Abstract
NK cells are innate immune lymphocytes important for early host defense against infectious pathogens and malignant transformation. MicroRNAs (miRNAs) are small RNA molecules that regulate a wide variety of cellular processes typically by specific complementary targeting of the 3′UTR of mRNAs. The Dicer1 gene encodes a conserved enzyme essential for miRNA processing, and Dicer1 deficiency leads to a global defect in miRNA biogenesis. Here, we report a mouse model of lymphocyte-restricted Dicer1 disruption in order to evaluate the role of Dicer1-dependent miRNAs in the development and function of NK cells. As expected, Dicer1-deficient NK cells had decreased total miRNA content. Further, miRNA-deficient NK cells exhibited reduced survival, as well as impaired maturation defined by cell surface phenotypic markers. However, Dicer1-deficient NK cells exhibited enhanced degranulation and interferon gamma (IFN-γ) production in vitro in response to cytokines, tumor target cells, and activating NK cell receptor ligation. Moreover, a similar phenotype of increased IFN-γ was evident during acute MCMV infection in vivo. MiRs-15a/15b/16 were identified as abundant miRNAs in NK cells that directly target the murine IFN-γ 3′UTR, thereby providing a potential mechanism for enhanced IFN-γ production. These data suggest that the function of miRNAs in NK cell biology is complex, with an important role in NK cell development, survival and/or homeostasis, while tempering peripheral NK cell activation. Further study of individual miRNAs in an NK cell specific fashion will provide insight into these complex miRNA regulatory effects in NK cell biology.
Introduction
Natural killer (NK) cells are an important component of the innate immune system, and have a key role in early host defense against pathogens and surveillance against malignant transformation (1–4). NK cells develop from precursors arising in the bone marrow (BM), and complete differentiation and maturation in peripheral lymphoid tissues under the direction of cytokines and transcription factors (5, 6), with IL-15 playing a central role (7, 8). During development from immature precursors, NK cells undergo an education process that results in tolerance to normal ‘self’ cells and prevents NK based autoimmunity. This tolerance is achieved through the expression of a repertoire of germline-encoded activating and inhibitory receptors that yield signals that are integrated and determine how the NK cell to responds to a target cell (9–11). NK cells also constitutively express a number of cytokine receptors, and NK responsiveness is also regulated by cytokines produced by accessory immune cells sensing pathogens (12). Thus, NK cell development and function in an immune response are distinct from adaptive T and B lymphocytes.
NK cells function to protect the host through two primary effector pathways – cytokine/chemokine production and cytotoxicity (13). Mature NK cells may be ‘primed’ by cytokines produced by accessory cells sensing pathogens in order to optimize their cytotoxic and cytokine-secretion potential upon subsequent receptor-based triggering (14–17). One aspect of NK cell priming by dendritic cell derived IL-15 includes the rapid translation of perforin and granzyme B effector proteins (14, 15). Secretion of cytokines (e.g. IFN-γ) and chemokines (e.g. MIP-α, MIP-β, RANTES) by NK cells may directly affect virus-infected or malignant cells, promote an anti-viral state in adjacent cells, recruit additional immune cells, as well as shape the subsequent adaptive immune response. In contrast, NK cytotoxicity is triggered primarily by receptor interaction with a target cell that results in granule exocytosis – the coordinated release of perforin and granzymes into a cytotoxic synapse – ultimately inducing apoptotic-like target cell death (18, 19). Lysosomal-associated membrane protein 1 (LAMP-1, CD107a) is retained within cytotoxic granules and is released on the NK cell surface membrane after ‘triggering’, providing a marker of recent degranulation (20). While a number of receptors, signaling molecules, and transcription factors have been identified that regulate NK cell development and activation, our understanding of the molecular mechanisms regulating these events remains incomplete.
MicroRNAs (miRNAs) are a family of hundreds of small, non-coding RNAs that regulate numerous cellular functions by targeting sites in the 3′UTR of mRNAs thereby suppressing translation or causing mRNA degradation (21). MiRNA genes are encoded in the genome, transcribed as long primary (pri-miRNA) transcripts that are subsequently ‘cropped’ by the Drosha/Dgcr8 complex into precursor miRNA (pre-miRNA) that have a characteristic stem loop structure (22). The pre-miRNA is exported to the cytoplasm where it is further processed by the Dicer complex (including Dicer1), yielding mature 19–26 nucleotide miRNAs. The mature miRNA is loaded into the RNA induced silencing complex (RISC), and thereby directs downregulation of protein levels (23). Genetic disruption of Dicer1 therefore results in decreased or absent levels of a large number of miRNAs due to interruption in miRNA biogenesis. Previous studies have shown the requirement of Dicer1 for the processing of miRNAs involved in the maturation and function of various cell types, including lymphocytes (24–29). In addition, specific miRNAs or miRNA clusters have been implicated in B and T cell development and modulate functional responses (30). However, only limited information is available on the expression and function of miRNAs in NK cells, their subsets, and developmental intermediates.
Mature murine NK cells have been shown to express more than 300 mature miRNAs using a combination of miRNA-SEQ with validation by real-time quantitative PCR and microarrays (31). Moreover, a subset of these miRNAs are modulated with short-term IL-15 activation, including miR-223 that was shown to target the murine granzyme B 3′UTR. A number of miRNAs were expressed in common with other lymphocytes (e.g. miR-181a, miR-21, miR-142–3p/5p, miR-16, miR-15b, miR-150, and let-7f), suggesting that these may regulate lymphocyte development and/or functional programs. A recent study has also identified miR-29 as a contributor to IFN-γ protein regulation in both innate NK cells and adaptive T cells (32). However, the functional role of the majority of expressed miRNAs in NK cells remains to be elucidated. What are the ramifications of global miRNA deficiency on developing and mature NK cells? Answering this question may provide evidence to support the role of miRNAs as regulators of specific aspects of NK cell development, survival, priming, triggering, or function. A prior study by Bezman et al. evaluated the phenotype of miRNA deficiency using a global, inducible estrogen receptor (ER)-Cre model that results in Dicer1 deletion in all cell types in the mouse after tamoxifen treatment. The authors concluded that miRNA-deficient NK cells have impaired survival, proliferation, as well as reduced functional capacity as measured by IFN-γ production and degranulation. These alterations resulted in compromised function during MCMV infection (33). However, the relative contribution of NK cell intrinsic miRNA deficiency to these phenotypes was challenging to assess using a global, non-selective miRNA deficiency model system.
In this study, we employed a lymphocyte-restricted Cre transgenic mouse (hCD2-Cre) expressed in 30–50% of NK cells (34), coupled with Dicer1 loxP-flanked alleles (24), to generate loss of Dicer1 in the early stages of NK development. We utilized this model to analyze the role of Dicer1-dependent miRNAs in the development, maturation, and function of NK cells. We observed an in vivo developmental and phenotypic maturation defect in miRNA deficient NK precursors and NK cells. However, despite this defect, Dicer1-deficient NK cells exhibited enhanced IFN-γ production and degranulation capacity to a number of stimuli ex vivo. Moreover, miRNA-deficient NK cells responding to MCMV in vivo exhibited similar enhancement of IFN−γ production. The miR-15/16 family was identified as potential novel contributors to IFN-γ protein suppression in NK cells. These findings are consistent with a complex role of miRNAs in NK cell biology that includes a critical role during NK development/homeostasis, and a separate role for miRNAs to dampen NK cell functional responses in mature NK cells.
Materials and Methods
Mice, cell lines, and viral infections
All mice were bred and maintained in specific pathogen-free (SPF) housing, and all experiments were conducted in accordance with the guidelines of and with the approval of the Washington University Animal Studies Committee. Mice were utilized between 8–12 weeks of age for all experiments. C57BL/6J, B6.RAG1−/−, B6.Cg-Tg(CD2-cre)4Kio/J [hCD2-Cre] (34), B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J [Rosa-LSL-YFP] (35), B6.dicerflox/flox [Dicerfl/fl] (24) were obtained from Jackson Labs (Bar Harbor, ME) or investigators at Washington University. YAC-1 cells line (a kind gift of W. Yokoyama) were maintained in K10 media (RPMI-1640, 10% FCS, 10mM HEPES, 1% NEAA, 1% sodium pyruvate, 1% L-glutamine, 1X penicillin/streptomycin). 293T cells (a kind gift of M. Sands) were grown in D10 (DMEM, 10% FCS, 10mM HEPES, 1% NEAA, 1% sodium pyruvate, 1% L-glutamine, 1X penicillin/streptomycin). Murine cytomegalovirus (MCMV) Smith strain was injected IP as described at a dose of 5×104 pfu/mouse (14).
