Abstract
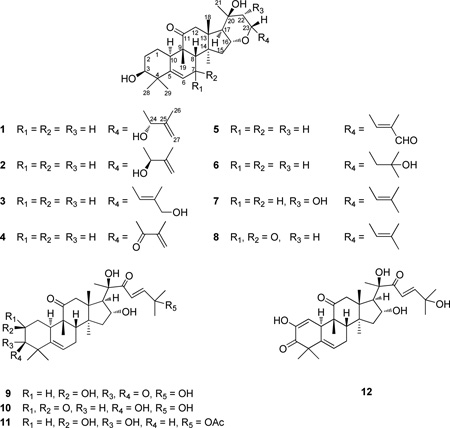
Eight new 16,23-epoxycucurbitacin derivatives, designated as elaeocarpucins A–H (1–8), and five known cucurbitacins (9–13) were isolated from the chloroform-soluble partitions of separate methanol extracts of the fruits and stem bark of Elaeocarpus chinensis collected in Vietnam. Isolation work was facilitated using a LC/MS dereplication procedure, and bioassay-guided fractionation was monitored using HT-29 human cancer cells. The structures of compounds 1–8 were determined on the basis of spectroscopic data interpretation, with the absolute configurations of isomers 1 and 2 established by the Mosher ester method. Compounds 1–13 were evaluated in vitro against the HT-29 cell line and using a mitochondrial transmembrane potential assay. Elaeocarpucin C (3), produced by partial synthesis from 16α,23α-epoxy-3β,20β-dihydroxy-10αH,23βH-cucurbit-5,24-dien-11-one (13), was found to be inactive when evaluated in an in vivo hollow fiber assay using three different cancer cell types (dose range 0.5–10 mg/kg/day, ip).
Elaeocarpus chinensis (Gardner & Champ.) Hook.f. ex Benth. (syn.: Friesia chinensis Gardner & Champ.), an evergreen tree of the family Elaeocarpaceae, is distributed mainly in subtropical or tropical areas of Asia, including southern mainland China, Laos, and Vietnam.1 Besides being grown for ornamental purposes, E. chinensis is used also as a traditional Chinese herbal medicine for the treatment of emmeniopathy as well as extravasated blood and inflammatory edema caused by traumatic injury.2 Elaeocarpus is a large genus comprised of 350–360 species distributed from Madagascar to Oceania, with the highest concentration occurring in Borneo and Papua New Guinea.3,4 Previous phytochemical work has resulted in the isolation of anthocyanins,5 cucurbitacin-type triterpenoids,6–12 flavonoids,13,14 other phenolic derivatives13, and indolizidine alkaloids,15–17 Among these compounds, cucurbitacins and their derivatives are tetracyclic triterpenoids obtained initially from plants of the family Cucurbitaceae, and are reported to have anticancer, antifertility, anti-inflammatory, cytotoxic, and purgative activities.18,19 Although the development of cucurbitacins as anticancer drug candidates has been hindered by their non-specific cytotoxicity, there is much interest in the relationship of structure to cytotoxicity within this compound class.18,19 Some cucurbitacins have been found to affect JAK-STAP and MAPK signaling pathways in cancer cells, and to show synergistic effects in combination with certain known anticancer therapeutic agents, such as doxorubicin and gemcitabine.20,21 Thus far, there have been no studies on the phytochemical constituents of E. chinensis.
As part of our ongoing program to discover new anticancer agents from varied natural sources,22,23 the CHCl3 extract of the fruits of Elaeocarpus chinensis was found to exhibit cytotoxic activity (IC50 0.4 µg/mL) against human colon cancer (HT-29) cells. Scrutiny of the NAPRALERTSM (Natural Products Alert) database24 indicated that more than 150 compounds have been isolated from the genus Elaeocarpus, with most of the cytotoxic compounds known being cucurbitacin-type triterpenes. In order to decide whether or not to further pursue this lead, the CHCl3-soluble extract of E. chinensis fruits was subjected to an LC-MS dereplication procedure, which revealed the probable presence of the known cytotoxic cucurbitacins, cucurbitacin D (9), 3-epi-isocucurbitacin D (10) and 25-O-acetylcucurbitacin F (11). In addition, certain unknown cytotoxic compounds corresponding to possible molecular formulas of C30H44O5 and C30H46O5 were evident. Accordingly, subsequent bioassay-guided fractionation was conducted using HT-29 cancer cells to monitor purification, and led to the isolation of six new 16,23-epoxycucurbitacins, elaeocarpucins A–F (1–6), together with five known cucubitacins inclusive of cucurbitacin D (9),25,26 3-epi-isocucurbitacin D (10),26 25-O-acetylcucurbitacin F (11),9,27 cucurbitacin I (12)28 and 16α,23α-epoxy-3β,20β-dihydroxy-10αH,23βH-cucurbit-5,24-dien-11-one (13).11 Moreover, from the less potently cytotoxic CHCl3 extract of the stem bark of the same plant, two additional new cucurbitacins, elaeocarpucins G (7) and H (8), were purified. Herein, we report the isolation and structure elucidation of the eight new compounds, 1–8, as well as the biological assessment of all isolates obtained in this investigation.
RESULTS AND DISCUSSION
Compound 1 was obtained as a white amorphous powder. Its molecular formula was assigned as C30H46O5 based on the [M + Na]+ ion peak at m/z 509.3225 (calcd 509.3243) in the HRESIMS. Observed in the 1H NMR spectrum were signals for seven tertiary methyl groups at δH 0.93 (3H, s, H-18), 1.02 (3H, s, H-28), 1.14 (3H, s, H-19), 1.17 (3H, s, H-29), 1.22 (3H, s, H-30), 1.32 (3H, s, H-21), and 1.72 (3H, s, H-26), while resonances at δH 3.48 (1H, brs, H-3), 4.40 (1H, ddd, J = 10.4, 10.4, 3.6 Hz, H-16), 4.01 (1H, ddd, J = 12.0, 3.0, 3.0 Hz, H-23), and 4.24 (1H, d, J = 2.8 Hz, H-24) were attributed to proton signals attached to four oxygenated methine carbons. In addition, three olefinic protons were recognized in the 1H NMR spectrum at δH 4.91 (1H, s, H-27a), 5.04 (1H, s, H-27b), and 5.67 (1H, d, J = 5.8 Hz, H-6). The 13C NMR spectrum of 1 showed 30 carbon signals, which were classified from DEPT and HSQC data into seven methyls, six methylenes, three methines, four quaternary carbons, five oxygen-bearing carbons (including four secondary and one tertiary), a trisubstituted double bond, a disubstituted terminal double bond, and a carbonyl group. The characteristic NMR data of compound 1 were comparable to those of 16α,23α-epoxy-3β,20β-dihydroxy-10αH,23βH-cucurbit-5,24-dien-11-one (13), a known 16α,23α-epoxycucurbitane analogue first isolated from Eleaocarpus hainanensis9 that was also identified in the present investigation. Comparison of the 1D- and 2D-NMR data of 1 with those of 13 revealed a major change evident in the side chain located at C-23, with a 2-methylprop-1-ene group in 13 being replaced by a 2-methylprop-2-en-1-ol moiety in 1. The signals of the latter unit occurred at δH 4.24 (1H, d, J = 2.8 Hz, H-24), 4.91 (1H, s, Ha-27), 5.04 (1H, s, Hb-27), and 1.72 (3H, s, H-26) in the 1H NMR spectrum as well as at δC 75.8 (CH, C-24), 142.7 (C, C-25), 19.5 (CH3, C-26), and 111.7 (CH2, C-27) in the 13C NMR spectrum. Moreover, key HMBC correlations from the terminal olefinic methylene protons of H-27 to C-24, C-25, and C-26, as well as H-24 and H-26 to C-27, supported the structure assigned for the side chain moiety. Thus, the planar structure of 1 could be proposed.
