Abstract
A series of chlorinated englerins (3–9), were isolated from Phyllanthus engleri and shown to selectively inhibit the growth of renal cancer cells. The compounds were shown to be extraction artifacts produced by exposure to chloroform decomposition products during their isolation. The most active compound, 3, was synthesized from englerin A (1).
Our group recently reported the isolation of two new sesquiterpenes, englerins A (1) and B (2), from the root bark and stem bark of Phyllanthus engleri Pax (Euphorbiaceae).1 Englerin A displayed remarkable potency and selectivity in its inhibition of renal cancer cell line growth. Consequently, englerin A (1) has been under intensive preclinical investigation at the National Cancer Institute, and several groups have published the synthesis of englerin A2–7 and analogues.8,9
At an early stage of this project, seven related bioactive compounds (3 – 9) were isolated, with these considered to be artifacts produced during the isolation procedure. Reported here are their structures and biological activity, and the synthesis from 1 of the most active compound.
Results and Discussion
An organic extract of stem bark was separated using batch elution from a diol-bonded phase medium to give five fractions of increasing polarity. The methylene chloride-soluble fraction was separated via flash chromatography over silica gel, using mixtures of chloroform and methanol, to yield three fractions, which potently inhibited the growth of UO-31 renal cancer cells, but were inactive against SF-295 CNS cancer cells. It was determined that it was at this stage when the natural compounds were modified by chloroform that had apparently spontaneously generated Cl2 on standing, since later attempts to purify the same fractions with newly purchased chloroform yielded only 1 and 2.
The modified fractions still possessed the selectivity of the parent extract towards renal cancer cells, and were therefore subjected to HPLC to yield a total of seven compounds, 3–9. The structures were elucidated by spectroscopic techniques in comparison to data obtained for 1.
HRMS established the molecular formula of 3 as C26H33O6Cl. The 1H and 13C NMR data (Tables 1 and 2) differed only slightly from 1, with the only differences being replacement of the C-2′, C-3′ vinyl doublet with a 1H singlet at 7.94 ppm, and alterations of the corresponding shifts for the three carbons of the cinnamate. This argued for substitution of chlorine at the C-2′ position, which was confirmed by HMBC correlation of H-3′ with C-4′, C-5′ and C-6′. The Z configuration of the double bond was established by analogy with the chemical shift of H-3′ being δ 7.91 for Z-ethyl α-chlorocinnamate,10 while for 3 it was δ 7.94. In addition, ACD predictions of shifts for this proton were δ 7.13 (E) and δ 7.85 (Z), respectively. The UV maximum at 284 nm was consistent with an intact cinnamate moiety, showing only a small shift from the 279 nm maximum of 1. Thus, 3 was assigned as [Z]-2′-chloroenglerin A.