Reagents and monoclonal antibodies
Endotoxin-free purified rmIL-15 and rmIL-12 were obtained from Peprotech (Rocky Hill, NJ) and reconstituted in sterile PBS with 0.1% bovine serum albumin. The following anti-mouse mAbs were obtained from BD Biosciences (San Jose, CA): IFN-γ (XMG1.2), NK1.1 (PK136), NKp46 (29A1.4), CD3 (145-2C11), CD45 (30-F11), CD27 (LG.3A10), CD19 (1D3), and Gr-1 (RB6-8C5), CD132 (4G3), pSTAT5 (clone 47). Additional mAbs were obtained from eBiosciences (San Diego, CA): CD107a (1D4B), CD122 (TM-β1) and CD11b (M1/70). Anti-NK1.1 (PK136) was also purified by the Washington University Antibody Production core from hybridoma supernatant. 2.4G2 (anti-CD16/32) hybridoma supernatant was used to block non-specific staining. CellTrace Violet was used to monitor cell division and was used according to the manufacturer’s instructions (Invitrogen/Molecular Probes, Eugene, OR). AccuCount beads were used for absolute cell number determinations via flow cytometry following the manufacturer’s instructions (Spherotech, Lake Forest, IL).
Cell isolation and sorting
Mouse tissues (spleen, bone marrow, blood, liver) were isolated from 8–12 week old mice as described (14, 17). Briefly, single cell suspensions were generated from spleen (mechanical disruption through a 70uM filter or glass tissue homogenizer), bone marrow (flushing 2 femurs with PBS), liver (mechanical disruption followed by Percoll gradient isolation), or blood (cardiac puncture). RBC lysis was performed using ACK buffer (150mM NH4Cl, 10mM K2CO3, 0.1mM EDTA), and viable cell numbers were determined by trypan blue exclusion. Blood is calculated as cells per mL of blood obtained. Isolation of highly purified NK cells (≥97% pure) was performed by flow cytometric sorting iCyte Reflection (iCyt, Champaign, IL) by gating on lymphocytes based on forward/side scatter, and then CD45+CD3− NK1.1+YFP+/− subsets.
RNA isolation and RT-qPCR
Sorted YFP+ or YFP− NK cells were immediately lysed in Trizol LS (Invitrogen, Carlsbad, CA) according to manufacturers’ instructions. Total RNA was extracted as previously described (36). cDNA was generated from RNA using either the TaqMan Reverse Transcription Kit with random hexamers or TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) with specific primers according to manufacturer’s instructions. Dicer1 excision in cDNA was detected by two methods: 1) internal to the loxP sites or 2) primers external to the loxP sites. Primer/probe sequences are available upon request. miRNA quantification was detected by TaqMan miRNA Kit with specific primers/probe. qRT-PCR was performed on an ABI 7300 using standard settings in a 20uL Reaction. All primers and probes were manufactured by IDT (Coralville, IA). All Dicer1 samples were normalized to 18S rRNA, and all miRNA samples were normalized to housekeeping small RNAs, sno135 (mouse) or RNU48 (human), using the ΔΔCT method. Samples were further normalized to the levels in either WT NK cells or Dicer1fl/fl YFP+ NK cells as appropriate.
Single Cell PCR
Bulk populations of YFP+ or YFP− NK cells were sorted as described above. These cells were processed essentially as described in (37). Briefly, individual cells were isolated and amplified using the GenomiPhi v2 Whole Genome Amplification Kit (GE Healthcare, Piscataway, NJ). The whole genome amplification product (1 uL) was then used in a multiplex PCR reaction to amplify the wild type, floxed, or excised Dicer1 alleles, and the resulting products were analyzed by agarose gel electrophoresis. Primers are available upon request.
Nanostring miRNA Expression Analysis
Total RNA was processed on a Nanostring (Seattle, WA) nCounter instrument using the mouse miRNA Expression Assay Kit according to the manufacturer’s instructions. Data obtained were then sequentially normalized to both housekeeping mRNA controls and positive miRNA-ligation reaction controls of the assay to adjust for total RNA content and ligation efficiency, respectively. This provides a normalized absolute count of 578 mature miRNAs present in the total RNA pool. Non-specific mature miRNA probes (e.g. miR-720) defined by lack of expression based on miRNA-SEQ were excluded from this analysis (31). MiRNAs from Dicer1fl/fl YFP+ NK cells that were detected at 2 SD above background were divided into categories and further analyzed for global miRNA fold change analysis by taking the geometric mean of each group of miRNAs.
Assessment of NK cell function and survival
Splenocytes (1×106) or sorted YFP+ NK cells (1x104) were cultured in 24-well plates (Corning, Corning, NY) containing either K10 + YAC-1 at a 10:1 E:T Ratio, K10 + 10 ng/mL IL-12 and 100 ng/mL IL-15, or K10 + plate-bound anti-NK1.1 (PK136) for 8 hours as described (17, 38); or in K10 + 5ng/mL IL-15 or 100ng/mL IL-15. Flow cytometry data were collected for cell surface markers including CD107a and intracellular IFN-γ on a Beckman Coulter Gallios flow cytometer. Data were analyzed using FlowJo (Tree Star Software, Ashland, OR) or Kaluza (Beckman Coulter, Miami, FL). In some experiments, data are presented as percent maximal CD107a or IFN-γ expression by normalizing to the condition with the highest intra-assay expression in YFP+ NK cells to account for inter-experiment variability. For proliferation assays splenocytes were stimulated as indicated after labeling in K10 with CellTrace Violet according to the manufacturer’s instructions, and data analyzed using FlowJo (Tree Star Software, Ashland, OR). For phospho-STAT5 assays, 1x106 splenocytes were cultured for 15 minutes in 5 ng/mL IL-15 before fixation and staining as described (39).
Assessment of miRNA targeting of 3′ UTRs using luciferase sensor plasmid assays
Overexpression of mature miRNAs and luciferase sensor-plasmids were performed as previously described (31). Briefly, psiCheck2 (Promega, Madison, WI) with the 3′ UTR of IFN-γ was cloned by amplification of the IFN-γ 3′ UTR from IFN-γ cDNA (a kind gift of M. Cooper). Two miR-15/16 binding sites were disrupted in two sequential rounds of mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Agilent/Stratagene, Santa Clara, CA) following the manufacturer’s instructions. MiRNA overexpression vectors were generated by sub-cloning the pre-miRNA gene plus 200bp flanking genomic sequence into the pMND overexpression vector (a kind gift of M. Sands) (31). All primers used for cloning are available upon request. 293T cells were co-transfected with 800 ng of each vector using Dharmafect Duo (Dharmacon, Lafayette, CO) and grown for 48 hours in D10. Dual-glo luciferase assay (Promega, Madison, WI) was then performed according to the manufacturer’s instructions on an LD400 luminescence detector (Beckman Coulter, Brea, CA). Renilla luciferase (experimental) was normalized to Firefly luciferase (control) followed by comparison of Renilla/Firefly ratios of the same psiCheck2 sensor plasmid co-transfected with an ‘empty’ GFP-only control vector. Over-expression was confirmed by GFP expression and RT-qPCR of miRNA samples.
Statistical analysis
Graphical analysis and statistics were performed with GraphPad Prism 5.0. Student’s t test, one-way ANOVA, and two-way ANOVA were used as appropriate, with p < 0.05 considered significant. *p<0.05, **p<0.005, ***p<0.001.