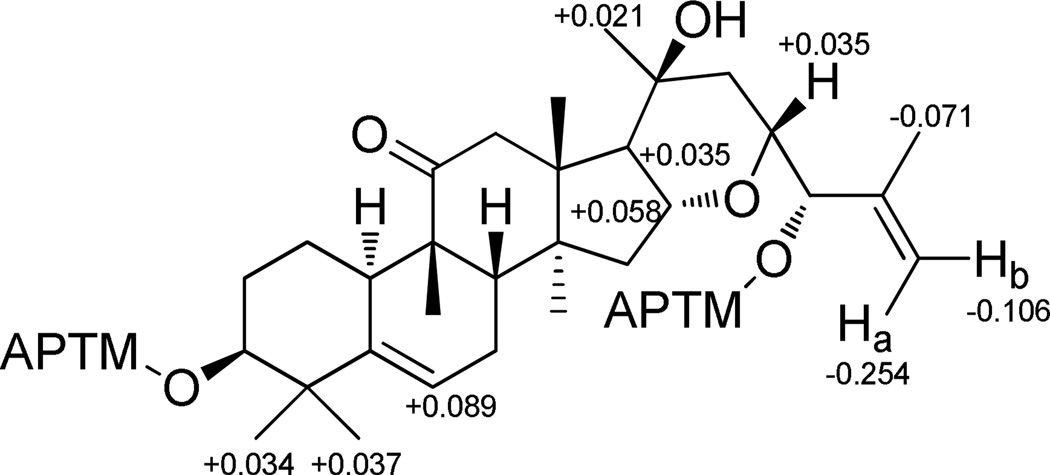
The Mosher ester procedure was employed to determine the absolute configuration of the OH groups located at C-3 and C-24 in compound 1. After treatment with (R)- and (S)-MTPA chloride, the secondary OH groups at C-3 and C-24 were both esterified, to afford the (S)- and (R)-MTPA derivatives, respectively. By analyzing the observed 1H NMR chemical shift difference values (ΔδS-R) of certain diagnostic key protons assigned unambiguously, the absolute configuration of C-3 and C-24 were both assigned as S (Figure 1). Furthermore, the observed NOESY cross peaks of H-10/H3-28 and H3-30, H-8/H3-18 and H3-19, H-17/H3-30, H-16/H3-18, H-23/H-15β, as well as H3-21/H-17 and H-12α, provided evidence that the relative configurations of the remaining chiral carbons of compound 1 were identical with those of previously reported related compounds.6–10 Hence, the structure of compound 1 was determined to be (3S,8S,9R,10R,13R,14S,16R,17R,21S,23R,24S)-16,23-epoxy-3,20,24-trihydroxycucurbit-5,25(27)-dien-11-one, and this substance has been accorded the trivial name, elaeocarpucin A.
Figure 1.
ΔδS-R values of MTPA esters of 1
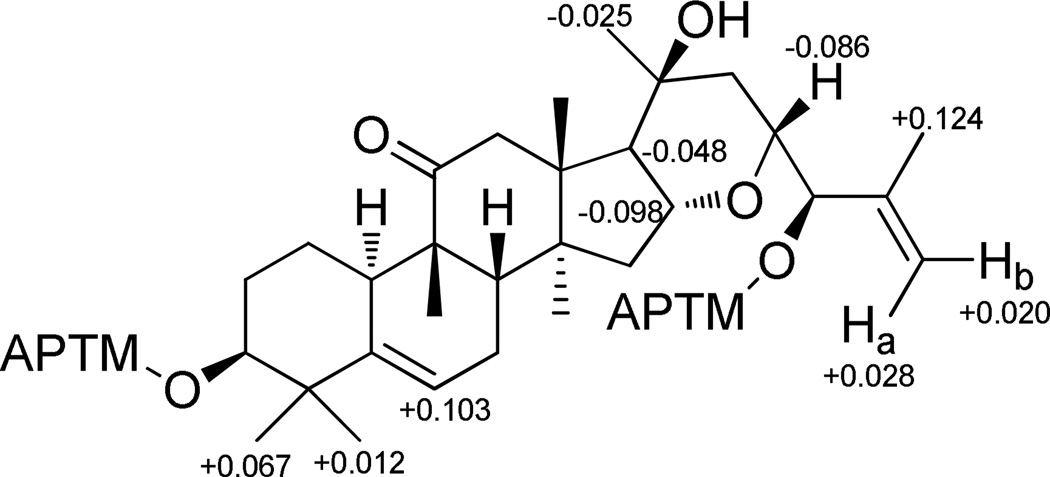
Compound 2 gave the same molecular formula, C30H46O5, as that of 1 by analysis of the HRESIMS. The NMR spectra of 1 and 2 were closely comparable, with the only differences evident in signals for the side chain at C-23. In the 1H NMR spectrum of 2, the H-23 and H-24 resonances appeared at δH 3.84 (ddd, J = 10.7, 8.0 and 2.7) and 3.87 (d, J = 7.2), respectively. Both were shifted upfield and showed a change of coupling pattern when compared with 1. Correspondingly, in the 13C NMR spectrum, a downfield shift of approximately 3.0 ppm for the carbon signal of C-24 (δC 78.8) was discernible. These observed differences suggested that the absolute configuration of C-24 might be R, which was confirmed subsequently by calculation of the 1H NMR chemical shift differences for the (S)- and (R)-MTPA esters of 2 produced as a result of the Mosher ester reaction (Figure 2). Thus, compound 2 (elaeocarpucin B) was determined structurally as the C-24 epimer of 1.
Figure 2.