Table 1.
13C NMR Data for 3–9 (125 MHz, d4-methanol)
| Carbon # | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|
| 1 | 48.9 | 48.9 | 48.9 | 48. 6 | 49.0 | 48.8 | 48.8 |
| 2 | 25.5 | 25.3 | 25.2 | 25.5 | 25.4 | 25.2 | 25.3 |
| 3 | 32.0 | 31.7 | 31.9 | 31.9 | 32.0 | 31.7 | 31.9 |
| 4 | 32.4 | 31.8 | 31.9 | 32.3 | 32.4 | 31.8 | 31.3 |
| 5 | 47.9 | 47.4 | 47.5 | 47.8 | 47.9 | 47.5 | 47.6 |
| 6 | 74.8 | 74.3 | 74.3 | 73.9 | 73.9 | 73.9 | 73.8 |
| 7 | 86.5 | 86.2 | 86.0 | 86. 5 | 86.5 | 86.3 | 86.2 |
| 8 | 40.8 | 40.1 | 39.9 | 40.3 | 40.2 | 40.2 | 39.9 |
| 9 | 76.6 | 76.4 | 76.4 | 76.5 | 76.6 | 76. 5 | 76.5 |
| 10 | 86.2 | 85.9 | 85.1 | 86.1 | 86.0 | 86.0 | 85.9 |
| 11 | 17.2 | 17.0 | 16.8 | 17.2 | 17.2 | 17.1 | 18.2 |
| 12 | 34.1 | 31.9 | 31.2 | 32.7 | 32.2 | 32.4 | 32.2 |
| 13 | 17.7 | 17.5 | 17.3 | 17.5 | 17.5 | 17.5 | 17.0 |
| 14 | 18.6 | 18.2 | 18.1 | 18.4 | 18.3 | 18.3 | 17.3 |
| 15 | 19.2 | 19.1 | 18.6 | 19.2 | 19.2 | 19.1 | 19.1 |
| 1′ | 163.5 | 167.2 | 167.1 | 169.1 | 169.1 | 168.3 | 168.2 |
| 2′ | 122.9 | 62.9 | 62.8 | 61.4 | 60.9 | 63.5 | 63.5 |
| 3′ | 138.9 | 64.7 | 64.6 | 76.5 | 76.3 | 76.1 | 76.0 |
| 4′ | 134.2 | 138.7 | 138.6 | 141.7 | 141.6 | 141.1 | 141.0 |
| 5′ | 131.8 | 129.3 | 129.3 | 128.4 | 128.3 | 128.7 | 128.7 |
| 6′ | 129.7 | 130.0 | 130.0 | 129.3 | 128.5 | 129.6 | 129.6 |
| 7′ | 131.6 | 130.6 | 130.7 | 129.4 | 129.4 | 129.7 | 129.8 |
| 8′ | 129.7 | 130.0 | 130.0 | 129.3 | 128.5 | 129.6 | 129.6 |
| 9′ | 131.8 | 129.3 | 129.3 | 128.4 | 128.3 | 128.7 | 128.7 |
| 1″ | 174.0 | 173.9 | 174.0 | 174.0 | 174.0 | 174.0 | 174.0 |
| 2″ | 61.1 | 61.0 | 61.0 | 61.0 | 61.0 | 61.0 | 61.0 |
Table 2.
1H NMR data for 3–9 (500 MHz, d4-methanol, δH (m, J in Hz)
| position | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|
| 1 | 1.76 (m) | 1.58 (m) | 1.61 (m) | 1.72 (m) | 1.74 (m) | 1.61 (m) | 1.62 (m) |
| 2a | 1.75 (m) | 1.63 (m) | 1.66 (m) | 1.72 (m) | 1.74 (m) | 1.18 (m) | 1.67 (m) |
| 2b | 1.34 (m) | 1.20 (m) | 1.25 (m) | 1.28 (m) | 1.33 (m) | 1.60 (m) | 1.21 (m) |
| 3a | 2.02 (m) | 1.77 (m) | 1.85 (m) | 1.98 (m) | 2.00 (m) | 1.76 (m) | 1.86 (m) |
| 3b | 1.28 (m) | 1.09 (m) | 1.16 (m) | 1.26 (m) | 1.29 (m) | 1.05 (m) | 1.18 (m) |
| 4 | 2.14 (m) | 1.34 (m) | 1.63 (m) | 2.19 (m) | 2.23 (m) | 1.32 (m) | 1.27 (m) |
| 5 | 1.75 (m) | 1.34 (m) | 1.39 (m) | 1.55 (m) | 1.64 (m) | 1.25 (m) | 1.47 (m) |
| 6 | 5.14 (d, 9.5) | 4.80 (d, 9.5) | 4.79 (d, 10.0) | 5.02 (d, 10.0) | 5.07 (d, 10.0) | 4.81 (d, 7.0) | 4.81 (d, 9.5) |