Results
hCD2-Cre transgenic/Rosa26-LSL-YFP mice are an NK cell Cre expression model
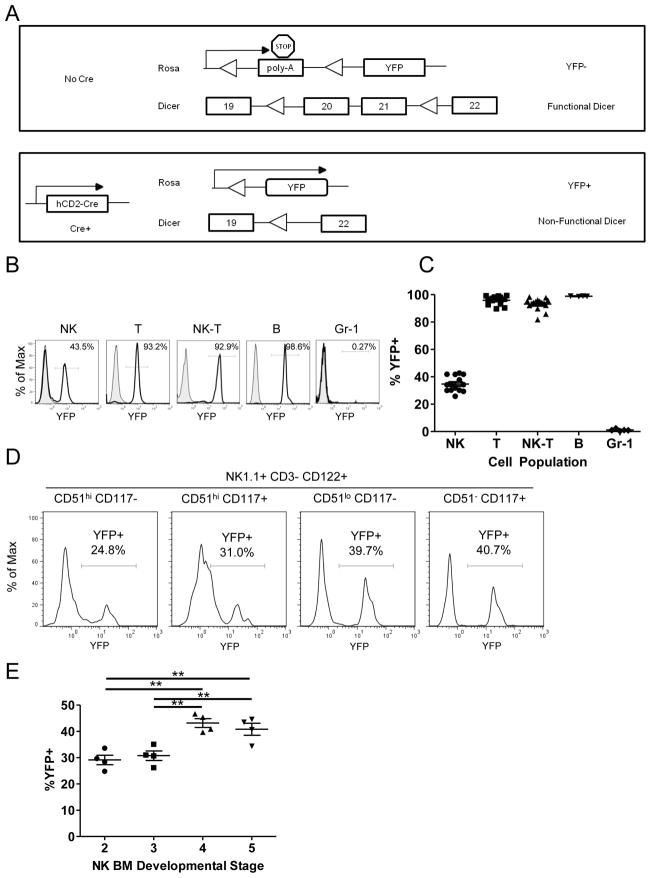
In order to investigate the role of miRNAs in NK cells, we utilized the lymphoid-restricted hCD2-Cre transgenic mouse (34) combined with the Cre reporter model Rosa26-LSL-YFP (35) (CD2-Cre/RosaYFP). In this model, all cells that experience Cre-mediated excision at the Rosa26 locus, and all daughter cells, express YFP protein (Fig. 1A). Using this model, YFP was found to be expressed in 25–50% of splenic NK cells, and expression was confirmed to be lymphocyte restricted with >95% of CD3+ T cells and CD19+ B cells, but < 1% of GR-1+ myeloid cells being YFP+ (Fig. 1B,C). Furthermore, YFP expression was detected in early NK cell precursors in the bone marrow as previously defined (5, 40), and gradually increased in proportion throughout development in the bone marrow, with the percentage of YFP+ NK maximal at the latest stage defined as CD51 (integrin αv)− CD117+ (Fig. 1D,E). In the spleen, YFP+ vs. YFP− NK cells exhibited no difference in the expression of Ly49, NKG2D, or NKp46 receptors (Supplemental Fig. 1A,B). We observed a modest enrichment in stage II/III NK cells (CD27+CD11b+) (Supplemental Fig. 1C) in YFP+ vs. YFP- NK cells. Since maturation differences could potentially bias the results of further analyses, we performed all experiments comparing YFP+ NK cells between Dicer1 genotypes, rather than YFP− and YFP+ NK cells in the same mouse. Collectively, these analyses establish the CD2-Cre/RosaYFP model, whereby YFP+ NK cells in this mouse provide a mechanism to evaluate the role of Dicer1 alterations in NK cells.
FIGURE 1.
hCD2-Cre transgenic Cre expression is lymphocyte-specific, present in NK cells at early stages of NK development, and maturing NK cells are YFP positive early in maturation. (A) Schema of Dicer1/Rosa26 excision in the absence (top) or presence (bottom) of the hCD2-Cre transgene. Cre expression is marked by YFP expression from the Rosa26 locus. LoxP sites are indicated with triangles, exons are indicated by boxes. (B) Representative flow histograms and (C) summary data demonstrating the YFP expression of lymphocyte subsets from the spleen in CD2-Cre/RosaYFP mice. Data are the percentage of YFP positivity in each cell type, including NK (NK1.1+CD3−), T (CD3+NK1.1−), NK-T (NK1.1+CD3+), B (CD19+CD3−NK1.1−), and myeloid (Gr-1+) cells and represent at least 3 independent experiments. (D) Representative histograms and (E) summary data of the percentage of YFP positivity in NK cell developmental intermediates in the bone marrow as defined by Kim et al. (5) with stage definitions by phenotype listed above each YFP histogram (representative of 2 independent experiments). Significance was calculated using one-way ANOVA. *p<0.05, **p<0.01, ***p<0.001.
Constitutive Dicer1-deficient NK cells have reduced miRNA content
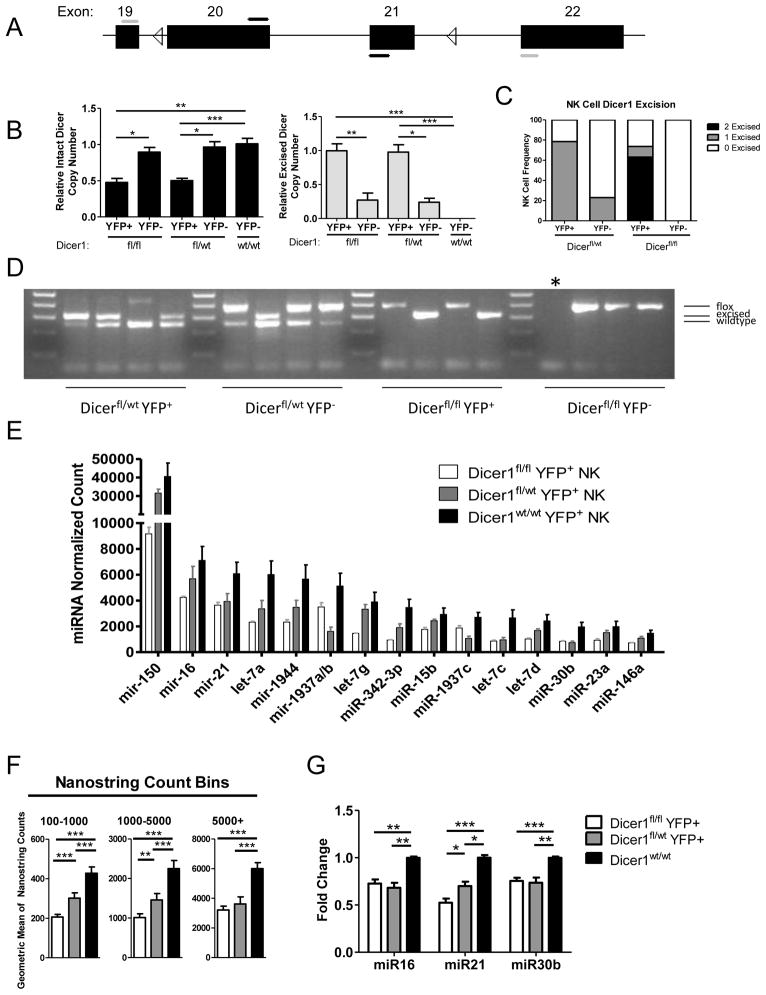
The CD2-Cre/RosaYFP mice were further crossed to mice with various genotypes of loxP-flanked Dicer1 thereby generating Dicer1fl/fl, Dicer1fl/wt, or Dicer1wt/wt mice. Since Cre excision at the Rosa26 locus occurred in only a proportion of mature NK cells, we first evaluated loxP-flanked Dicer1 excision status in YFP+ and YFP− NK cells. Utilizing qRT-PCR to identify excised versus wild type Dicer1 mRNA (Fig. 2A) in sorted bulk NK cell populations, we compared the levels of Dicer1 excision in the Dicer1fl/fl mice to the Dicer1fl/wt mice. Surprisingly, we found that as whole the YFP+ NK cells in both genotypes had comparable degrees of excision detected by this assay (Fig. 2B). Complete excision in the Dicer1fl/fl NK cells is expected to result in no detection with the internal primers and maximal detection with the external primers. To better investigate Dicer1 excision at the level of an individual NK cell rather than a bulk population, we performed single cell PCR on sorted YFP+ and YFP− NK cells (Fig. 2C,D). This identifies wild type, floxed, and excised Dicer1 alleles using a multiplexed PCR approach. We found that 79% of Dicer1fl/wt and 74% of Dicer1fl/fl NK cells had excised Dicer1 alleles. In Dicer1fl/wt the single floxed allele was eliminated. In Dicer1fl/fl mice we detected Dicer1Δ/Δ (63%), Dicer1Δ/fl (11%), and Dicer1fl/fl (26%) alleles in YFP+ NK cells. Thus, 86% of NK cells with evidence of Dicer1 excision had removed both Dicer1 alleles, confirming the efficiency of the hCD2-Cre model in YFP+ NK cells at the single cell level. A fraction of YFP+ NK cells in Dicer1fl/fl mice had not excised either allele of Dicer1, consistent with the qRT-PCR results. Furthermore, by both assays we found that Dicer1 floxed alleles were also excised in a small fraction of YFP− NK cells (Fig. 2B,C), possibly due to loss of YFP in Dicer1fl/fl cells undergoing apoptosis or imperfect Rosa26-YFP excision. This therefore provided a second rationale for comparing YFP+ NK cells between Dicer1 genotypes in order to eliminate this additional potential confounder.
