pooled data from 3 independent siRNA experiments, mRNA expression in control wells transfected with control siRNA and stimulated with IFN-α was set as 100%, and expression in all other conditions was set relative to that control (Fig. 4F). Statistically significant suppression of CXCL9 expression by Fb in control wells (p = 0.0175, paired Student t test) was lost when USP18 expression was knocked down (p = 0.1963, paired Student t test) (Fig. 4F). The partial effect of USP18 RNAi on restoration of the IFN response may be related to incomplete suppression of USP18 expression, but also likely reflects redundancy in inhibition of IFNAR responses by several Fb-induced inhibitors of cytokine signaling (4). Our data suggests that β2 integrin-induced USP18 plays a partial role in inducing a refractory state to type I IFNs, which is likely mediated by the coordinated action of several signaling inhibitors. The known suppression of IFNAR-Jak1 interactions by USP18 (36) can explain the diminished IFN-α-induced activation of Jak1 that was observed in Fb-treated macrophages.
Figure 4. β2integrin ligation by fibrinogen induces USP18, a MAPK-dependent negative regulator of IFN-α signaling.
Primary human macrophages were plated onto control wells or fibrinogen (Fb) coated wells.
A, Cumulative data from macrophages plated for 4 h. mRNA expression was measured by qPCR and normalized relative to GAPDH mRNA. Results are presented as mean ± SEM of 6 independent donors. Statistical analysis was performed using the 2-tailed, paired Student t test, p = 0.0198.
B, Macrophages were pretreated with either the vehicle control DMSO or two MAPK inhibitors: p38 inhibitor SB203580 (15 μM) and MEK inhibitor U0126 (15 μM) for 30 min. Macrophages were plated for 4 h, and IFN-α (1000 U/mL) was added for 15 min. Whole cell lysates were immunoblotted with Abs against USP18 and p38α. Data are representative of 2 independent experiments.
C, Macrophages were pretreated with either the vehicle control DMSO or Syk inhibitor piceatannol (80 μM) for 30 min. Macrophages were plated for 4 h, and IFN-α (1000 U/mL) was added for 15 min. Whole cell lysates were immunoblotted with Abs against USP18 and p38α. Data are representative of at least 3 independent experiments.
D – E, Macrophages were nucleofected with non-specific (control) or USP18-specific short interfering RNAs (siRNA), and 3 days later were plated onto control or Fb-coated wells and stimulated with IFN-α for 2 hrs. D, Immunoblot shows knockdown efficiency. E, mRNA expression was measured by qPCR and normalized relative to GAPDH mRNA. Error bars represent SD of triplicate wells, and data are representative of 3 independent experiments. F, Cumulative data from 3 donors is presented as mean ± SEM. Transcript expression in IFN-α-stimulated control cells was set as 100%, and expression in all other conditions was set relative to that control. * p = 0.0175, paired Student t test; NS = not significant (p = 0.1963).
Concurrent ligation of ITAM-coupled receptors and TLR4 inhibits the TLR4-induced autocrine IFN response
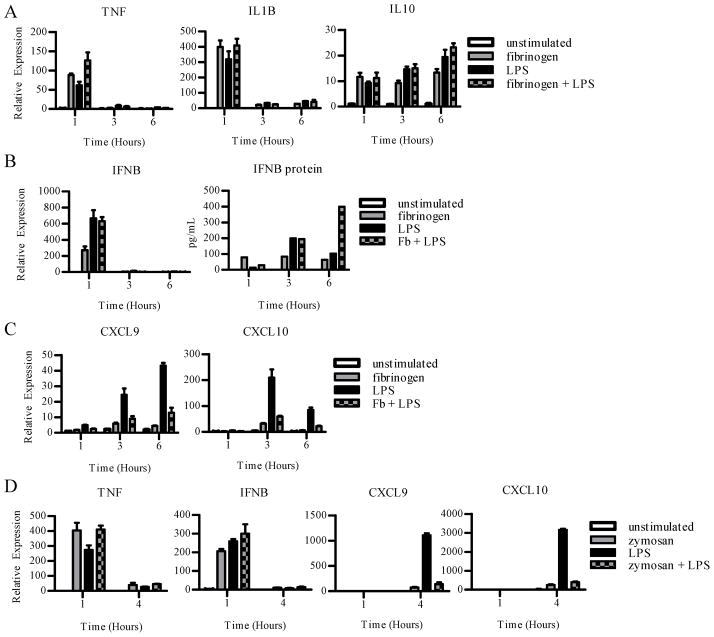
Previous work has shown that preligation of ITAM-coupled receptors globally inhibits TLR4 signaling (4). However, at sites of innate and inflammatory responses, macrophages are often exposed to ligands of ITAM-associated receptors and TLRs concurrently. Thus, we wished to test the effects of simultaneous engagement of ITAM-associated receptors on TLR4 responses, and whether ITAM signaling would have differential effects on TLR-induced direct signaling via NF-κB versus indirect signaling via autocrine IFN-β. We first tested the effects of simultaneous stimulation of primary human macrophages with Fb and LPS, which mimics concurrent stimulation of TLR4 and β2 integrins by, respectively, microbial products and fibrin(ogen) and complement split