| 8a | 2.73 (dd, 14.5, 8.0) | 2.44 (dd, 14.5, 8.0) | 2.43 (dd, 14.5, 8.0) | 2.45 (dd, 15.0, 8.0) | 2.57 (dd, 15.0, 8.0) | 2.42 (dd, 15.0, 8.0) | 2.40 (dd, 14.5, 8.0) |
| 8b | 1.89 (dd, 14.5, 3.0) | 1.80 (dd, 14.5, 3.0) | 1.82 (dd, 14.5, 3.0) | 1.81 (dd, 15.0, 3.0) | 1.90 (dd, 15.0, 3.0) | 1.81 (dd, 15.0, 3.0) | 1.75 (dd, 14.5, 3.0) |
| 9 | 5.27 (dd, 8.0, 3.0) | 5.12 (dd, 8.0, 3.0) | 5.15 (dd, 8.0, 3.0) | 5.18 (dd, 8.0, 3.0) | 5.23 (dd, 8.0, 3.0) | 5.11 (dd, 8.0, 3.0) | 5.15 (dd, 8.0, 3.0) |
| 11 | 0.93 (d, 7.0) | 0.56 (d, 6.5) | 0.72 (d, 7.0) | 0.89 (d, 7.0) | 0.93 (d, 7.0) | 0.62 (d, 7.0) | 0.64 (d, 7.0) |
| 12 | 1.87 (m) | 1.46 (m) | 1.27 (m) | 1.87 (m) | 1.88 (m) | 1.66 (m) | 1.82 (m) |
| 13 | 0.97 (d, 7.0) | 0.84 (d, 7.0) | 0.64 (d, 7.0) | 0.95 (d, 7.5) | 0.99 (d, 6.5) | 0.89 (d, 7.0) | 0.77 (d, 6.5) |
| 14 | 1.02 (d, 6.5) | 0.89 (d, 7.0) | 0.80 (d, 6.5) | 0.98 (d, 7.0) | 1.01 (d, 6.5) | 0.92 (d, 6.5) | 0.78 (d, 7.0) |
| 15 | 1.19 9 (s) | 1.10 (s) | 1.11 (s) | 1.16 (s) | 1.19 (s) | 1.11 (s) | 1.11 (s) |
| 2′ | - | 5.02 (d, 9.5) | 4.98 (d, 10.0) | 4.40 (d, 9.0) | 4.36 (d, 9.0) | 4.58 (d, 8.5) | 4.54 (d, 9.0) |
| 3′ | 7.94 (s) | 5.31 (d, 9.5) | 5.27 (d, 10.0) | 4.88 (d, 9.0) | 4.90 (d, 9.0) | 4.93 (d, 8.5) | 4.92 (d, 9.0) |
| 5′ | 7.87 (brd, 7.5) | 7.47 (brdd, 8.0, 3.0) | 7.48 (brdd, 7.5, 2.0) | 7.43 (d, 7.0) | 7.44 (dd, 8.0, 2.0) | 7.39 (dd, 8.0, 2.0) | 7.40 (dd, 8.0, 1.5) |
| 6′ | 7.45 (m) | 7.37 (m) | 7.37 | 7.36 (dd, 8.0, 7.0) | 7.36 (dd, 8.0, 7.0) | 7.36 (dd, 8.0, 7.0) | 7.35 (m) |
| 7′ | 7.45 (m) | 7.37 (m) | 7.37 | 7.31 (brd, 8.0) | 7.33 (d, 7.0) | 7.31 (d, 7.0) | 7.35 (m) |
| 8′ | 7.45 (m) | 7.37 (m) | 7.37 | 7.36 (dd, 8.0, 7.0) | 7.36 (dd, 8.0, 7.0) | 7.36 (dd, 8.0, 7.0) | 7.35 (m) |
| 9′ | 7.87 (brd, 7.5) | 7.47 (brdd, 8.0, 3.0) | 7.48 (brdd, 7.5, 2.0) | 7.43 (d, 7.0) | 7.44 (dd, 8.0, 2.0) | 7.39 (dd, 8.0, 2.0) | 7.40 (dd, 8.0, 1.5) |
| 2″ | 4.15 (s) | 4.12 (s) | 4.12 (s) | 4.13 (s) | 4.13 (s) | 4.12 (s) | 4.11 (s) |
Compounds 4 and 5 gave identical molecular formulas of C26H34O6Cl2, as determined by HRMS. The isotope abundances for the M+H+1, M+H+2 and M+H+3 ions were indicative of a formula with two chlorine atoms. As with 3, the core sesquiterpene signals in the 1H and 13C NMR spectra were very similar to 1, with the only substantive differences at C-1′, C-2′ and C-3′, where the 13C NMR signals for C-2′ and C-3′ were changed to δ 62.9 and 64.7 for 4 and δ 62.8 and 64.6 for 5, respectively, with a pair of corresponding 1H 1H NMR doublets at δ 5.02 and δ 5.31 for 4 and δ 4.98 and δ 5.27 for 5 also observed, each with ca. 10 Hz couplings. This suggested compounds 4 and 5 to be a pair of epimeric dichloro- derivatives, namely, 2′,3′-dichlorodihydroenglerin A. The absolute configuration of the C-2′ and C-3′ centers was not determined for either of these compounds.