FIGURE 2.
hCD2-Cre transgene expression in NK cells results in Dicer1 excision and loss of mature miRNA expression. (A) Representation of the genomic loxP (triangles)-flanked Dicer1 allele exons (boxes). Primers designed to amplify intact Dicer1 (black) or excised Dicer1 (gray) from cDNA are shown. (B) Real-time RT-qPCR analysis of intact Dicer1 mRNA (left, black bars) or excised Dicer1 mRNA (right, gray bars) in various Dicer1 genotypes of sorted YFP+ NK cells. For qPCR all samples were normalized to 18s rRNA using the ΔΔCT method, and then compared to the level of Dicer1 mRNA in WT NK cells (left) or Dicer1fl/fl YFP+ NK cells (right). Data shown are the mean ± SEM of 2 experiments. (C) Single cell PCR Dicer1 allele frequency. Analysis was supported by at least 12 cells for all cell types except Dicer1fl/fl YFP-NK cells, for which 6 cells were used. (D) Representative gel of Single Cell PCR. Each lane represents a PCR reaction of a single isolated NK cell. Informative product sizes are listed to the right. Dicer1wt = 259bp, Dicer1Δ = 309bp, Dicer1fl = 390bp. Ladder is in increments of 100bp from 100bp to 500bp. * Indicates a failed PCR reaction. Both negative and positive controls provided the expected results (not shown). (E) MiRNA expression in YFP+ Dicer1fl/fl, Dicer1fl/wt, Dicer1wt/wt NK cells measured by Nanostring. Absolute expression profiles of the top 15 miRNAs expressed in Dicer1wt/wt as analyzed by Nanostring showing decreased miRNA expression in Dicer1-deficient NK cells. (F) Summary of miRNA expression changes in Dicer1fl/fl, Dicer1fl/wt, and Dicer1wt/wt YFP+ NK Cells. The geometric mean of Nanostring count groups (>5000, 1000–5000, 100–1000) was compared between Dicer1 genotypes. (G) Confirmation of selected miRNA expression utilizing qRT-PCR of miR-16, miR-21, and miR-30b. Data shown are the mean ± SEM of 3 independent experiments. Significance was calculated using Student’s t-test. *p<0.05, **p<0.01, ***p<0.001.
To confirm the functional impact of excision of Dicer1 in the Dicer1fl/fl and Dicer1fl/wt mice, we next determined the expression of mature miRNAs within various Dicer1 genotypes. We examined YFP+ NK cells for mature miRNA expression by Nanostring miRNA assays (Fig. 2E,F), and found that there was a consistent loss of miRNA content in the Dicer1fl/fl and Dicer1fl/wt NK cells. We further confirmed this loss for three miRNAs representing a range of expression in NK cells (miR-16, miR-21, and miR-30b) (31) via quantitative real time RT-qPCR (Fig. 2G). Consistent with a minor proportion of YFP+ Dicer1fl/fl NK cells lacking Dicer1 excision, miRNA expression in the Dicer1fl/fl NK cell population was reduced but not absent. Taken together, these data suggest that examining both Dicer1fl/wt and Dicer1fl/fl YFP+ NK cells will be useful to elucidate the phenotype of miRNA-deficient NK cells.
MiRNA-deficient NK cells exhibit defects in survival and maturation
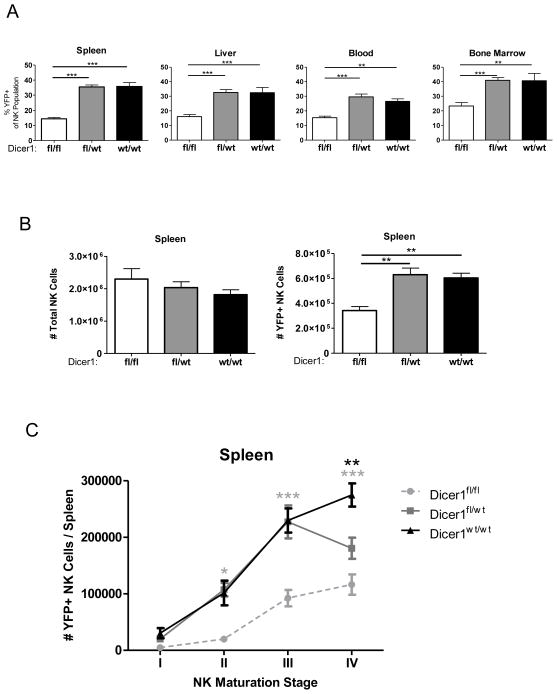
To evaluate effects of Dicer1-deficiency on NK cells in vivo, we first analyzed the percentage of YFP+ NK cells in the spleen, liver, blood, and bone marrow of CD2-Cre/RosaYFP mice with various Dicer1 genotypes. We observed a reduced proportion of YFP+ NK cells in Dicer1fl/fl mice (spleen: 14.42 +/− 3.66%) compared to Dicer1fl/wt (spleen: 35.44 +/− 7.18%) and Dicer1wt/wt mice (spleen: 35.74 +/− 7.68%) (Fig. 3A), further indicating a loss of YFP+ NK cells in the Dicer1fl/fl mice. In addition, while the absolute number of total NK cells was equivalent in all three genotypes (Fig. 3B), there was a significant reduction in the splenic YFP+ NK cell compartment in Dicer1fl/fl mice. While the bone marrow showed a similar pattern, the liver and blood showed no significant difference in the numbers of total NK and YFP+ NK cells for all three genotypes (Supplemental Fig. 1D).
FIGURE 3.
MiRNA-deficient NK cells exhibit an in vivo survival defect. Mononuclear cells were isolated from spleen, liver, blood, and bone marrow. NK cells (CD45+NK1.1+ NKp46+CD3−) were analyzed for YFP expression. (A) Percent YFP positive NK cells in indicated tissues for each genotype. (B) Total viable cells were enumerated, the percentage of YFP+/− NK cells was analyzed by flow cytometry, and absolute total NK and YFP+ NK cell numbers were calculated. (C) NK cells were further analyzed for CD27 and CD11b expression, and the absolute number of cells was calculated as above for maturation stages I (CD27−CD11b−), II (CD27+CD11b−), III (CD27+CD11b+), and IV (CD27−CD11b+) as described (26–29). Significance was calculated using 2-way ANOVA and is presented as Dicer1fl/fl (gray) or Dicer1fl/wt (black) v. Dicer1wt/wt. Data shown are the mean ± SEM of 5 experiments (A–B) or 3 experiments (C). Significance in (A–B) was defined using 1-way ANOVA with a Neuman-Keuls post-test. *p<0.05, **p<0.01, ***p<0.001.
Given the remarkable loss of YFP+ NK cells in the spleen of Dicer1fl/fl mice, we examined whether YFP+ NK loss was associated with peripheral maturation stage. NK cell maturation has been defined in the periphery of mice using co-expression of CD27 and CD11b surface markers (40, 41), and we evaluated YFP expression in these NK cell maturation subsets (Fig. 3C and Supplemental Fig. 1E). There was a defect in Dicer1fl/fl NK cells beginning at Stage II (CD27highCD11blow) and continuing through Stage IV (CD27lowCD11bhigh), while a defect in the Dicer1fl/wt YFP+ NK cells was only evident at Stage IV. This supports a significant, but less severe, effect of miRNA loss on NK cell maturation and/or survival in the Dicer1fl/wt YFP+ NK cells as compared to the Dicer1fl/fl YFP+ NK cells. Thus, Dicer1 loss in NK cells leads to a lower absolute number and percentage of YFP+ NK cells, an effect which is cell-intrinsic and evident at early stages of maturation for Dicer1fl/fl mice, and late stages for Dicer1fl/wt mice.