products at sites of infection or inflammation. Stimulation of human macrophages with LPS or Fb alone induced expression of direct NF-κB target genes TNF and IL1B, and concurrent stimulation with LPS and Fb showed minimal differences relative to stimulation with either ligand alone (Fig. 5A). Consistent with previous reports (37), there was a trend towards increased IL10 gene expression after concurrent LPS and Fb stimulation (Fig. 5A). These results show that concurrent ligation ofβ2 integrins does not alter the core TLR4-mediated inflammatory program, although these stimuli may synergize when used at lower concentrations (21).
Figure 5. Concurrent ligation of ITAM-coupled receptors and TLR4 inhibits induction of the TLR4-induced IFN-β-mediated autocrine loop.
Primary human macrophages were added to control or fibrinogen (Fb) coated wells with or without LPS (10ng/mL) (A–C). White bars represent unstimulated controls cells at each timepoint. mRNA expression was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA.
A, Expression of TNF, IL1B and IL10 mRNAs was measured. Data are representative of 3 independent experiments.
B, Induction of autocrine IFN-β mRNA expression was measured by qPCR, and IFN-β protein from culture supernatants was measured by ELISA. Data are representative of 3 independent experiments.
C, Induction of type I IFN dependent CXCL9 and CXCL10 mRNAs downstream of LPS stimulation was measured. Data are representative of 3 independent experiments.
D, Cells were treated with zymosan (100μg/mL), LPS (10 ng/mL), or zymosan plus LPS. Induction of LPS-dependent and type I IFN dependent mRNAs were measured. Data are representative of 3 independent experiments.
Next, we tested the effects of concurrent stimulation with Fb on TLR4-induced IFN-β production. In contrast to the experiments in Figs. 1–3 where polymyxin B was used to block potential contaminating endotoxin in the Fb preparation, in these experiments (Fig. 5) the experimental design necessitated omitting polymyxin B. Both Fb and LPS stimulation, used individually, induced IFN-β mRNA (Fig. 5B); Fb-induced expression of IFNB was suppressed by polymyxin B and thus was likely secondary to endotoxin contamination (data not shown); polymyxin B did not affect the expression of TNF, IL6, or IL10. Concurrent stimulation with Fb and LPS induced IFN-β mRNA comparably to stimulation with LPS alone (Fig. 5B, left) and induced comparable or higher amounts of IFN-β protein, depending on the time point (Fig. 5B, right). These results show that the induction phase of the TLR4-induced IFN-β-mediated autocrine loop was intact after concurrent β2 integrin and TLR4 stimulation. We then tested if Fb inhibits autocrine IFN-β signaling. Despite induction of similar (or increased) amounts of IFN-β, Fb strongly suppressed TLR4-induced expression of the IFN-β-dependent CXCL9 and CXCL10 genes (Fig. 5C). To corroborate our findings, we wished to test whether other ITAM-coupled receptors could block the TLR4-induced IFN-β-mediated autocrine loop. We tested the effects of ligation of Dectin-1, which recognizes fungal β glucans and contains a hemITAM motif (38). Dectin-1 was ligated using zymosan, a β-glucan-containing yeast cell wall preparation widely used to model microbial pathogens. Similar to Fb, concurrent zymosan stimulation minimally affected TLR4-induced TNF and IFNB expression but strongly inhibited induction of the IFN-β response genes CXCL9 and CXCL10 (Fig. 5D). The induction of IFNB expression by Dectin-1 alone was most likely secondary to stimulatory DNA in the zymosan preparation, as previously described (39). Overall, the results show thatITAM-associated receptors fundamentally altered the TLR4 response by selectively blocking autocrine IFN-β signaling. One consequence of this block in IFN response is decreased inflammatory chemokine gene expression.