ΔδS-R values of MTPA esters of 2
Compound 3 was obtained as a pale yellow powder. The HRESIMS gave a sodiated molecular ion peak at m/z 509.3255 [M + Na]+, consistent with a molecular formula of C30H46O5, the same as those of both compounds 1 and 2. The 1H and 13C NMR spectra of 3 were very similar to those of compound 13. On comparison of the 1H NMR data of these two compounds, the H3-27 signal at δH 1.68 in compound 13 was absent, while an oxygenated methylene resonance appeared at δH 4.02 (2H, s). This suggested that the C-27 methyl group in compound 13 is substituted by a primary alcohol group in compound 3. Correspondingly, in the 13C NMR spectrum of 3, the observed downfield shift of 2.7 ppm for C-25 and an upfield shift of 4.0 ppm for C-26 were consistent with the substitution of an OH group at C-27. The HMBC spectrum of 3 showed key correlations of H-24 with C-23, C-25 and C-27, as well as H-27 and H-26 with C-25 and C-24, which supported the above functional group assignment. The trans- configuration of the C-24, C-25 double bond was deduced based on the key NOESY correlation between H-24 and H-27. Other observed NOE effects indicated the relative configuration of the remaining part of the molecule of 3 to be identical with that of 1 and other known 16α,23α-epoxycucurbitacins. Thus, the structure of compound 3 (elaeocarpucin C) was determined as (3S,8S,9R,10R,13R,14S,16R,17R,21S,23R,24E)-16,23-epoxy-3,20,27-trihydroxycucurbit-5,24-dien-11-one.
Compound 4 was obtained as white amorphous powder. The HRESIMS of 4 gave a sodiated molecular ion peak at m/z 507.3073 [M + Na]+, suggesting an elemental formula of C30H44O5, representing one more degree of unsaturation than in compounds 1–3. In the 1H NMR spectrum of 4, the proton signal of an oxygenated CH, assigned as H-24 in compound 1 was absent, and the corresponding oxygenated carbon was substituted by a carbonyl group at δC 198.9 in the 13C NMR spectrum. These differences suggested that the OH group at C-24 in 1 is oxidized to a carbonyl group in 4. Moreover, downfield shifts of 0.97 ppm for H-23, 1.09 ppm for Ha-27, 0.82 ppm for Hb-27, and 0.17 ppm for H-26, due to the deshielding effect caused by the nearby carbonyl group at C-24, were observed in the 1H NMR spectrum. Data from the HSQC, HMBC, and NOESY 2D-NMR spectra were consistent with the above deduction. Thus, the structure of compound 4 (elaeocarpucin D) was elucidated as (3S,8S,9R,10R,13R,14S,16R,17R,21S,23R)-16,23-epoxy-3,20-dihydroxycucurbit-5,25(27)-dien-11,24-dione.
The molecular formula of compound 5 was deduced as C30H44O5 based on its HRESIMS, the same as that of compound 4. In the 1H NMR spectrum, a proton singlet appeared at δH 9.40 and this signal exhibited a correlation in the HSQC spectrum with a carbonyl group signal at δC 195.2, which implied the presence of a formyl group. In the HMBC spectrum, key correlations were observed between the aldehyde proton with the methyl group carbon at δC 9.9 (C-26), and two carbon signals of a double bond at δC 151.8 and 139.0 (C-24 and C-25), respectively, which suggested that the formyl group was at C-25. Comparison of the NMR data of 5 with those of compound 13 showed a downfield 1H NMR shift of 0.54 ppm for H-24, and downfield 13C NMR shifts of 26.7 and 3.1 ppm for C-24 and C-25, respectively. These were consistent with the substitution of a formyl group at C-27 in 5. The trans- configuration of the C-24-C-25 double bond was deduced from the key NOE correlation between the aldehyde proton (H-27) and H-24. Thus, the structure of compound 5 (elaeocarpucin E) was deduced as (3S,8S,9R,10R,13R,14S,16R,17R,21S,23R,24E)-16,23-epoxy-3,20-dihydroxycucurbit-27-aldehyde-5,24-dien-11-one.
Compound 6 gave a molecular formula of C30H48O5, as determined by HRESIMS, with one degree of unsaturation less than compound 1. In comparison to the NMR spectra with those of compound 1, besides one trisubstituted double bond ascribed to C-5-C-6, no other double bond signal was found, which implied that the side chain of 6 is saturated. This inference was confirmed by the observed 1H-1H COSY correlations of H-23 (δH 4.22, 1H, dddd, J = 11.1, 11.1, 2.4 and 2.4 Hz) with two CH2 groups, H-22 (δH 1.31 and 1.45, each 1H) and H-24 (δH 1.42 and 1.87, each 1H). In addition to the 13C NMR signal at δC 72.3 of C-20, another quaternary oxygenated carbon appeared at δC 70.5 and exhibited HMBC correlations with the H3-26 (δH 1.20, 3H, s) and H3-27 (δH 1.27, 3H, s) signals, respectively, which indicated a tertiary OH group at C-25. Analysis of the NOESY spectrum suggested the relative configuration of the remainder of the molecule of 6 to be identical with that of 1. Thus, the structure of 6 (elaeocarpucin F) was determined as (3S,8S,9R,10R,13R,14S,16R,17R,21S,23S)-16,23-epoxy-3,20,24-trihydroxycucurbit-5-en-11-one.
Compound 7 was obtained as a white amorphous powder, and its molecular formula was deduced as C30H46O5 from the HRESIMS. The NMR spectra of 7 were very similar to those of compound 13, with the only difference being the replacement of the CH2 group at C-22 by an oxygenated CH. In the COSY spectrum, a broad singlet of an oxygenated proton at δH 2.95 (H-22) exhibited a weak (due to the small J value of coupling constant between H-22 and H-23) but discernible enhancement with H-23 at δH 4.67 (1H, d, J = 7.8 Hz), and its corresponding carbon signal at δC 77.6 showed a HMBC correlation with the methyl proton signal at δH 1.38 (3H, s, H-21). Thus, it was inferred that an OH group is positioned at C-22 in compound 7. Correspondingly, a shift to higher field of around 6 ppm was observed for C-7 in the 13C NMR spectrum due to the γ-effect caused by this substituent. In the NOESY spectrum, key NOE cross peaks of H-22/H-23, H-23/H-16, H-16/H-18 and H-21/H-17 were observed, which indicated α-orientation of the OH group. Accordingly, the structure of compound 7 (elaeocarpucin G) was assigned as (3S,8S,9R,10R,13R,14S,16R,17R,21R,22R,23S)-16,23-epoxy-3,20,22-trihydroxycucurbit-5,24-dien-11-one.