Dicer-deficient NK cells exhibit reduced IL-15-induced survival and proliferation
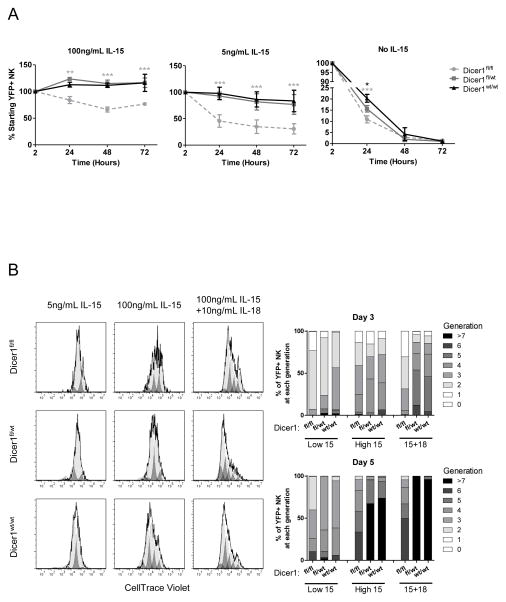
We next investigated whether the defects of Dicer1-deficient NK cells could be rescued in vitro by IL-15, an essential NK cell survival factor. To examine the survival of YFP+ NK cells in vitro, bulk splenocytes were cultured in K10 medium with varying concentrations of rmIL-15. After 24, 48, and 72 hours in medium only (without IL-15) there was a marked decrease in the absolute number of YFP+ NK cells in all Dicer1 genotypes as expected, although a significantly greater survival of the Dicer1wt/wt compared to either the Dicer1fl/wt or Dicer1fl/fl at 24 hours (Fig. 4A). With low dose (5 ng/mL) IL-15, there was prolonged survival of the YFP+ NK cells in the Dicer1fl/wt and Dicer1wt/wt mice, but this effect was significantly decreased in the Dicer1fl/fl mice. With high dose (100 ng/mL) IL-15 a similar phenotype was observed; however, high dose IL-15 partially rescued Dicer1fl/fl YFP+ NK cell numbers, compared to low dose IL-15. These results were also confirmed using sorted YFP+ NK cells (Supplemental Fig. 2A). These results indicate that Dicer1-deficient NK cells have a cell-intrinsic defect in cell survival which is partially rescued with high doses of IL-15.
FIGURE 4.
MiRNA-deficient NK cells exhibit defective survival and proliferation in vitro. (A) Splenocytes from CD2-Cre/RosaYFP reporter mice and the indicated Dicer1 genotypes were plated in 24 well plates in K10 medium supplemented with 0, 5, or 100 ng/mL of rmIL-15. The percentage and absolute number of YFP+ NK cells was determined by flow cytometry using counting beads after 2, 24, 48, and 72 hours of culture. Data shown is the mean ± SEM YFP+ NK cell number normalized to the 2-hour time-point from 3 independent experiments. Significance was calculated using 2-way ANOVA and is presented as Dicer1fl/fl (gray) or Dicer1fl/wt (black) v. Dicer1wt/wt. *p<0.05, **p<0.01, ***p<0.001. (B) Splenocytes from CD2-Cre/RosaYFP reporter mice and the indicated Dicer1 genotypes were labeled with CellTrace Violet and plated in K10 medium supplemented with 5 ng/mL (low 15), 100 ng/mL (high IL-15) rmIL-15, or 100 ng/mL rmIL-15 plus 10 ng/mL rmIL-18 (15+18). Cells were harvested after 3 or 5 days and YFP+ NK cells were analyzed for proliferation indicated by dilution of CellTrace Violet dye.
As a potential mechanism responsible for the IL-15-induced survival defect, we investigated IL-15R protein expression (CD122, CD132), as well as downstream phospho-STAT5 (Supplemental Fig. 2B). We found no decrease in the CD122 or CD132 positive percentage (or MFI) of Dicer1fl/fl, compared to Dicer1fl/wt or Dicer1wt/wt, on YFP+ NK cells. Similarly, there was no decrease in phospho-STAT5 after short-term IL-15 stimulation. In fact, we observed a modest increase in CD132 expression (96 ± 0.6% vs. 91 ± 1.9%, p<0.05) and IL-15-induced phospho-STAT5 (91 ± 1.1% vs. 84 ± 0.7%, p<0.001) in Dicer1fl/fl compared to Dicer1wt/wt YFP+ NK cells. Thus, it is unlikely that reduced IL-15R expression or pSTAT5 signals are responsible for the observed survival defect.
We next evaluated the ability of IL-15 and IL-18 to induce proliferation in NK cells from these mice. Splenic NK cells were labeled with CellTrace Violet (a CFSE-analogue) and cultured in 5 ng/mL IL-15, 100 ng/mL IL-15, or 100 ng/mL IL-15 + 10 ng/mL IL-18 to induce proliferation (Fig. 4B). After 3 days, Dicer1fl/fl YFP+ NK cells had reduced proliferation in 100 ng/mL IL-15 + 10 ng/mL IL-18 (p<0.05) compared to Dicer1wt/wt NK cells, as measured by dye dilution with cell division. After 5 days, Dicer1fl/fl YFP+ NK cells had reduced proliferation in both 100ng/mL IL-15 (p<0.01) and 100ng/mL IL-15 + 10ng/mL IL-18 (p<0.01) compared to Dicer1wt/wt NK cells. There was normal proliferation in the Dicer1fl/wt NK cells. Therefore, Dicer1-deficient NK cells exhibit an in vitro survival and proliferative defect.
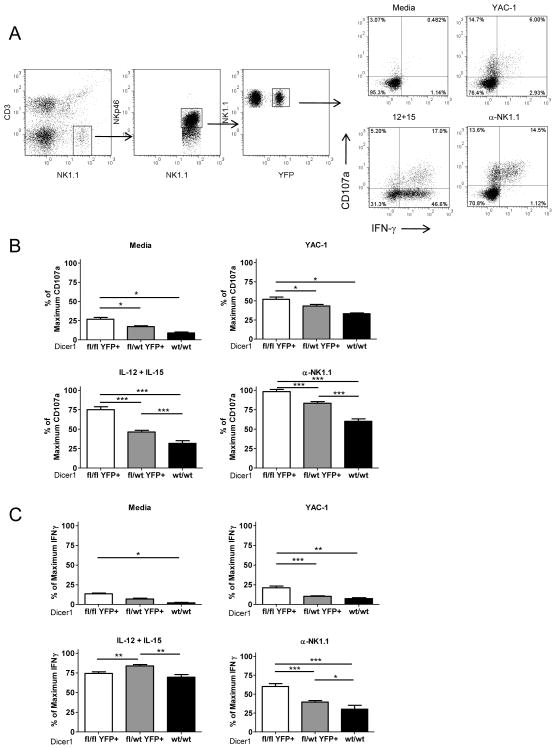
Dicer-deficient NK cells exhibit enhanced degranulation and IFN-γ production ex vivo
We evaluated the functional capacity of Dicer1-deficient NK cells in vitro. Splenocytes from these mice were cultured in either K10 medium alone or stimulated with YAC-1 cells (NK cell stimulatory cell line); 10 ng/mL IL-12 + 100 ng/mL IL-15, an IFN-γ inducing cytokine stimulus; or plate-bound anti-NK1.1 (PK136) to generate a potent activating NK cell receptor signal (Fig. 5A). After 7 hours of culture both CD107a (a marker of degranulation) (Fig. 5B) and IFN-γ production (Fig. 5C) were assessed by flow cytometry. Notably, the Dicer1fl/fl mice had a significantly higher percentage of CD107a+ (degranulated) YFP+ cells when cultured ex vivo, even in the absence of stimulation, or when stimulated via cytokines, NK cell receptor ligation, or tumor cell interaction. In addition, the Dicer1fl/wt YFP+ NK cells also had significantly higher degranulation than the Dicer1wt/wt NK cells in the IL-12+IL-15 and anti-NK1.1 cross linking conditions, and a similar trend that did not reach statistical significance for medium or YAC-1 co-culture. IFN-γ responses were similarly enhanced in Dicer1fl/fl YFP+ NK cells in medium, YAC-1 co-culture and NK1.1 ligation, compared to Dicer1wt/wt NK cells. In the setting of IL-12 + IL-15 stimulation, there was a significant increase in IFN-γ in Dicer1fl/wt compared to Dicer1wt/wt and Dicer1fl/fl NK cells; however, the Dicer1fl/fl did not exhibit enhanced IFN-γ with this cytokine combination. In addition, to confirm that this phenotype was cell-intrinsic, sorted YFP+ NK cells were cultured in media alone or with plate-bound anti-NK1.1 (Supplemental Fig. 2C). Sorted YFP+ Dicer1fl/fl cells stimulated with anti-NK1.1 produced significantly more IFN-γ than their YFP+ Dicer1wt/wt counterparts (93 ± 7.3 v. 39 ± 18, p<0.05). Collectively, these data show that miRNA-deficient NK cells have a cell-intrinsic enhanced capacity to produce IFN-γ and degranulate in response to multiple stimuli in vitro.
FIGURE 5.