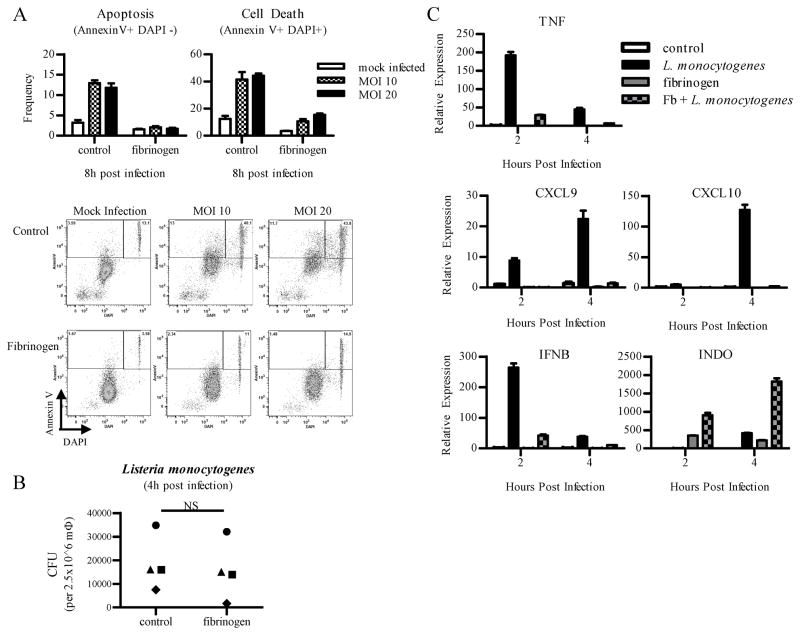
ITAM signaling protects primary human Mφ from L. monocytogenes-induced apoptosis
Type I IFNs promote apoptosis of infected cells, which can be detrimental for host defense against pathogens such as Listeria monocytogenes (22, 40). Thus, we tested whetherβ2 integrin ligation that makes cells resistant to IFNAR signaling would affect macrophage survival after infection with L. monocytogenes, which induces IFN-β that promotes macrophage apoptosis (26). Macrophages were first plated onto Fb coated wells for 18 h prior to infection with live L. monocytogenes (wild-type strain 10403S). Apoptosis was assessed using flow cytometry to measure annexin V binding to phosphatidylserine. DAPI nuclear staining was used to identify necrotic cells, and dead cells were defined as AnnexinV+DAPI+. In 4 independent experiments, Fb nearly completely suppressed L. monocytogenes-induced apoptosis and cell death of infected macrophages (Fig. 6A). This dramatic difference in cell death could not be explained by differences in infection and intracellular bacterial replication, as intracellular bacterial numbers were comparable between control and Fb-treated macrophages in individual donors (Fig. 6B). As expected, infection with L. monocytogenes induced expression of TNF, CXCL9, CXCL10, and IFNB (Fig. 6C). In accordance with our results that Fb blocks IFN responses, activation of these genes was attenuated in cells that were stimulated with Fb (Fig. 6C). Similar to previous studies, induction of TNF was abrograted by Fb, since preligation of β2 integrins inhibits TNF production downstream of TLR ligands (4). Decreased expression of CXCL9 and CXCL10 is likely explained by a combination of diminished production of IFN-β and suppression of IFNAR signaling, and the decreased IFN response contributes to enhanced cell survival. As a control, induction of INDO, which encodes indoleamine 2,3-dioxygenase, was synergistically induced by Fb and L. monocytogenes infection, showing that Fb did not globally block macrophage responses to L. monocytogenes. These results show that engagement of ITAM-associated β2 integrins alters macrophage responses to infection with L. monocytogenes by attenuating inflammatory and IFN responses while promoting macrophage survival and not compromising their anti-microbial functions.
Figure 6. Primary human Mφ pre-ligated with fibrinogen are protected from L. monocytogenes-induced apoptosis.