Compounds 6 – 9 were found to all share the same molecular formula, C26H35O7Cl, based on HRMS measurements. The chemical shifts of the carbon signals for C-3′ were observed from ca. δ 64 to ca. δ 76 for these four compounds. This information, coupled with the addition of water compared to 3, indicated that these compounds are epimeric chlorohydrins. As with the previous compound 3, HMBC correlations of H-3′ with carbons C-4′, C-5′ and C-6′ established that the hydroxy group was at C-3′, thereby locating the chlorine at C-2′. The absolute configurations of these epimers were not determined.
All of the compounds were tested in the NCI 60-cell assay, and most showed some selectivity for renal cancer cell lines, with 3 having the best potency and renal cancer selectivity, being approximately 2.5-fold less active than 1. The mean GI50 values and individual GI50 values for eight renal cancer cell lines are tabulated in Table 3. It is notable that the pairs of epimers 4 and 5, as well as 6 and 7, showed significant differences in potency for many of the cell lines. Compounds 8 and 9 were less potent and did not meet criteria in the one-dose NCI 60-cell test for further testing in the full screen.
Table 3.
Selected NCI 60 Cell Line Data for Compounds 3 – 7.
| compound | 1 n = 3 |
3 n = 1 |
4 n = 1 |
5 n = 1 |
6 n = 1 |
7 n = 1 |
|---|---|---|---|---|---|---|
| mean GI50 (μM) | 3.7 | 5.6 | 8.3 | 12 | 6.9 | 12 |
| range at GI50 (log units) | 3.30 | 3.48 | 2.65 | 3.42 | 3.42 | 2.43 |
| individual renal cancer cell line GI50 values (μM) | ||||||
| 786-0 | 1.1 | 10 | 12 | 12 | 1.7 | 11 |
| A498 | 0.011 | 0.028 | 0.14 | 0.067 | 0.18 | 0.66 |
| ACHN | 0.017 | 0.041 | 0.14 | 0.049 | 0.32 | 1.4 |
| CAKI-1 | 16 | 0.39 | 0.42 | 0.16 | 0.46 | 5.0 |
| RXF 393 | 0.011 | 0.059 | n.t.a | n.t. a | 0.51 | 0.39 |
| SN12C | 1.1 | 0.56 | 0.93 | 1.0 | 0.63 | 4.7 |
| TK-10 | 1.9 | 21 | 18 | 25 | 17 | 27 |
| UO-31 | 0.015 | 0.035 | 0.39 | 0.16 | 0.25 | 1.9 |
Assay failure; no data for this cell line.
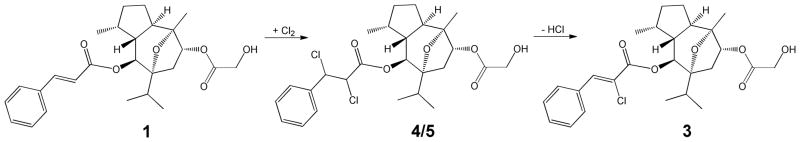
The identity and likely origin of 3 was confirmed by synthesis from englerin A (1), using a known method, wherein molecular chlorine is generated from HCl by treatment with oxone. Following dichlorination, dehydrohalogenation with Et3N yielded 3 in modest yield.10 While it is not possible to specify the conditions that inadvertently led to dehydrohalogenation, it is likely that molecular chlorine was generated in stored CHCl3 and reacted with the biosynthetic product englerin A (1) to form the dichloro products 4 and 5, which then yielded 3 (Scheme 1). The generation of molecular chlorine by photo-oxidation has long been recognized due to chloroform’s importance in 19th century anesthesia, with phosgene and hydrochloric acid among the other major products generated.11–13
Scheme I.