The molecular formula of compound 8 was determined as C30H44O5 from the protonated molecular ion peak at m/z 485.3261 [M + H]+ in the HRESIMS. The NMR data of 8 were again similar to those of 13. When comparing the 13C NMR spectra of these two compounds, the signal of a CH2 group at δC 24.0 ascribed to C-7 in compound 13 was absent, with a carbonyl group signal appearing at δC 199.6 instead, thus suggesting a carbonyl group at C-7 in 8. In the 1H NMR spectrum, downfield shifts of approximately 0.5 ppm for H-6 and 0.6 ppm for H-8 were observed due to deshielding effects caused by the carbonyl group at C-7. These were consistent with the downfield shifts of around 27 ppm and 5 ppm for C-5 (δC 167.0) and C-6 (δC 125.6), respectively, as well as ca. 15 ppm for C-8 (δC 58.1) in the 13C NMR spectrum. Key HMBC correlations of H-3, H-10, H-28 and H-29 to C-5, and H-8 to C-7, were observed to support the structure proposed. Further analysis of the NOESY experiment revealed the consistent relative configuration of 8 with other cucurbitacin analogues isolated in this investigation. Thus, the structure of compound 8 (elaeocarpucin H) was deduced as (3S,8S,9S,10R,13R,14S,16R,17R,21S,23R)16,23-epoxy-3,20-dihydroxycucurbit-5,24-dien-7,11-dione.
All isolates (1–13) were evaluated their cytotoxic activity against HT-29 human colon cancer cells. The known cucurbitacins, cucurbitacin D (9), 3-epi-isocucurbitacin D (10), 25-O-acetylcucurbitacin F (11) and cucurbitacin I (12), were found to be the most active in inhibiting the proliferation of HT-29 cancer cells, with IC50 values ranging from 0.039 to 0.54 µM. Of the eight new 16,23-epoxycucurbitacins, elaeocarpucin C (3) was found to display potent cytotoxicity against HT-29 cells with an IC50 value of 0.41 µM, while elaeocarpucins D (4), G (7), and H (8) were less active against this same cell line. Thus, a 24(25)-en-27-ol functionality on the side chain of these new compounds seems to be required for potent cytotoxicity. In general, when the C-17-C-23 unit is contained in an epoxide ring, the resultant cytotoxicity is reduced when compared with known compounds such as 3-epi-isocucurbitacin D (10) (Table 3).
Table 3.
Cytotoxicity of Compounds Isolated from E. chinensisa
Compounds 1, 2, 5, 6 and 13 were not cytotoxic against HT-29 cells (IC50 > 10 µM), using a standard protocol.31
Results are expressed as IC50 values (µM).
Used as a positive control substance.
Compounds 1–3, 6, 9, 10, and 13 were also evaluated in a HT-29 cell-based mitochondrial transmembrane potential (MTP) assay, but none of these substances was found to be active (IC50 >20 µM).
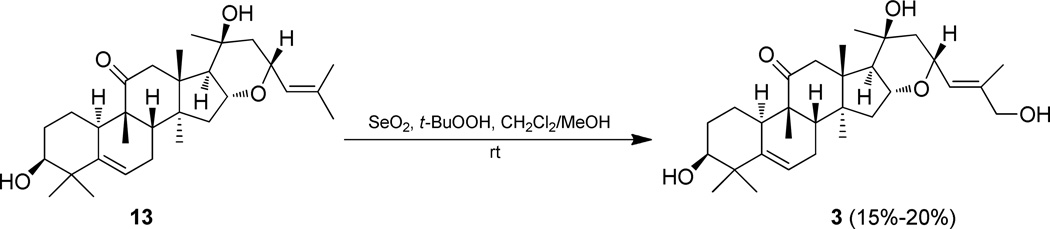
The initial cytotoxic activity of compound 3 encouraged further biological evaluation of this compound. A sufficient amount of 3 (>25 mg) was generated from the known inactive compound 13 by selectively oxidizing the allylic methyl group (C-27) into a primary alcohol (Figure 3), for evaluation in the in vivo hollow fiber assay. This method may be used as a secondary discriminator to prioritize compounds possessing promising in vitro activity for potential further testing in a relevant in vivo xenograft model.29–32 The human cancer cell lines evaluated using ip administration comprised MDA-MB-435 (melanoma), MCF-7 (breast), and HT-29 (colon) for the in vivo hollow fiber assay. However, no inhibition of proliferation by 3 was observed over the course of the study for any of the cancer cell types, which were administrated at a dose range of 0.5 to 10 mg /kg/day.
Figure 3.
Semi-synthesis of compound 3 from compound 13
EXPERIMENTAL SECTION
General Experimental Procedures
Melting points were measured using a Fisher Scientific melting point apparatus and were uncorrected. Optical rotations were recorded on a Perkin-Elmer 343 automatic polarimeter. UV spectra were measured with a Perkin-Elmer Lambda 10 UV/vis spectrometer. IR spectra were obtained on a Thermo Scientific Nicolet 6700 FT-IR spectrometer. NMR spectroscopic data were run at room temperature on Bruker Avance DRX-400 or 600 MHz spectrometers, and the data were processed using MestReNova 6.0 software (Mestrelab Research SL, Santiago de Compostela, Spain). Accurate mass values were performed on a Micromass LCT ESI spectrometer. Sodium iodide was used for mass calibration for a calibration range of m/z 100–2000. LC-MS experiments were performed on a liquid chromatographic/autosampler system that consisted of a Waters Alliance 2690 Separations Module (Waters, Milford, MA) and a Micromass LC-TOF™ II mass spectrometer (Micromass, Wythenshawe, UK) equipped with an orthogonal electrospray source (Z-spray). Column chromatography was carried out with silica gel (230–400 Mesh; Sorbent Technologies, Atlanta, GA). Analytical TLC was conducted on precoated 250 µm thickness silica gel UV254 aluminum-backed plates (Sorbent Technologies). Waters Atlantis ® (4.6 × 150 mm) and semi-preparative (10 × 150 mm) C18 (5 µm) columns were used for analytical and semi-preparative HPLC, respectively, as conducted on a Waters system comprised of a 600 controller, a 717 Plus autosampler, and a 2487 dual wavelength absorbance detector.
Plant Material
The fruits and stems of E. chinensis were collected in Honba Forest Reserve (12° 06.953’ N; 109° 0.072’E; Alt. 275 m), Khanh Hoa Province, Vietnam by T. N. N., Vuong Tan Tu, and D. D. S. in November, 2008, who also identified this plant. A voucher specimen (original collection DDS et al. 13583; recollection 114330) has been deposited in the John G. Searle Herbarium of the Field Museum of Natural History (under accession number FM 2287877), Chicago, Illinois.
LC-MS Dereplication Procedure
LC-UV conditions
Sample concentration: 10 mg/mL MeOH solution; mobile phase: gradient elution of MeOH/H2O (0–10 min, from 50:50 to 70:30; 11–30 min, 100% MeOH); UV detection wavelength: 220 nm; flow rate: 0.75 mL/min. Injection volume: 45 µL for the 96-well plate with sample concentration of ca. 20 µg/mL, and 11.3 µL for the 96-well plate sample concentration of ca. 5 µg/mL, respectively.