MiRNA-deficient NK cells have increased functional capacity defined by in vitro degranulation and IFN-γ production. Splenocytes were cultured for 7h in K10 medium only, or stimulated with YAC-1 tumor cells, IL-12+IL-15, or anti-NK1.1 (plate bound PK136). (A) Example flow cytometry gating scheme that identifies NK1.1+NKp46+CD3−YFP+ NK cells expressing cell surface CD107a or intracellular IFN-γ protein in Dicer1wt/wt splenocytes. Summary data of the percent positive NK cells for CD107a (B) or IFN-γ (C) expression after 7h with the indicated stimulus. The mean ± SEM of 5 independent experiments is shown expressed as the percent maximal expression within each experiment to account for expected inter-assay variability. Significance was calculated using 1-way ANOVA. *p<0.05, **p<0.01, ***p<0.001.
We also investigated the impact of maturation stage defined by CD27/CD11b on degranulation (CD107a), and observed that stage II-III NK cells in the Dicer1fl/fl mice were primarily responsible for the enhanced degranulation (Supplemental Fig. 3A). Prior studies have shown that stage III NK cells exhibit a higher degranulation capacity compared to the more mature stage IV subset, which may allow increased degranulation due to miRNA defects in these cells to be more readily apparent (40, 41). In addition, this effect was not due to altered Ly49/NKG2D receptor expression (Supplemental Fig. 3B). Thus, neither maturation stage skewing nor Ly49/NKG2D expression changes seem responsible for the observed phenotype. Granzyme B protein levels were found to be variable in NK cells from these mice, with a trend toward increased expression in Dicer1fl/fl (Supplemental Fig. 3C). This implies that miRNA loss, even the smaller decrement in Dicer1fl/wt mice, may control the activation threshold of NK cells.
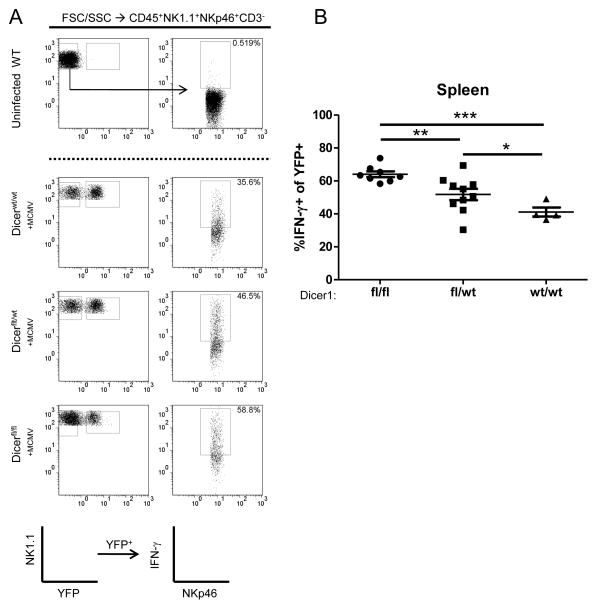
Dicer1-deficient NK cells exhibit enhanced IFN-γ production in vivo during MCMV infection
To evaluate the impact of miRNA-deficiency in NK cells during a physiologic in vivo response, we infected Dicer1fl/fl, Dicer1fl/wt, and Dicer1wt/wt with 5x104 PFU of MCMV and evaluated IFN-γ production 36 hours post-infection, the expected peak IFN-γ time point. Consistent with the in vitro results, splenic YFP+ NK cells from Dicer1fl/fl mice had significantly higher IFN-γ protein production compared to Dicer1wt/wt mice (Fig. 6A,B). Further, Dicer1fl/wt mice demonstrated an intermediate phenotype, with IFN-γ production between Dicer1fl/fl and Dicer1wt/wt. These data suggest that miRNA-deficient NK cells have an enhanced ability to produce IFN-γ in vivo during an ongoing anti-viral response.
FIGURE 6.
miRNA-deficient NK cells exhibit enhanced IFN-γ production and degranulation in response to MCMV in vivo. Mice were infected IP with 5x104 PFU of MCMV, and spleens were analyzed for IFN-γ expression by flow cytometry after 36 hours. (A) Representative flow cytometry of YFP expression and IFN-γ staining, previously gated on CD45+NK1.1+NKp46+CD3− lymphocytes. (B) Summary data of IFN-γ staining. Data shown are the mean ± SEM of two independent experiments. Significance was calculated using 1-way ANOVA. *p<0.05, **p<0.01, ***p<0.001.
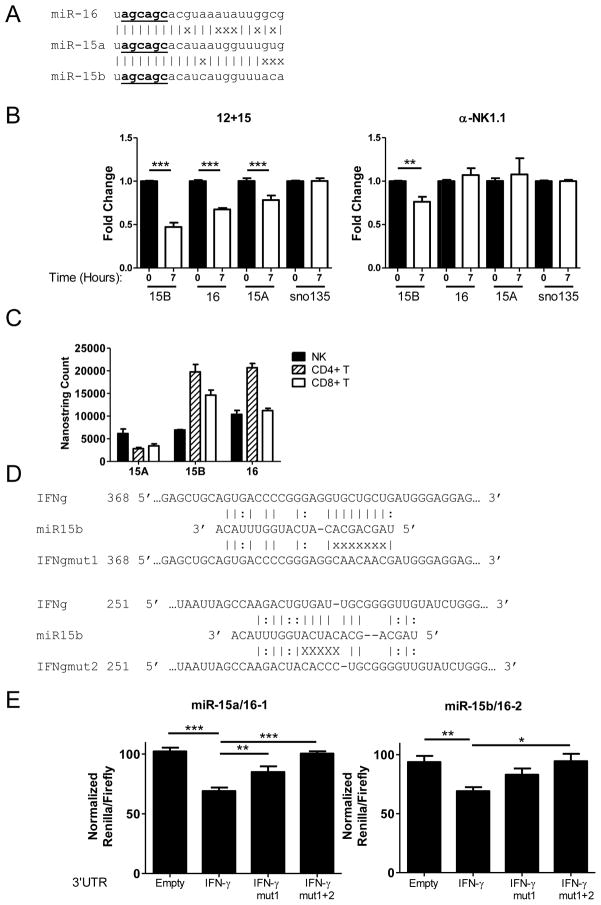
The miR-15/16 family specifically targets the IFN-γ 3′ UTR
As IFN-γ production was significantly increased in miRNA-deficient NK cells both in vitro and in vivo, we evaluated the role of individual miRNAs for their potential to directly target IFN-γ protein production via binding to the murine IFN-γ 3′UTR. A number of miRNAs are predicted (42) by algorithms to bind to the murine IFN-γ 3′UTR, including miRs−15a/15b/16, a closely related family of microRNAs (Fig. 7A). Culture of sorted NK cells for 7 hours with stimuli that induce IFN-γ protein expression, 100 ng/mL IL-15 + 10 ng/mL IL-12 or plate-bound anti-NK1.1, resulted in a significant reduction of miR-15b in both conditions, and a significant reduction in miR-15a and miR-16 in the 12+15 condition (Fig. 7B). These miRNAs are among the most highly expressed miRNAs in IFN-γ producing lymphocytes (Fig. 7C), and have decreased expression in Dicer1fl/fl and Dicer1fl/wt mice (Fig. 2E). We therefore evaluated the miR-15/16 family, along with a number of candidates using in vitro over-expression of individual miRNAs with concurrent 3′UTR luciferase ‘sensor plasmid’ as a readout of targeting (Supplemental Fig. 4A). Some miRNAs, such as miR-27a and miR-29a, were found to repress the empty vector, and were discarded due to a lack of specificity. The miR15/16 family was not found to target the empty psiCheck2 vector (Supplemental Fig. 4A), but was strongly predicted to target the IFN-γ 3′ UTR in two sites (Fig. 7D). In co-transfection assays in which Renilla luciferase is fused to the 3′ UTR of IFN-γ, overexpression of miR-15a/16-1 or miR-15b/16-2 in 293T cells (Supplemental Fig. 4B) dramatically decreased the Renilla signal compared to negative controls (GFP only ‘empty’ vector or overexpression of an irrelevant miRNA) (Fig. 7E). This indicates that miRs-15a/15b/16 regulate the IFN-γ 3′UTR. To interrogate the specificity of the IFN-γ 3′ UTR repression, we specifically mutated the top predicted site (IFNgmut-1) or the top two (IFNgmut-1+2) predicted sites (Fig. 7D). This resulted in a sequential and substantial loss and abrogation of IFN-γ 3′UTR repression by miR15a/15b/16, indicating that the targeting was direct. Thus, these biochemical results, plus the expression pattern in resting/stimulated WT NK cells and Dicer1fl/fl vs. Dicer1wt/wt mice, are consistent with miR-15/16 targeting of the 3′UTR as a possible mechanism for increased IFN-γ production in our model.