In support of this mechanism, new 1H NMR peaks were observed in samples of 4 and 5 stored dry at −20º C for extended periods, which precisely matched those of 3, indicating that spontaneous conversion of both 4 and 5 to 3 can occur. These new peaks were ca. 30% of the area of the parent. A UV absorbance maximum at 284 nm was also observed in the stored samples of 4 and 5, which may be accounted for by the formation of 3. It is also possible that the bioactivity observed for 4 and 5 may be due to in situ production of 3 in cell culture. In contrast, the chlorohydrins 6 – 9, as well as 3, were stable under the storage conditions used.
Our group is attempting to produce other 2′-halogenated products via this route, since direct iodination or fluorination could be used to synthesize isotopically labeled tracer compounds that may be useful in understanding the distribution and metabolism of englerin A (1) in animals and humans.
Experimental Section
General Experimental Procedures
Solvents were of HPLC grade, with chloroform stabilized with a hydrocarbon (amylene) stabilizer. Optical rotations ([α]D) were obtained on a Perkin-Elmer 241 polarimeter in a 100 by 2 mm cell (units 10−1 deg cm2 g−1), while ultraviolet (UV) absorption spectra were obtained using a Varian Cary 50 Bio UV-Visible spectrophotometer. NMR experiments were performed on a Varian INOVA 500 MHz NMR spectrometer. 1H and 13C NMR spectra were referenced to the deuterated solvent peaks. High resolution mass spectrometric measurements were acquired on an Agilent 6520 Accurate Mass Q-TOF instrument with internal reference masses at 121.05087 and 922.00979, both to within 5 ppm.
High-performance liquid chromatography (HPLC) was performed using a Varian ProStar 210/215 solvent delivery module equipped with a Varian ProStar 325 UV-Vis detector, operating under Star 6.41 chromatography workstation software. HPLC-DAD-MS(±) analysis was carried out on an Agilent 1100 series separation module binary pump system with a vacuum degasser, thermostat control (40 °C) column compartment, well-plate autosampler, diode array detector, and quadrupole mass detector. Equipment was under the control of Agilent ChemStation© (Revision A.10.02) software. Standard HPLC-DAD-MS gradient conditions were 0.5 mL/min gradient elution from 90% H2O (5% CH3CO2H)/MeCN to 100% MeCN over 20 min followed by a 5 min flush with 100% MeCN, using a Phenomenex Jupiter C18 150 × 2 mm 5 micron column.
Plant Material
Specimens of Phyllanthus engleri were collected on February 6, 1989 by Roy Gereau and James Lovett of the Missouri Botanical Garden and C.O. Kyalawa and Z.H. Mbwambo near Ilembula, Njombe District, Iringa Region, Tanzania (longitude 8° 50′ S, latitude 34° 31′ E), at an elevation of 1380 m. The plant (voucher specimen Q66T-3055, http://www.tropicos.org/Specimen/149907) was identified definitively by A. Radcliffe-Smith of the Royal Botanical Garden, Kew, Richmond, UK. The specimen collected was a 3 m tall tree of 15 cm dbh in a Brachystegia-Combretum woodland. Leaf (377 g dry wt.), stem bark (387 g), stem wood (255 g), root bark (201 g), and root wood (312 g) samples were collected separately and dried in the field.
Extraction and Isolation
Dried plant material was ground and extracted using the standard NCI extraction proptocol.14 The root bark yielded 14.85 g of an organic solvent extract (N042029). A 2.61 g portion of this material was dissolved in CH2Cl2-CH3OH 1:1 and coated on 27 g of diol bonded phase media using a rotary evaporator. The medium was resuspended in hexane and evaporated to remove all of the initial solvent to produce a dry flowable powder, which was packed in a Buchner funnel over an equal amount of uncoated diol medium, then eluted batchwise with 150 mL of hexane, CH2Cl2, EtOAc, acetone, and CH3OH in succession. Evaporation gave 612 mg of a CH2Cl2 fraction.