Cytotoxicity assay screening
Fractions were collected into two 96-well plates (250 µL/well × 90 and negative control/well × 6) with sample concentrations of 20 µg/mL and 5 µg/mL, respectively, and was tested for the HT-29 cell growth inhibition activity, according to an established protocol.33
LC-MS conditions
HPLC conditions: mobile phase: a gradient elution of MeOH/H2O (0–10 min, from 50:50 to 70:30; 11–30 min, 100% MeOH); injection volume: 45 µL (10 mg/mL). The mobile phase flow rate was maintained at 0.75 mL/min and was split post column using a microsplitter valve (Upchurch Scientific, Oak Harbor, WA) to ca. 20 µL/min for introduction to the ESI source. Optimal ESI conditions: capillary voltage, 3000 V; source temperature, 110 °C; cone voltage, 55 V. Q1 was set to optimally pass ions from m/z 100–2000 and all ions transmitted into the pusher region of the TOF analyzer were scanned over m/z (100–1000 range) with a 1 sec integration time. Data were acquired in a continuum mode during the LC run.
Data analysis
Using a combination search of proposed molecular formulas corresponding to the major active peaks and the key word “Eleaocarpus” in the SciFinder® database (Chemical Abstracts Service, Columbus, OH), the peaks with unknown molecular formulas were designated for further fractionation.
Extraction and Isolation
The air-dried and milled fruits (480 g) of E. chinensis were extracted by maceration with MeOH (3 × 2 L) at room temperature for two days each. After removing the solvent under reduced pressure, the combined and concentrated MeOH extract was suspended in a mixture of 80% MeOH/H2O (1 L), then partitioned with hexane and CHCl3 in turn, to afford hexane- (20 g) and CHClS (7 g)-soluble extracts. The CHCl3-soluble extract, with an IC50 value of 0.4 µg/mL against HT-29 cells, was subjected to a LC-MS dereplication procedure, in which the effluent from the HPLC chromatography was split, with part passed into a mass spectrometer and part collected in a 96-well plate. The latter was subjected to cytotoxicity screening using HT-29 cancer cells. ESIMS analysis indicated that the active peaks were at m/z 560, 516, 484 and 486, of which those at 516 and 560 amu corresponded to the presence of three known cytotoxic cucurbitacins, cucurbitacin D (9), 3-epi-isocucurbitacin D (10), and 25-O-acetylcucurbitacin F (11). However, peaks at m/z 484 and 486, with possible molecular formulas of C30H44O5 and C30H46O5, respectively, did not seem to match those of any known cucurbitacin triterpenes. Accordingly, bioassay-guided fractionation was used to facilitate the isolation process.
The CHCl3-soluble extract was subjected to chromatography over a silica gel column and eluted with a CH2Cl2−acetone gradient to afford ten fractions (F1–F10). Fractions, F3, F4, and F5 were active against HT-29 cells with IC50 values of 0.3, < 0.16, and 0.2 µg/mL, respectively. Fraction F3 (220 mg) was chromatographed over an open C18 column (2.2 × 20 cm) using MeOH-H2O mixtures (70:30 to 100% MeOH) for elution, to give three subfractions (F301–F303). F302 was purified by HPLC on a semi-preparative RP-18 column, using MeOH-H2O (60:40) as solvent, to afford 1 (10 mg), 2 (9.0 mg), 6 (7.0 mg) and a mixture of two compounds, which was subjected to further separation by HPLC, using CH3CN-H2O (33:67) for elution, to give 4 (4.0 mg) and 5 (1.0 mg), respectively. Fraction F4 (250 mg) was fractionated over an open C18 column (2.2 × 20 cm), eluted with MeOH-H2O (70:30 to 100% MeOH) to afford five subfractions (F401–F405). Cucurbitacin D (9, 35 mg) was obtained as crystals from a MeOH-H2O (ca. 70:30) solution of F401. Further purification of combined fractions F402-4 was conducted on a semi-preparative RP-18 HPLC column, using MeOH-H2O (60:40) as solvent, to yield 3-epi-isocucurbitacin D (10, 2.5 mg), cucurbitacin I (12, 1.0 mg), and 25-O-acetylcucurbitacin F (11, 3.5 mg). Elaeocarpucin C (3, 4.0 mg) was purified from fraction F5 (270 mg) by repeated separation on a semi-preparative RP-18 HPLC column, using MeOH-H2O (60:40) and CH3CN-H2O (35:65) sequentially for elution. In addition, compound 13 (10 mg) was recrystallized from the inactive fraction F2 using acetone as solvent. In order to obtain a sufficient amount of 13 as starting material to support the semi-synthesis of elaeocarpucin C (3), the residue of F2 was chromatographed over a silica gel column and eluted with CH2Cl2−acetone mixtures (20:1 to 5:1) to afford an additional 200 mg quantity of this compound.
The stems of E. chinensis were also investigated in the present study. A CHCl3-soluble extract (11 g) was prepared from the air-dried and then powdered stems (900 g) by following the extraction and partition procedures described above for the fruits. However, this was less cytotoxic (IC50 10.1 µg/mL, HT-29 cells) than the CHCl3-soluble extract of the fruits. This extract was fractionated over a silica gel column using CH2Cl2−acetone mixtures of increasing polarity to yield eight fractions (F1′–F8′). All fractions were analyzed using HPLC and TLC, and F4′ was found to be rich in cucurbitacins, and was determined to contain cucurbitacin D (9), 3-epi-isocucurbitacin D (10), 25-O-acetylcucurbitacin F (11), and unknown cucurbitacin analogues. Separation of F4′ over a semi-preparative RP-18 HPLC column using MeOH-H2O (60:40) led to the purification of compounds 7 (2.0 mg) and 8 (1.8 mg).
Elaeocarpucin A (1): white powder; mp 258–260 °C; [α]20D +144.0 (c 0.07, MeOH); UV (MeOH) λmax (log ε) 207 (3.73) nm; IR (film) νmax 3445, 1687, 1456, 1375, 1215, 1097, 1075, 1028, 755 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 509.3225 [M + Na]+ (calcd for C30H46O5Na, 509.3243).
Table 1.