FIGURE 7.
The miR-15/16 family is highly expressed in NK cells, decreases upon cytokine activation, and directly represses the murine IFN-γ 3′UTR. (A) Schematic of the relationship of the miR-15/16 family members. (|) indicates a base pair match, whereas (x) indicates base pair differences. Underlined nucleotides indicate the miRNA “seed” sequence. (B) RT-qPCR of sorted WT NK cells activated at baseline (black) or activated for 7 hours (white) in 100ng/mL IL-15 + 10ng/mL IL-12 (left) or with plate-bound PK136 (right). (C) Nanostring miRNA expression analysis comparing the levels of miR-15/16 in sorted NK, CD4+ T, and CD8+ T lymphocytes illustrating that miR-15/16 is abundant in IFN-γ producing cells. (D) Schematic of two separate predicted mir-15/16 binding sites within the IFN-γ 3′UTR, with Watson-Crick (|) and wobble (:) base pairing indicated. In the mutated IFN-γ 3′UTR sequence, “x” indicates bp location altered to disrupt predicted miR-15/16 binding. Nucleotide numbering indicates position from the 5′ end of the IFN-γ 3′ UTR. (E) 293T cells were co-transfected with the psiCheck2 sensor plasmid under the control of the indicated 3′UTR and a pMND vector over-expressing GFP and either miR-15a/16-1 or miR-15b/16-2. Compared to a GFP-only expression vector, or irrelevant miRNA (miR-21, not shown), mmu-miR-15a/16-1 (left) and mmu-miR-15b/16-2 (right) repress translation via direct targeting of the IFN-γ 3′ UTR. This repression is abrogated when both binding sites are mutated, indicating that miR-15/16 repression is specific. Data shown are the mean ± SEM of at least 2 (B,C) or 3 (E) independent experiments. Significance was calculated using 1-way ANOVA. *p<0.05, **p<0.01, ***p<0.001.
Discussion
In this study we utilized a combination of genetic mouse models to achieve ‘selective’ miRNA deficiency in NK cells, which are identifiable due to their expression of YFP. Based upon the Rosa26-LSL-YFP reporter allele, Cre expression and Dicer1 excision consistently occurred early in NK cell development, and persisted in mature peripheral NK cells. This model demonstrated that Dicer1-dependent miRNAs play a role in NK cell development, survival, and proliferation -when miRNAs are globally decreased NK cell development and maturation are impaired. These results suggest that specific miRNAs or groups of miRNAs are important intrinsic regulators of NK cell development and homeostasis. However, our results also revealed that miRNA-deficient NK cells had enhanced functionality, which included increased IFN-γ production and degranulation (as a surrogate for cytotoxicity) in vitro, and enhanced IFN-γ responses in vivo during MCMV infection. This phenotype indicates that miRNAs also play a role in dampening NK cell responses in mature peripheral NK cells. Indeed, we identified a family of related miRNAs, miR-15a/15b/16, that are decreased in Dicer1-deficient mice and during NK cell activation, and directly target the IFN-γ 3′UTR. Thus, miRNA regulation of the NK cell molecular programs is complex, with effects in development and homeostasis, as well as regulating NK cell functional responses.
Limited studies have been performed to date evaluating the expression and significance of miRNAs in NK cell biology. We have previously defined the expression of miRNAs in resting mature murine NK cells and following IL-15 activation using multiple profiling platforms, including miRNA-SEQ, and identified novel miRNAs in NK cells (31). Consistent with our current hypothesis regarding IFN-γ regulation, miRs-15a/15b/16 were highly expressed in resting NK cells. In the prior study, miR-223 was also identified as a potential direct regulator of another NK cell effector molecule, granzyme B, in resting NK cells. In our current study, granzyme B protein levels had a high degree of variability, with a trend toward higher granzyme B protein in Dicer1fl/fl NK cells, compared with Dicer1fl/wt or Dicer1wt/wt. Since our Dicer1-deficiency model results in the decrease of a large number of miRNAs simultaneously, there may be indirect effects on other pathways important for granzyme B expression in NK cells, or it may disrupt miRNAs that both positively and negatively regulate granzyme B expression. Current experiments utilizing miR-223−/− mice (43) are underway to further evaluate the regulation of NK cell function by miR-223.
Bezman et al. recently reported the phenotype of NK cells in the context of drug-induced Dicer1 or Dgcr8 (a critical component of the Drosha complex) loss, utilizing an estrogen receptor (ER)-Cre model to excise Dicer1fl/fl or Dgcr8fl/fl alleles (33). Such an approach reduces miRNAs in all cells of the mouse (including NK cells) after 5 days of tamoxifen treatment. This study identified a phenotype similar to our model for miRNA-deficiency in regards to NK cell survival; however, there were a number of phenotypic differences that warrant discussion in the context of the disparate models. The hCD2-Cre in our study reduces miRNA expression in NK cells from a uniform point during NK cell development, with consistent loss of miRNAs in mature NK cells. In contrast, the ER-Cre model results in a ‘window’ of Cre mediated excision in both mature NK cells and presumably their precursors following tamoxifen treatment, and thus provides a potentially heterogeneous NK cell population with regards to the timing of Dicer1 or Dgcr8 loss during NK cell development and maturation. In addition, ER-Cre eliminates Dicer1 and Dgcr8 in all mouse cells providing no selectivity for NK cells, and therefore using this approach it may be challenging to differentiate the effects of NK cell intrinsic vs. extrinsic miRNA deficiency. These differences between the model systems may be responsible for the contrasting phenotypes we are reporting for mature NK cell function. The ER-Cre model described a ‘hypofunctional’ NK cell phenotype with impaired IFN-γ production and degranulation in response to activating NK cell receptors in vitro - the opposite of the phenotype we observed using the hCD2-Cre model. In addition, the authors report increased NKG2D receptor expression in miRNA-deficient NK cells, which we did not observe in the hCD2-Cre model. In the setting of MCMV infection, the ER-Cre approach identified no change in IFN-γ production by splenic NK cells during MCMV infection. However, in the hCD2-Cre model there was a clear and statistically significant increase in IFN-γ protein in splenic NK cells 36 hours post-infection. Similar to our phenotype with hCD2-Cre driving Dicer1-loss in NK cells, CD8+ T cell responses in Dicer1−/− mice achieved by a distal Lck-Cre identified enhanced activation with faster kinetics in miRNA-deficient CD8+ T cells (44). Recently, Eckelhart, et. al. and Narni-Mancinelli, et. al. have independently reported NK cell-specific NKp46-Cre mice (45, 46). Future studies utilizing these models that direct Cre expression only in NK cells or NK cell precursors will provide additional information on the functional ramifications of miRNA biogenesis or specific miRNAs in vivo.
The data presented support a role for miRNAs in the appropriate survival, maturation, and proliferation of NK cells. As the turnover of Dicer1 protein and miRNAs is rapid (26–29), and NK cell maturation takes weeks (47), Dicer1 protein and therefore Dicer1-dependent small RNAs are likely reduced at an early stage of NK development. By using a model in which Dicer1 excision occurs contemporaneously and at an early stage of NK cell development, we could analyze the role of Dicer1 in the survival and maturation of NK cells. By analyzing various organs, we observed that Dicer1fl/fl NK cells failed to reach a defined stage of peripheral NK cell development: the CD27+CD11blow stage, which is associated with cellular proliferation (40), consistent with the crucial function of miRNAs in cell division (48). In contrast, Dicer1fl/wt NK cells were underrepresented in the final stages of NK maturation, possibly indicating a failure in proper senescence (49, 50) and consistent with the intermediate level of miRNAs in these cells as analyzed by Nanostring. As the numbers of YFP+ NK cells were only significantly decreased in the spleen and bone marrow, we cannot formally rule out that these alterations may be due to a homing defect of the Dicer1fl/fl NK cells. However, this seems less likely given the defect in the NK YFP+ percentage of all peripheral organs. Furthermore, there was a significant decrease in the ability of miRNA-deficient NK cells to survive ex vivo in IL-15, which was not explained by reduced IL-15R protein expression or signals via phospho-STAT5. MiRNAs previously reported to have a role in regulating development and proliferation, miR-150 (51), miR-16 (52), and miR-21 (53), are highly expressed in NK cells, and are likely candidates to be involved in the repression of genes required for proper development and proliferation in maturing NK cells.