Primary human macrophages were plated onto control wells or fibrinogen (Fb) coated wells for 18 h. Culture media was changed, and cells were infected with live L. monocytogenes (wild-type strain 10403S) at MOI 10, 20 (A) or MOI 5 (B, C).
A, Flow cytometry was used to assess apoptosis, measured by annexin V binding to phosphatidylserine in the DAPI negative population. In vitro infections were performed in triplicate wells, and results are presented as mean frequency ± SD of triplicate wells. Cell death was measured by positive staining for both annexin V and DAPI. Data are representative of at least 3 independent experiments.
B, Viable bacteria from macrophages were quantified 4 h post infection. 4 independent experiments are shown, with each symbol representing a different donor. Statistical analysis was done using the 2-tailed, paired Student t test, and intracellular burden between control-infected and Fb-infected macrophages was not statistically different.
C, Induction of multiple infection induced mRNAs was measured by qPCR, and results are presented as mean ± SD of triplicate wells normalized relative to GAPDH mRNA. Data are representative of at least 3 independent experiments.
Discussion
One important function of ITAM-associated receptors is to modulate macrophage responses to heterologous receptors (41). Quantitative regulation of the amplitude of TLR and cytokine receptor signaling by ITAM-associated receptors has been previously reported (11, 21). In this study we found that ITAM-associated receptors, including β2 integrins, Fc receptors, and likely Dectin-1, inhibit IFNAR signaling. Inhibition by β2 integrins was mediated by a MAPK-dependent pathway that leads to induction of signaling inhibitors that include USP18. This represents to our knowledge the first implication of USP18 in mediating cross-inhibition of IFNAR by a different receptor. We have also found that ITAM-associated receptors qualitatively alter the nature of TLR4 responses by suppressing autocrine type I IFN signaling, and alter macrophage responses to L. monocytogenes infection to attenuate inflammatory mediator production and IFN responses while enhancing macrophage survival and preserving antimicrobial function. These findings provide a mechanism of cross-inhibition of IFNAR signaling by ITAM-coupledβ2 integrins and insights into how ligation of ITAM-associated receptors can modulate innate responses to pathogens.
Mechanisms that cross-inhibit cytokine-activated Jak-STAT signaling fall into two broad categories (21). One category corresponds to rapidly induced inhibitory signals that directly modify cytokine receptors or associated signaling molecules. The second category corresponds to induction of inhibitory molecules that target Jak-STAT signaling; the predominant inducible inhibitors of Jak-STAT signaling are the SOCS proteins. The mechanism of cross-inhibition of IFNAR signaling by ITAM-associated β2 integrins that we have described falls into the second category, as inhibition exhibited delayed kinetics and was associated with MAPK-dependent induction of several inhibitors of IFNAR-mediated Jak-STAT signaling, including SOCS3 and USP18. Of these inhibitors, USP18 played a more prominent role than did SOCS3, although it is most likely that several ITAM-induced inhibitors coordinately suppressed IFNAR signaling in a partially redundant manner. This is reminiscent of cross-inhibition of TLR signaling by prior engagement of ITAM-associated receptors that is mediated by MAPK-dependent induction of a distinct set of inhibitors that includes A20 and Hes1. Thus, ITAM-coupledβ2 integrins induce expression of a broad set of inhibitory molecules that modulate macrophage responses. Induction of inhibitory signaling is dependent on MAPKs and highlights the newly described suppressive aspects of MAPK signaling (42, 43) that complement the well-established activating effects of MAPKs (44).