1H NMR Chemical Shifts of Compounds 1–8a
| position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.47, m | 1.47, m | 1.42, m | 1.43, m | 1.49, m | 1.49, m | 1.48, m | 1.66b 1.78b |
| 2α | 1.73, m | 1.74, m | 1.73, m | 1.73, m | 1.74, m | 1.74, m | 1.75, m | 1.78, m |
| 2β | 1.67, m | 1.66, m | 1.68, m | 1.68, m | 1.68, m | 1.65, m | 1.68, m | |
| 3 | 3.48, brs | 3.48, brs | 3.48, brs | 3.48, brs | 3.48, brs | 3.48, brs | 3.48, brs | 3.65, brs |
| 6 | 5.67, d (5.8) | 5.67, d (5.8) | 5.67, d (5.8) | 5.67, d (5.6) | 5.68, d (6.0) | 5.67, d (5.8) | 5.67, d (5.6) | 6.17, d (brs) |
| 7α | 1.88, m | 1.89, m | 1.91, m | 1.90, m | 1.91, m | 1.90, m | 1.93, m | |
| 7β | 2.46, m | 2.44, m | 2.44, m | 2.44, m | 2.44, m | 2.44, m | 2.44, m | |
| 8 | 1.96b | 1.94b | 1.96 b | 1.92, d (8.3) | 1.96, d (8.0) | 1.91b | 1.94b | 2.54, brs |
| 10 | 2.25, m | 2.25, m | 2.25, m | 2.25, m | 2.25, m | 2.24, m | 2.24, m | 2.67, m |
| 12α | 3.03, d (14.8) | 3.03, d (14.8) | 3.03, d (14.8) | 3.03, d (14.8) | 3.04, d (14.4) | 3.01, d (14.4) | 3.04, d (14.8) | 3.04, d (14.8) |
| 12β | 2.43, d (14.8) | 2.42, d (14.8) | 2.43, d (14.8) | 2.42, d (14.8) | 2.44, d (14.4) | 2.42, d (14.4) | 2.42 (14.8) | 2.55 (14.8) |
| 15α | 1.48, m | 1.48, m | 1.47, m | 1.50, m | 1.48, m | 1.42, m | 1.48, m | 1.36, m |
| 15β | 1.85, m | 1.84, m | 1.85, m | 1.84, m | 1.85, m | 1.82, m | 1.85, m | 2.40, m |
| 16 | 4.40, ddd (10.4, 10.4, 3.6) | 4.33, ddd 10.4, 10.4, 3.7) | 4.38, ddd (10.1, 10.1, 3.9) | 4.42, ddd (10.1, 10.1, 3.9) | 4.43, ddd (10.4, 10.4, 3.8) | 4.37, ddd (10.4, 10.4, 3.6) | 4.37, ddd (10.0, 10.0, 3.6) | 4.37, ddd (10.4, 10.4, 3.6) |
| 17 | 1.95b | 1.95b | 1.99, d (10.5) | 2.00, d (10.5) | 1.95, d (10.1) | 1.95, d (10.4) | 2.34, d (10.1) | 1.90, d (10.6) |
| 18 | 0.93, s | 0.93, s | 0.94, s | 0.94, s | 0.96, s | 0.93, s | 0.95, s | 0.98, s |
| 19 | 1.14, s | 1.14, s | 1.13, s | 1.14, s | 1.15, s | 1.13, s | 1.14, s | 1.17, s |
| 21 | 1.32, s | 1.31, s | 1.32, s | 1.36, s | 1.36, s | 1.31, s | 1.38, s | 1.32, s |
| 22α | 1.57, dd (12.0, 14.0) | 1.36, m | 1.52, m | 1.67, m | 1.52, m | 1.45, m | 2.95, br s | 1.49, m |
| 22β | 1.25, m | 1.26, m | 1.38, m | 1.52, m | 1.41, m | 1.31, m | 1.43 | |
| 23 | 4.01, ddd (12.0, 3.0, 3.0) | 3.84, ddd (10.7, 8.0, 2.7) | 4.61, ddd (11.1, 8.5, 2.8) | 4.98, dd (11.8, 2.8) | 4.86, ddd (11.1, 7.2, 2.7) | 4.22, dddd (11.1, 11.1, 2.4, 2.4) | 4.67, d (7.8) | 4.22, ddd (11.2, 8.0, 2.4) |
| 24 | 4.24, d (2.8) | 3.87, d (7.2) | 5.44, d (8.3) | - | 5.66, dd (7.6, 1.4) | 1.87, m 1.42, m |
5.34, br d (8.0) | 5.12, d (8.0) |
| 26 | 1.72, s | 1.72, s | 1.72, s | 1.89, s | 1.77, d (1.2) | 1.20, s | 1.72 (br s) | 1.69, s |
| 27a | 4.91, s | 4.92, s | 4.02, s | 6.10, s | 9.40, s | 1.27, s | 1.77 (s) | 1.71, s |
| 27b | 5.04, s | 5.04, s | 5.86, s | |||||
| 28 | 1.02, s | 1.02, s | 1.02, s | 1.01, s | 1.02, s | 1.01, s | 1.01, s | 1.14, s |
| 29 | 1.17, s | 1.17, s | 1.17, s | 1.17, s | 1.18, s | 1.17, s | 1.17, s | 1.28, s |
| 30 | 1.22, s | 1.22, s | 1.21, s | 1.24, s | 1.25, s | 1.20, s | 1.24, s | 1.24, s |
Measured at 400 MHz; obtained in CDCl3 with TMS as internal standard; J values (Hz) are given in parentheses. Assignments are based on 1H-1H COSY, HSQC, and HMBC spectroscopic data.
Overlapping signals.
Table 2.
| position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | 20.5 | 20.5 | 20.5 | 20.5 | 20.7 | 21.0 | 20.5 | 20.5 |
| 2 | 28.4 | 28.5 | 28.5 | 28.6 | 28.7 | 28.6 | 28.5 | 28.3 |
| 3 | 76.0 | 76.1 | 76.0 | 76.1 | 76.2 | 76.0 | 76.1 | 76.1 |
| 4 | 42.0 | 41.7 | 41.7 | 41.7 | 41.8 | 42.0 | 41.7 | 43.0 |
| 5 | 139.6 | 139.7 | 139.6 | 139.6 | 139.8 | 139.6 | 139.6 | 167.0 |
| 6 | 120.3 | 120.4 | 120.4 | 120.3 | 120.5 | 120.4 | 120.3 | 125.6 |
| 7 | 23.9 | 23.9 | 23.9 | 24.0 | 24.3 | 24.9 | 23.9 | 199.6 |
| 8 | 42.7 | 42.7 | 42.7 | 42.7 | 42.8 | 42.7 | 42.7 | 58.1 |
| 9 | 49.3 | 49.4 | 49.3 | 49.3 | 49.5 | 49.3 | 49.3 | 49.5 |
| 10 | 35.1 | 35.2 | 35.1 | 35.2 | 35.3 | 35.1 | 35.2 | 37.4 |
| 11 | 213.4 | 213.4 | 213.4 | 213.2 | 213.5 | 213.4 | 213.5 | 211.0 |
| 12 | 48.2 | 48.3 | 48.3 | 48.2 | 48.4 | 48.2 | 48.2 | 48.3 |
| 13 | 48.5 | 48.5 | 48.3 | 48.4 | 48.6 | 48.4 | 48.5 | 47.8 |
| 14 | 47.9 | 48.0 | 48.1 | 47.9 | 48.2 | 47.9 | 47.5 | 47.5 |
| 15 | 40.6 | 40.6 | 40.7 | 40.3 | 40.7 | 40.7 | 40.5 | 40.5 |
| 16 | 76.7 | 76.6 | 77.3 | 77.0 | 76.6 | 76.5 | 76.6 | 76.5 |
| 17 | 55.2 | 55.3 | 54.9 | 54.9 | 55.1 | 55.0 | 48.7 | 54.7 |
| 18 | 19.8 | 19.8 | 19.9 | 19.9 | 20.1 | 19.8 | 20.3 | 19.9 |
| 19 | 20.3 | 20.3 | 20.3 | 20.4 | 20.5 | 20.3 | 20.4 | 21.1 |
| 20 | 72.0 | 72.1 | 72.4 | 72.3 | 72.5 | 72.3 | 74.8 | 72.2 |
| 21 | 29.3 | 29.3 | 29.3 | 29.2 | 29.4 | 29.2 | 25.7 | 29.7 |
| 22 | 41.1 | 45.0 | 48.7 | 45.3 | 47.2 | 49.5 | 77.6 | 49.1 |
| 23 | 76.5 | 76.7 | 72.5 | 77.2 | 73.2 | 74.2 | 75.4 | 72.8 |
| 24 | 75.8 | 78.8 | 124.8 | 198.9 | 151.8 | 46.9 | 120.8 | 124.8 |
| 25 | 142.7 | 143.7 | 138.6 | 143.0 | 139.0 | 70.5 | 138.2 | 136.6 |
| 26 | 19.5 | 17.7 | 14.2 | 18.1 | 9.9 | 28.0 | 18.8 | 18.5 |
| 27 | 111.7 | 114.7 | 67.9 | 126.4 | 195.2 | 31.0 | 26.0 | 25.8 |
| 28 | 27.2 | 27.2 | 27.2 | 27.2 | 27.3 | 27.1 | 27.1 | 27.8 |
| 29 | 25.3 | 25.4 | 25.3 | 25.4 | 25.6 | 25.3 | 25.3 | 24.8 |
| 30 | 21.2 | 21.1 | 21.1 | 21.2 | 21.3 | 21.1 | 21.3 | 21.1 |
Measured at 100 MHz; obtained in CDCl3 with TMS as internal standard. Assignments are based on HSQC, and HMBC NMR spectra.