NK cell activation involves an intricate relationship between activating and inhibitory signaling, with an activation threshold determined by a variety of interactions, many of which have yet to be fully understood (54). Previous reports have shown a specific role for miR-181a in tuning the sensitivity of T cells (55, 56) by targeting the phosphatases PTPN22 and SHP-2. The increased NK cell functionality in our hCD2-Cre model implies the net effect of reduced Dicer1-dependent miRNAs in NK cells is loss of activating signal repression, possibly via a loss of targeting of DAP12, other ITAM-containing receptors or their downstream signaling molecules, as well as cytokine receptor signaling pathways. As NK cell activation can be extremely sensitive to minute changes in activating and inhibitory receptor levels (57), it is a prime candidate for miRNA-based “fine tuning”. It is likely that because both degranulation and IFN-γ production are increased in the Dicer1fl/fl NK cells, molecule(s) upstream of both degranulation and cytokine production may be targets of miRNAs. The impact of miRNAs on the signals that ultimately are integrated from activating and inhibitory cell surface molecules are a potential mechanism contributing to whether an NK cell responds once triggered.
IFN-γ regulation is multi-faceted, and includes control of gene transcription as well as post-transcriptional events (58). We identified loss of miR-15/16 family miRNAs as potential players responsible for the increased IFN-γ production in the hCD2-Cre/RosaYFP Dicer1fl/fl mice. These related miRNAs are highly expressed in resting NK cells, target the 3′ UTR of murine IFN-γ, and are decreased in Dicer1fl/fl and Dicer1fl/wt NK cells. It has been previously suggested that IFN-γ is regulated in humans by a psuedoknot element in its 5′ UTR (59) and by miR-29 (32, 60). While we also initially identified miR-29 as a potential IFN-γ regulator using in silico algorithms, it did not target the mouse 3′UTR in our luciferase-based assays. Consistent with our targeting data, miR-15/16 has a much higher relative expression than miR-29 in NK cells, and is highly expressed in NK and T cells, providing further biological rationale for the regulation of IFN-γ. Moreover, the 3′ UTR of IFN-γ is conserved between human and mouse, and miR-15/16 is predicted to target human IFN-γ, indicating a potential conserved mechanism between species. The miR-15/16 family is known to be highly expressed in lymphocytes as defined by miRNA-SEQ (31), suggesting that it has an integral role in lymphocyte development and/or function. Additionally, the miR-15/16 loci has been implicated in a variety of cellular roles in lymphocytes (61–63). This study’s finding that miR15/16 targets IFN-γ, a crucial immune response molecule, provides further evidence for miR15’s vital role in the immune response. Future studies will involve specifically targeting miR-15/16 family members in NK cells.
MiRNAs have been found to have a role in nearly all cell types, and to exert their regulation on a wide variety of cellular functions. In this study, we identify the role of miRNAs in modulating the proliferation, survival, and the essential functions of NK cells. MiRNA-deficient NK cells have decreased survival and proliferation, but increased degranulation and IFN-γ production, implying a central role for miRNAs in the control of physiologic NK cell responses. The role of miRNAs in dampening NK cell function may be found to have a broad impact on the fields of autoimmunity, viral defense, tumor immunotherapy, and hematopoietic stem cell transplantation, in which roles for NK cells are well established (33). Future studies, which will require more subtle manipulations of individual miRNAs or miRNA clusters, will further define the mechanism by which miRNAs control the development and activation threshold of NK cells.
Supplementary Material
Acknowledgments
We thank Drs. Skip Virgin and Wayne Yokoyama for providing mice, Drs. Mark Sands and Megan Cooper for providing reagents, Bill Eades and the Siteman Cancer Center High Speed Cell Sorting Core for cell sorting, and the Rheumatology Antibody Production Core for producing purified anti-NK1.1 (PK136) mAb. We also thank Dr. Megan Cooper and members of the Fehniger laboratory for critical review of the manuscript, Tammi Vickery and Patsy Alldredge for assistance with Nanostring assays, Dr. Stephen Oh for advice on phopho-STAT5 flow cytometry, and Theresa Geurs for technical assistance with MCMV infections.
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–29. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Santo JP. Natural killer cells: diversity in search of a niche. Nat Immunol. 2008;9:473–5. doi: 10.1038/ni.f.201. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Iizuka K, Kang HSP, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–8. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 6.Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol. 2003;3:413–25. doi: 10.1038/nri1088. [DOI] [PubMed] [Google Scholar]
- 7.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 8.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–79. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson HA, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Advances in Immunology. 2009:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 10.Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. 2010;142:847–56. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr Opin Immunol. 2001;13:458–64. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 13.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 14.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–17. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaix J, Tessmer MS, Hoebe K, Fuséri N, Ryffel B, Dalod M, Alexopoulou L, Beutler B, Brossay L, Vivier E, Walzer T. Cutting edge: Priming of NK cells by IL-18. J Immunol. 2008;181:1627–31. doi: 10.4049/jimmunol.181.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, Ley TJ, Virgin HW, Fehniger TA. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–83. doi: 10.1182/blood-2009-09-245464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orange JS. Formation and function of the lytic NK-cell immunological synapse. Nat Rev Immunol. 2008;8:713–25. doi: 10.1038/nri2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu Rev Immunol. 2002;20:323–70. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 20.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 23.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–73. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedeli M, Napolitano A, Wong MPM, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, Casorati G. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol. 2009;183:2506–12. doi: 10.4049/jimmunol.0901361. [DOI] [PubMed] [Google Scholar]
- 26.Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koralov SB, Muljo Sa, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Kuipers H, Schnorfeil FM, Fehling HJ, Bartels H, Brocker T. Dicer- Dependent MicroRNAs Control Maturation, Function, and Maintenance of Langerhans Cells In Vivo. J Immunol. 2010;185:400–409. doi: 10.4049/jimmunol.0903912. [DOI] [PubMed] [Google Scholar]
- 29.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belver L, Papavasiliou FN, Ramiro AR. MicroRNA control of lymphocyte differentiation and function. Curr Opin Immunol. 2011;23:368–73. doi: 10.1016/j.coi.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, Heinz ME, Crosby SD, Nagarajan R, Ramsingh G, Link DC, Ley TJ, Mardis ER. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 33.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DGT, Lanier LL. Distinct Requirements of MicroRNAs in NK Cell Activation, Survival, and Function. J Immunol. 2010;185:3835–46. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–25. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 35.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tighe S, Held MA. Isolation of total RNA from transgenic mouse melanoma subsets using fluorescence-activated cell sorting. Meth Mol Biol. 2010;632:27–44. doi: 10.1007/978-1-60761-663-4_2. [DOI] [PubMed] [Google Scholar]
- 37.Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, Sermon K. Whole-genome multiple displacement amplification from single cells. Nat Protocols. 2006;1:1965–70. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 38.Ruitenberg JJ, Ghanekar SA, Brockstedt DG, Maecker HT. Simultaneous detection of murine antigen-specific intracellular cytokines and CD107a/CD107b by flow cytometry. Nat Methods 2007 [Google Scholar]
- 39.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4:132–42. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 40.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–96. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 41.Hayakawa Y, Smyth MJ. CD27 Dissects Mature NK Cells into Two Subsets with Distinct Responsiveness and Migratory Capacity. J Immunol. 2006;176:1517–24. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 44.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A. 2010;107:21629–34. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckelhart E, Warsch W, Zebedin E, Simma O, Stoiber D, Kolbe T, Rülicke T, Mueller M, Casanova E, Sexl V. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117:1565–73. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 46.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, Vivier E. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31:1900–9. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 48.Bueno MJ, de Castro Perez I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 49.Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, Martindale JL, Rinker-Schaeffer CW, Gorospe M. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2:ra69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 51.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–5. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 54.Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, Chevrier N, Walzer T, Kerdiles YM, Marguet D, Vivier E, Ugolini S. Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Sci Signal. 2011;4:ra21. doi: 10.1126/scisignal.2001608. [DOI] [PubMed] [Google Scholar]
- 55.Li QJ, Chau J, Ebert PJR, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Ebert PJR, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–9. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmes TD, El-Sherbiny YM, Davison A, Clough SL, Blair GE, Cook GP. A human NK cell activation/inhibition threshold allows small changes in the target cell surface phenotype to dramatically alter susceptibility to NK cells. J Immunol. 2011;186:1538–45. doi: 10.4049/jimmunol.1000951. [DOI] [PubMed] [Google Scholar]
- 58.Young HR. Regulation of Interferon-g Gene Expression. J Interf Cytok Res. 1996;568:563–568. doi: 10.1089/jir.1996.16.563. [DOI] [PubMed] [Google Scholar]
- 59.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–32. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 60.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 63.Ofir M, Hacohen D, Ginsberg D. miR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res. 2011;9:440–7. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.