Activation of MAPKs by ITAM-associated receptors is well established to depend on ITAM signaling (21), and a role for ITAM signaling in inhibition of IFN responses is further supported by the observed loss of inhibition of IFN signaling by Fb when Syk was inhibited. Although all ITAM-associated receptor ligands that were tested, Fb, CD11b antibodies, zymosan, and immune complexes, similarly inhibited IFN responses, there were some differences in underlying mechanisms. For example, Fb, but not zymosan or immune complexes, effectively induced USP18 expression (data not shown). In addition, MAPK signaling (which was necessary for Fb-induced expression of USP18) played a more prominent role in mediating inhibition of IFN responses by Fb than by zymosan or immune complexes. These differences between Fb and zymosan or immune complexes may be explained by different avidity of ligation of ITAM-associated receptors that are expressed at different levels on the cell surface; avidity of ITAM receptor ligation has been shown to qualitatively and quantitatively alter downstream signaling (21, 45). An alternative explanation for differences between Fb, zymosan and immune complexes is engagement of additional but distinct signaling pathways that cooperate with ITAM signaling to suppress IFN responses. For example,β2 integrins activate additional non-ITAM-dependent signaling pathways (46), zymosan activates receptors in addition to Dectin-1 (11), and immune complexes activate the inhibitory FcγRIIb that induces ITIM signaling, and DC-SIGN, which signals by non-ITAM-mediated pathways, in addition to ITAM-associated FcγRs (47). Another interesting point is the specificity of robust inhibition of IFNAR signaling by ITAM-associated receptors but not by TLRs, as TLR2 only mildly cross-inhibited IFNAR signaling (data not shown) and TLR4 induced, rather than inhibited, autocrine IFN responses. These differences between ITAM-associated receptors and TLRs are most likely explained by more effective activation of calcium-mediated and inhibitory signaling pathways by ITAM-associated receptors that lead to enhanced expression of signaling inhibitors (4, 21, 48).
During an innate immune or inflammatory response, ITAM-associated receptors will be engaged by endogenous ligands and by microbial products (also known as pathogen-associated molecular patterns or PAMPs). The major known endogenous ligands present at high concentrations at infectious/inflammatory sites are immune complexes, fibrin(ogen) and complement fragments that ligate, respectively Fc receptors and β2 integrins. Our results suggest that ligation of these ITAM-associated receptors on monocytes that migrate into inflammatory sites will attenuate inflammatory responses on subsequent encounter of pathogens or type I IFNs, thereby preventing excessive cell activation and tissue damage, while promoting cell survival and maintaining host defense. This idea is supported by our results showing that Fb attenuates inflammatory responses to L. monocytogenes infection while promoting cell survival. In addition to sensing endogenous inflammatory factors, ITAM-associated receptors such as C-type lectin receptors sense PAMPs and thus can be coordinately ligated along with various PRRs when macrophages encounter pathogens. Our data suggest that co-temporaneous ligation of ITAM-associated receptors and IFN-inducing PRRs (such as cell surface TLR3 and TLR4, endocytic TLRs 7–9, and cytoplasmic sensors of microbial RNA or DNA) by select pathogens will qualitatively change the macrophage response by selectively attenuating IFN responses. Our attempts to directly test this idea using microbial pathogens available to us that engage both ITAM-associated receptors and IFN-inducing PRRs were not successful likely because of difficulties in selectively manipulating ITAM signaling in the primary human macrophage system that we used. However, we obtained evidence to support this idea using an alternative approach of co-stimulation of β2 integrins and TLR4, Dectin-1 and TLR4, andβ2 integrins and the as yet unknown PRR that senses L. monocytogenes and induces IFN production. ITAM-associated receptors selectively attenuated autocrine IFN signaling while preserving activation of inflammatory NF-κB target genes.
Selective attenuation of IFN responses in the context of pathogen encounter can have beneficial or detrimental effects on host defense, depending on context and the specific pathogen. Type I IFNs are detrimental to host survival in certain bacterial infections, such as Francisella tularensis and Listeria monocytogenes (22). Our results suggest that ITAM-mediated attenuation of IFN signaling would be protective in infections by these pathogens as it promotes macrophage survival that is important for pathogen clearance. Attenuation of IFN-dependent chemokine production can also be beneficial by preventing excessive inflammation and attendant toxicity. However, because ITAM-associated receptor signaling can attenuate inflammatory cytokine production, ITAM signaling may also have deleterious effects on the host response to L. monocytogenes. The latter possibility is supported by evidence that mice lacking DAP12 have increased protection from L. monocytogenes (18, 40) and that CD11b-deficient mice can better clear L. monocytogenes at early timepoints (5). The data collectively suggest a context-dependent role for ITAMs in innate immunity, possibly determined by avidity of receptor ligation or availability of ligands. For example F. tularensis and L. monocytogenes are not known to directly activate ITAM-associated receptors, so a protective ITAM-activating signal would need to be delivered by opsonization with antibodies or complement, or by endogenous ligands such as Fb. Abrogation of IFN signaling can also compromise antiviral responses and decrease the efficacy of antigen presentation, which will be detrimental to the host. Thus, the overall effects of modulation of PRR responses and selective attenuation of autocrine IFN responses by ITAM-associated receptors on host defense will vary according to the pathogen. ITAM-TLR-IFN crosstalk and the ability to effectively prevent excessive IFN signaling may also be important in autoimmune diseases, where allelic variants in FcγRs and the CD11b subunit of β2 integrins have been linked to autoimmune diseases such as SLE that are characterized by TLR-driven type I IFN production. In this context, hypomorphic alleles encoding FcγR and CD11b variants may predispose to autoimmunity by enabling unrestrained and excessive IFN responses.