Multiplicity obtained from the DEPT spectrum.
Elaeocarpucin B (2): white powder; mp 270–272 °C; [α]20D +94.0 (c 0.08, MeOH); UV (MeOH) λmax (log ε) 207 (3.63) nm; IR (film) νmax 3442, 1687, 1457, 1375, 1216, 1097, 1075, 1022, 754 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 509.3264 [M + Na]+ (calcd for C30H46O5Na, 509.3243).
Elaeocarpucin C (3): Pale yellow amorphous powder; [α]20D +91.0 (c 0.04, MeOH); UV (MeOH) λmax (log ε) 206 (3.87), 219 (3.48) nm; IR (film) νmax 3435, 1685, 1465, 1375, 1215, 1067, 755 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 509.3255 [M + Na]+ (calcd for C30H46O5Na, 509.3243).
Elaeocarpucin D (4): white amorphous powder; [α]20D +136.0 (c 0.09, MeOH); UV (MeOH) λmax (log ε) 213 (3.84) nm; IR (film) νmax 3471, 1685, 1462, 1375, 1096, 754 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 507.3073 [M + Na]+ (calcd for C30H44O5Na, 507.3086).
Elaeocarpucin E (5): white amorphous powder; [α]20D +82.0 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 218 (4.12) nm; IR (film) νmax 3458, 1703, 1688, 1460, 1213, 1376, 1072, 1021, 755 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 507.3112 [M + Na]+ (calcd for C30H44O5Na, 507.3086).
Elaeocarpucin F (6): white amorphous powder; [α]20D +79.0 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 206 (3.57) nm; IR (film) νmax 3432, 1688, 1462, 1213, 1391, 1376, 1162, 1072, 755 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 511.3399 [M + Na]+ (calcd for C30H44O5Na, 511.3388).
Elaeocarpucin G (7): white amorphous powder; [α]20D +55.0 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 206 (3.50) nm; IR (film) νmax 3476, 2948, 2917, 2845, 1687, 1462, 1380, 1059, 755 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 487.3423 [M + H]+ (calcd for C30H47O5, 487.3423).
Elaeocarpucin H (8): white powder; mp 244–246 °C; [α]20D +68.0 (c 0.17, MeOH); UV (MeOH) λmax (log ε) 228 (3.95) nm; IR (film) νmax 3429, 2968, 2925, 2855, 1695, 1647, 1458, 1377, 1026, 756 cm−1; 1H NMR (400 MHz, CDCl3) and 13C NMR (100 MHz, CDCl3) data, see Tables 1 and 2; HRESIMS m/z 485.3261 [M + H]+ (calcd for C30H45O5, 485.3267).
Preparation of the (R) and (S)-MTPA Ester Derivatives of Compound 1
Portions of compound 1 (1.0 mg of each) were added into two NMR tubes, and dried under a vacuum overnight at room temperature. Deuterated pyridine (1 mL) was transferred to each tube to give a clear solution. (S)-(+)-α-Methoxy-α-(trifluoromethyl)phenylacetyl (MTPA) chloride (10 µL) or (R)-MTPA chloride (10 µL), was injected into the NMR tubes separately under a N2 gas steam and mixed quickly with the dissolved sample. The NMR tubes with reagents were sealed and stored overnight in a dryer until the reaction was completed, with 1H NMR spectroscopy used to monitor the reaction. The 1H NMR chemical shifts of the (R)-MTPA ester and the (S)-MTPA ester of 1 were recorded directly after each reaction and were assigned based on COSY and NOESY experiments, with ambiguous and overlapping signals not used for the ΔδS-R calculation.34,35 1H NMR data of R-MTPA ester of 1 (400 MHz, pyridine-d5): δ 5.923 (1H, d, J = 4.4 Hz, H-24), 5.547 (1H, d, J = 5.6 Hz, H-6), 5.255 (1H, s, H-27a), 5.139 (1H, brs, H-3), 5.069 (1H, s, H-27b), 4.752 (1H, ddd, J = 9.4, 9.4, 2.3 Hz, H-16), 4.512 (1H, m, H-23), 1.830 (3H, s, H-26), 2.072 (1H, d, J = 9.4 Hz, H-17), 1.442 (3H, s, H-21), 1.282 (3H, s, H-30), 1.258 (3H, s, H-18), 1.216 (3H, s, H-29), 1.132 (3H, s, H-19), 1.097 (3H, s, H-28). 1H NMR data of S-MTPA ester of 1 (400 MHz, pyridine-d5): δ 5.889 (1H, d, J = 2.7 Hz, H-24), 5.636 (1H, d, J = 5.6 Hz, H-6), 5.077 (1H, brs, H-3), 5.001 (1H, s, H-27a), 4.963 (1H, s, H-27b), 4.810 (1H, ddd, J = 9.4, 9.4, 2.3 Hz, H-16), 4.547 (1H, m, H-23), 2.107 (1H, d, J = 9.4 Hz, H-17), 1.759 (3H, s, H-26), 1.463 (3H, s, H-21), 1.327 (3H, s, H-30), 1.253 (3H, s, H-29), 1.221 (3H, s, H-18), 1.131 (3H, s, H-28), 1.123 (3H, s, H-19).