A subsample of 515 mg of the CH2Cl2 fraction was chromatographed on a 5 × 14 cm flash column of silica gel. The sample was dissolved in CHCl3 and eluted with increasing amounts of CH3OH in CHCl3 [CHCl3, CHCl3-CH3OH 4:1(v/v), CHCl3-CH3OH 1:1, CH3OH]. Fractions were combined on the basis of TLC and fraction E (194 mg) was found to have excellent selective activity in the two-cell assay (IC50 0.18 μg/mL in UO-31, 93 μg/mL in SF-295). Adjacent fractions had lesser activity (D: 187 mg, IC50 0.91 μg/mL in UO-31, >100 μg/mL in SF-295; F: 57 mg, IC50 14 μg/mL in UO-31, >100 μg/mL in SF-295) Recovery of biological activity was judged acceptable within the limits of assay precision. To guard against loss of activity in the next step, C18 HPLC, an experiment was conducted wherein 10 mg of the active fraction from a parallel separation fraction from the stem bark was applied to a prewashed Bondesil C18 SPE cartridge and eluted with CH3OH, 50% CH3OH-THF, then THF. The fractions were tested in the tracking assay and the methanol eluate was found active against UO-31 cells (IC50 0.03 μg/mL), but not against SF-295 cells (IC50 >100 μg/mL) while the THF eluates were entirely inactive against either cell line. The parent fraction, tested in tandem, was also extremely active against UO-31 but not SF-295 (IC50s 0.04 μg/mL, >100 μg/mL, respectively). This established that the bioactive compounds could be eluted from a C18 column and that the conditions of separation were unlikely to alter the active compounds in such a way that activity would be destroyed.
The active fraction E was then chromatographed by HPLC using a Varian Microsorb 60-8 C18 column (250 × 21.4 mm) using a CH3OH/H2O gradient starting at 75% CH3OH for 5 min, linearly to 85% at 32 min, to 100% CH3OH at 36 min, held and returned to original conditions at 40 min, with a flow rate of 32 mL/min. Injections were made with 40 mg of sample in 400 μL DMSO per injection. Detection was at 225 nm. Consistent separations were obtained, with three groups of peaks separated (see Supporting Information). The first, earliest eluting peaks appeared at 7–12 min, while a larger series of more abundant peaks eluted from 20–30 min, and the third group eluted after 35 min. This late group of peaks showed no cytotoxic activity, and by 1H NMR spectroscopy appeared to be composed of phytosterols, consistent with the report of phyllanthol from the plant many years ago.15 The peaks in the two earlier eluting clusters were subjected to HPLC-MS and 1H NMR spectroscopic analysis, and appeared to be a related series of C26 compounds.
Eluting at 27 min was compound 3 (8.9 mg), well separated from other peaks (chromatogram in Supporting Information). At 25 min, the first of a poorly resolved set of three compounds was further purified by rechromatography in the same solvent without the 100% CH3OH step in the gradient to give 4.3 mg of 4. The middle peak was rechromatographed on a 10 × 250 mm C8 Dynamax column, isocratic at 80% aq. MeCN for 25 min, to 100% MeCN at 30 min to give 4.8 mg of 5, eluting at 16 min. The earliest cluster of peaks in the primary HPLC separation yielded 6 (3.6 mg, 10 min), 7 (3.5 mg, 12 min), 8 (3.8 mg, 8 min) and 9 (4.0 mg, 9 min).
[Z]-2′-Chloroenglerin A (3)
NSC#746567: white solid; [α]D -60 (c 0.1, EtOH); UV (EtOH) λmax (log ε) 284 (4.14) nm; 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2, in CDCl3, see Supporting Information; ESIMS m/z 477 [M+H]+ 477 (100%), 478 (27%), 479 (35%), 480 (9%); HRESIMS m/z 477.2054 (calcd for C26H34ClO6, 477.2038).
2′,3′-Dichlorodihydroenglerin A (epimer 1) (4)
NSC#746565: white solid; 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; in d6-DMSO, see Supporting Information; ESIMS m/z 513 [M+H]+ 513 (100%), 514 (29%), 515 (65%), 516 (19%); HRESIMS m/z 513.1771 (calcd for C26H35Cl2O6, 513.1805).