In conclusion, ITAM-associated receptors modulate TLR and cytokine receptor signaling in both a quantitative and qualitative manner. Preligation of ITAM-associated receptors globally and quantitatively attenuates TLR responses and serves to restrain inflammation and associated pathology. On the other hand, co-ligation of ITAM-associated receptors and TLRs selectively attenuates autocrine IFN signaling and qualitatively changes TLR responses to make them appropriate for a given pathogen and the inflammatory micro-environment that contains various endogenous ligands for ITAM-associated receptors.
Supplementary Material
Acknowledgments
We thank Xiaoyu Hu and Anna Yarilina for helpful discussions and critical reading of the manuscript.
Abbreviations used in this paper
- Fb
fibrin(ogen)
Footnotes
This work was supported by grants from the NIH (to L.B.I.). L.H. was supported by the NIH Grant T32 AIO7621 (to the Graduate Program in Immunology and Microbial Pathogenesis, Weill Cornell Graduate School of Medical Sciences).
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hamerman JA, Ni M, Killebrew JR, Chu CL, Lowell CA. The expanding roles of ITAM adapters FcRgamma and DAP12 in myeloid cells. Immunol Rev. 2009;232:42–58. doi: 10.1111/j.1600-065X.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamerman JA, Lanier LL. Inhibition of immune responses by ITAM-bearing receptors. Sci STKE. 2006;2006:re1. doi: 10.1126/stke.3202006re1. [DOI] [PubMed] [Google Scholar]
- 3.Pasquier B, Launay P, Kanamaru Y, Moura IC, Pfirsch S, Ruffié C, Hénin D, Benhamou M, Pretolani M, Blank U, Monteiro RC. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Gordon RA, Huynh L, Su X, Park Min K-H, Han J, Arthur JS, Kalliolias GD, Ivashkiv LB. Indirect inhibition of Toll-like receptor and type I interferon responses by ITAM-coupled receptors and integrins. Immunity. 2010;32:518–530. doi: 10.1016/j.immuni.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 6.Pfirsch-Maisonnas S, Aloulou M, Xu T, Claver J, Kanamaru Y, Tiwari M, Launay P, Monteiro RC, Blank U. Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized “inhibisome” clusters. Sci Signal. 2011;4:ra24. doi: 10.1126/scisignal.2001309. [DOI] [PubMed] [Google Scholar]
- 7.Mócsai A, Abram CL, Jakus Z, Hu Y, Lanier LL, Lowell CA. Integrin signaling in neutrophils and macrophages uses adaptors containing immunoreceptor tyrosine-based activation motifs. Nat Immunol. 2006;7:1326–1333. doi: 10.1038/ni1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abram CL, Lowell CA. The expanding role for ITAM-based signaling pathways in immune cells. Sci STKE. 2007;2007:re2. doi: 10.1126/stke.3772007re2. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol. 2008;9:1179–1188. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 10.Sancho D, Joffre OP, Keller AM, Rogers NC, Martinez D, Hernanz-Falcon P, Rosewell I, e Sousa CR. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osorio F, Reis e Sousa C. Myeloid c-type lectin receptors in pathogen recognition and host defense. Immunity. 2011;34:651–664. doi: 10.1016/j.immuni.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Turnbull IR, McDunn JE, Takai T, Townsend RR, Cobb JP, Colonna M. DAP12 (KARAP) amplifies inflammation and increases mortality from endotoxemia and septic peritonitis. J Exp Med. 2005;202:363–369. doi: 10.1084/jem.20050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Boulé MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 17.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177:2051–2055. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 18.Hamerman JA, Tchao NK, Lowell CA, Lanier LL. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat Immunol. 2005;6:579–586. doi: 10.1038/ni1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 20.Cao W, Rosen DB, Ito T, Bover L, Bao M, Watanabe G, Yao Z, Zhang L, Lanier LL, Liu YJ. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J Exp Med. 2006;203:1399–1405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10:340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton MJ, Antignano F, von Rossum A, Boucher JL, Bennewith KL, Krystal G. TLR agonists that induce IFN-beta abrogate resident macrophage suppression of T cells. J Immunol. 2010;185:4545–4553. doi: 10.4049/jimmunol.1002045. [DOI] [PubMed] [Google Scholar]