2′,3′-Dichlorodihydroenglerin A (epimer 2) (5)
NSC#746566: white solid; 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; in d6-DMSO, see Supporting Information; ESIMS m/z 513 [M+H]+ 513 (100%), 514 (33%), 515 (80%), 516 (15%); HRESIMS m/z 513.1814 (calcd for C26H35Cl2O6, 513.1805).
2′-Chloro-3′-hydroxydihydroenglerin A (epimer 1) (6)
NSC#746563: white solid; [α]D -47 (c 0.3, EtOH); 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; in d6-DMSO, see Supporting Information; ESIMS m/z 495 [M+H]+ 495 (100%), 496 (31%), 497 (35%), 498 (11%); HRESIMS m/z 495.2138 (calcd for C26H35ClO7, 495.2144).
2′-Chloro-3′-hydroxydihydroenglerin A (epimer 2) (7)
NSC#746564: white solid; [α]D -10 (c 0.3, EtOH); 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; in d6-DMSO, see Supporting Information; ESIMS m/z 495 [M+H]+ 495(100%), 496(30%), 497(35%), 498(10%); HRESIMS m/z 495.2134 (calcd for C26H35ClO7 495.2144).
2′-Chloro-3′-hydroxydihydroenglerin A (epimer 3) (8)
NSC#746858: white solid; [α]D -37 (c 0.3, EtOH); 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; ESIMS m/z 495 [M+H]+ 495 (100%), 496 (26%), 497 (36%), 498 (10%); HRESIMS m/z 495.2149 (calcd for C26H35ClO7, 495.2144).
2′-Chloro-3′-hydroxydihydroenglerin A (epimer 4) (9)
NSC#746859: white solid; [α]D -28 (c 0.4, EtOH); 1H and 13C NMR data in d4-MeOH, see Tables 1 and 2; ESIMS m/z 495 [M+H]+ 495 (100%), 496 (25%), 497 (34%), 498 (8%); HRESIMS m/z 495.2112 (calcd for C26H35ClO7, 495.2144).
Synthesis of (Z)-2′-Chloroenglerin A (3)
To a mixture of englerin A (1) (26 mg, 58 μmol) and oxone (178 mg, 292 μmol) in CH2Cl2 (0.5 mL) was added 2 N aqueous HCl (64 μL, 128 μmol of aq. HCl), in a single portion at room temperature. After stirring for 2 h at r.t., Et3N (49 μL, 350 μmol) was added, and the solution was left on the magnetic stir pad overnight, ~12 h. The reaction mixture was partitioned against 10 mL of CH2Cl2 and 10 mL of water, the aqueous layer was washed two more times with 10 mL of CH2Cl2. The CH2Cl2 layers were combined and partitioned against a concentrated brine solution. The organic layer was dried over anhydrous Na2SO4 and the supernatant was dried under vacuum. The crude product was passed through a silica gel column to give a fraction containing a mixture of Z and E isomers of 2′-chloro-englerin A as a clear oil (0.14 g, 40% yield, Z:E ratio of 9:1 by HPLC/ELSD). Purification of (Z)-2′-chloroenglerin A by reversed-phase C18 preparative HPLC, using methanol-water, gave 2 mg of the Z-isomer 3 as well as several products with molecular mass corresponding to the chlorohydrins 6–9.
(Z)-2′-Chloroenglerin A (3)
1H NMR (d4-MeOH) δ 7.96 (1H, s, H-3′), 7.89 (2H, m, H-5′, H-9′), 7.46 (3H, m, H-6′, H-7′, H-8′), 5.29 (1H, dd, J = 3.0, 8.0 Hz, H-9), 5.15 (1H, d, J = 9.3 Hz, H-6), 4.16 (2H, s, H-2″), 2.75 (1H, dd, J = 8.0, 14.6 Hz, H-8a), 2.15 (1H, m, H-4), 2.03 (1H, m, H-3a), 1.90 (1H, m, H-8b), 1.88 (1H, m, H-12), 1.76 (3H, m, H-1, H-2a, H-5), 1.32 (2H, m, H-2b, H-3b), 1.20 (3H, s, H-15), 1.03 (3H, d, J = 6.8 Hz, H-14), 0.98 (3H, d, J = 7.1 Hz, H-13), 0.94 (3H, d, J = 7.1 Hz, H-11); 13C NMR (d4-MeOH) δ 174.0 (C-1″), 163.5 (C-1′), 138.9 (C-3′), 134.2 (C-4′), 131.8 (C-9′), 131.8 (C-5′), 131.6 (C-7′), 129.7 (C-8′), 129.7 (C-6′), 122.8 (C-2′), 86.5 (C-7), 86.2 (C-10), 76.6 (C-9), 74.7 (C-6), 61.0 (C-2″), 49.0 (C-1), 47.8 (C-5) 40.8 (C-8), 34.1 (C-12), 32.4 (C-4), 32.0 (C-3), 25.5 (C-2), 19.2 (C-15), 18.6 (C-13), 17.7 (C-14), 17.2 (C-11).
Biological Assays
Fractionation was guided by a 48 h cell growth assay using either XTT or SRB endpoints with the UO-31 or A498 renal cancer cell lines. NCI 60-cell tests were conducted as described previously.16
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. This research was supported, in part, by both the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and,in part, by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We gratefully acknowledge Joel Schneider and Paul Grothaus for helpful comments, and Heidi Bokesch (MTL), Sergey Tarasov and Marzena Dyba (Biophysics Resource Core, Structural Biophysics Laboratory, CCR) for acquiring high-resolution mass spectra.
Footnotes
Dedicated to Dr. Gordon M. Cragg, formerly Chief, Natural Products Branch, National Cancer Institute, Frederick, Maryland, for his pioneering work on the development of natural product anticancer agents.
Supporting Information Available. Full NMR data for 3–9, NCI 60-cell testing data for 3–9, and an HPLC chromatogram. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ratnayake R, Covell DG, Ransom TT, Gustafson KR, Beutler JA. Org Lett. 2009;11:57–60. doi: 10.1021/ol802339w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Nakashige M, Chain WJ. J Am Chem Soc. 2011;133:6553–6556. doi: 10.1021/ja201921j. [DOI] [PubMed] [Google Scholar]
- 3.Molawai K, Delpont N, Echavarren AM. Angew Chem Int Ed Engl. 2010;49:3517–3519. doi: 10.1002/anie.201000890. [DOI] [PubMed] [Google Scholar]
- 4.Nicolaou KC, Kang Q, Ng SY, Chen DY. J Am Chem Soc. 2010;132:8219–8222. doi: 10.1021/ja102927n. [DOI] [PubMed] [Google Scholar]
- 5.Sun BF, Wang CL, Ding R, Xu JY, Lin GQ. Tetrahedron Lett. 2011;52:2155–2158. [Google Scholar]
- 6.Willot M, Radtke L, Könning D, Fröhlich R, Gessner VH, Strohmann C, Christmann M. Angew Chem Int Ed Engl. 2009;48:9105–9108. doi: 10.1002/anie.200905032. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Chen X, Ma D. Angew Chem Int Ed Engl. 2010;49:3513–3515. doi: 10.1002/anie.201000888. [DOI] [PubMed] [Google Scholar]
- 8.Chan KP, Chen DY. ChemMedChem. 2011;6:420–423. doi: 10.1002/cmdc.201000544. [DOI] [PubMed] [Google Scholar]
- 9.Radtke L, Willot M, Sun H, Ziegler S, Sauerland S, Strohmann C, Frohlich R, Habenberger P, Waldmann H, Christmann M. Angew Chem Int Ed Engl. 2011;50:1–6. [Google Scholar]
- 10.Kim KM, Park IH. Synthesis. 2004:2641–2644. [Google Scholar]
- 11.Morson TNR. Pharm J. 1848;8:69. [Google Scholar]
- 12.Baskerville C, Hamor WA. J Ind Eng Chem. 1912;4:278–288. [Google Scholar]
- 13.Chapman AT. J Am Chem Soc. 1935;57:419–422. [Google Scholar]
- 14.McCloud TG. Molecules. 2010;15:4526–4563. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton DHR, Page JE, Warnhoff EW. J Chem Soc. 1954:2715–2719. [Google Scholar]
- 16.Shoemaker RH. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. See also http://dtp.nci.nih.gov/branches/btb/ivclsp.html. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.