- 24.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 25.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Stockinger S, Materna T, Stoiber D, Bayr L, Steinborn R, Kolbe T, Unger H, Chakraborty T, Levy DE, Müller M, Decker T. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 27.Pathak MK, Yi T. Sodium stibogluconate is a potent inhibitor of protein tyrosine phosphatases and augments cytokine responses in hemopoietic cell lines. J Immunol. 2001;167:3391–3397. doi: 10.4049/jimmunol.167.6.3391. [DOI] [PubMed] [Google Scholar]
- 28.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70:655–665. doi: 10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- 29.Shi C, Velázquez P, Hohl TM, Leiner I, Dustin ML, Pamer EG. Monocyte trafficking to hepatic sites of bacterial infection is chemokine independent and directed by focal intercellular adhesion molecule-1 expression. J Immunol. 2010;184:6266–6274. doi: 10.4049/jimmunol.0904160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z, Shen Y, Yang W, Mecklenbrauker I, Neel BG, Ivashkiv LB. Inhibition of IFN-alpha signaling by a PKC- and protein tyrosine phosphatase SHP-2-dependent pathway. Proc Natl Acad Sci USA. 2005;102:10267–10272. doi: 10.1073/pnas.0408854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 32.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 33.Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, de la Torre JC, Zhang DE. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 35.Knobeloch KP, Utermöhlen O, Kisser A, Prinz M, Horak I. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol Cell Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KGS, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerber JS, Mosser DM. Reversing lipopolysaccharide toxicity by ligating the macrophage Fc gamma receptors. J Immunol. 2001;166:6861–6868. doi: 10.4049/jimmunol.166.11.6861. [DOI] [PubMed] [Google Scholar]
- 38.Brown GD, Herre J, Williams DL, Willment JA, Marshall ASJ, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biondo C, Signorino G, Costa A, Midiri A, Gerace E, Galbo R, Bellantoni A, Malara A, Beninati C, Teti G, Mancuso G. Recognition of yeast nucleic acids triggers a host-protective type I interferon response. Eur J Immunol. 2011;41:1969–1979. doi: 10.1002/eji.201141490. [DOI] [PubMed] [Google Scholar]
- 40.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 41.Turnbull IR, Colonna M. Activating and inhibitory functions of DAP12. Nat Rev Immunol. 2007;7:155–161. doi: 10.1038/nri2014. [DOI] [PubMed] [Google Scholar]
- 42.Ananieva O, Darragh J, Johansen C, Carr JM, McIlrath J, Park JM, Wingate A, Monk CE, Toth R, Santos SG, Iversen L, Arthur JSC. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028–1036. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- 43.Kim C, Sano Y, Todorova K, Carlson BA, Arpa L, Celada A, Lawrence T, Otsu K, Brissette JL, Arthur JSC, Park JM. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat Immunol. 2008;9:1019–1027. doi: 10.1038/ni.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schett G, Zwerina J, Firestein G. The p38 mitogen-activated protein kinase (MAPK) pathway in rheumatoid arthritis. Ann Rheum Dis. 2008;67:909–916. doi: 10.1136/ard.2007.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivashkiv LB. A signal-switch hypothesis for cross-regulation of cytokine and TLR signalling pathways. Nat Rev Immunol. 2008;8:816–822. doi: 10.1038/nri2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abram CL, Lowell CA. The ins and outs of leukocyte integrin signaling. Annu Rev Immunol. 2009;27:339–362. doi: 10.1146/annurev.immunol.021908.132554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 48.Kelly EK, Wang L, Ivashkiv LB. Calcium-activated pathways and oxidative burst mediate zymosan-induced signaling and IL-10 production in human macrophages. J Immunol. 2010;184:5545–5552. doi: 10.4049/jimmunol.0901293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.