Preparation of the (R) and (S)-MTPA Ester Derivatives of Compound 2
The (R)-MTPA ester and the (S)-MTPA ester of 2 were produced by following the same Mosher reaction procedure applied to compound 1. 1H NMR data of R-MTPA ester of 1 (400 MHz, pyridine-d5): δ 5.855 (1H, d, J = 8.2 Hz, H-24), 5.546 (1H, d, J = 5.2 Hz, H-6), 5.216 (1H, s, H-27a), 5.140 (1H, brs, H-3), 5.014 (1H, s, H-27b), 4.745 (1H, m, H-16), 4.467 (1H, m, H-23), 2.119 (1H, d, J = 9.6 Hz, H-17), 1.615 (3H, s, H-26), 1.483 (3H, s, H-21), 1.271 (3H, s, H-29), 1.265 (3H, s, H-30), 1.229 (3H, s, H-18), 1.138 (3H, s, H-19), 1.109 (3H, s, H-28); 1H NMR data of S-MTPA ester of 1 (400 MHz, pyridine-d5): δ 5.924 (1H, d, J = 8.0 Hz, H-24), 5.649 (1H, d, J = 4.8 Hz, H-6), 5.244 (1H, s, H-27a), 5.082 (1H, brs, H-3), 5.034 (1H, s, H-27b), 4.647 (1H, m, H-16), 4.381 (1H, m, H-23), 2.071 (1H, d, J = 9.7 Hz, H-17), 1.739 (3H, s, H-26), 1.458 (3H, s, H-21), 1.283 (3H, s, H-29), 1.264 (3H, s, H-30), 1.176 (3H, s, H-28), 1.161 (3H, s, H-18), 1.198 (3H, s, H-19).
Generation of Compound 3 from Compound 13
Selenium dioxide (SeO2, 500 mg) was dissolved in 2.5 mL distilled water, and then 12.5 mL MeOH was added to give a clear solution. Next, 10 g of silica gel was added to this solution to form a slurry, with the solvent evaporated under reduced pressure to afford a silica gel powder containing 5% selenium dioxide.36 A portion of this pretreated silica gel (1 g) was suspended in 7 mL CH2Cl2 with 0.5 mL t-BuOOH (5.0–6.0 M in decane) and stirred for 15 min at room temperature. Compound 13 (200 mg) was dissolved in a mixture of CH2Cl2-MeOH (4:1, 20 mL), and the solution obtained was added dropwise to the above-mentioned oxidizing reagent. The mixture was sealed and stirred overnight at room temperature, with the product analyzed by TLC (CH2Cl2-acetone 5:1; Rf 0.2). After the reaction, the mixture was filtered and the residue was washed with CHCl3. The filtrate was partitioned with water, and the organic phase was evaporated under reduced pressure after washing with saturated NaCl water, to give a mixture of compound 3 and unchanged compound 13. This mixture was subjected to chromatography on an open reversed-phase C18 column, using a gradient of MeOH/H2O (70:30 to 100% MeOH) for elution, to afford 32 mg of 3 and 150 mg of 13. The yield of this reaction was around 15–20%, and the unchanged 13 could be recycled (Figure 3). When performing this selective oxidation procedure, modification of the reaction by raising the temperature used, prolonging the reaction time, or increasing the amount of oxidant, did not increase the yield of the desired primary alcohol (3), but led to the generation of an α,β-unsaturated aldehyde derivative, as an undesired side product, which was identified as compound 5.
Cytotoxicity Assay
Compounds 1–13 were evaluated against human colon cancer cells (HT-29), according to a previously described protocol.33
Mitochondrial Transmembrane Potential Assay (MTP Assay)
A JC-1 mitochondrial membrane potential assay kit obtained from Cayman Chemicals was used to detect the ΔΨ. Experiments were conducted according to the protocol established previously.37,38
In Vivo Hollow Fiber Assay
The potential in vivo anticancer activity of elaeocarpucin C (3) against HT29, MCF-7, and MDA-MB-435 human cancer cells was evaluated in the murine hollow fiber model (HT29: 1 × 106; MCF-7: 5 × 106 and MDA-MB-435: 1 × 106 per fiber), according to an established procedure. Based on the modified MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay results, a dose range from 0.5 to 10 mg elaeocarpucin C (3) per kg per day for four days was used. The vehicle was 5% EtOH and 5% Tween 80 in physiologic saline, and paclitaxel was used as the positive control (2.0 mg/kg per day for four days).32,39
Supplementary Material
Figure 4.
ACKNOWLEDGMENT
This study was supported by grant P01 CA125066 (awarded to A. D. Kinghorn) from NCI, NIH. Elaeocarpus chinensis samples were collected under the terms of agreement between the University of Illinois at Chicago and the Institute of Ecology and Biological Resources of the Vietnam Academy of Science and Technology, Hanoi, Vietnam. We acknowledge Mr. John Fowble, College of Pharmacy, The Ohio State University (OSU), and Dr. Chun-Hua Yuan, Campus Chemical Instrument Center, OSU, for facilitating the acquisition of the 400 and 600 MHz NMR spectra. We thank Ms. Nan Kleinholz, Mr. Mark Apsega and Dr. Kari Green-Church, Campus Chemical Instrument Center, OSU, for the mass spectrometric measurements. We are very grateful to Dr. Gordon M. Cragg, formerly of NCI-Frederick, for serving for several years as NCI Program Director of a forerunner of the above-cited program project, namely, grant U19 CA52956 from NCI, NIH.
Footnotes
ASSOCIATED CONTENT
Supporting Information. 1H, 13C and 2D NMR spectra of compounds 1–8, 1H NMR of (R)- and (S)-MTPA esters of 1 and 2, and hollow fiber assay data for 3. This material is available free-of-charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Flora Reipublicae Popularis Sinicae. Vol. 49. Beijing: Science Press; 1989. Flora of China Editorial Committee; p. 22. [Google Scholar]
- 2.Iconographia Cormophytorum Sinicorum. Vol. 2. Beijing: Science Press; 1989. Institute of Botany, the Chinese Academy of Sciences; p. 787. [Google Scholar]
- 3.Zmarzty S. Kew Bull. 2001;56:405–447. [Google Scholar]
- 4.Tang Y, Phengklai C. Elaeocarpaceae. In: Wu Z, Raven PH, Hong D, editors. Flora of China. vol 12. 2007. [Accessed on 10/27/2011]. pp. 223–239. Available online at http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=10299. [Google Scholar]
- 5.Lowry JB. Phytochemistry. 1970;9:2411. [Google Scholar]
- 6.Bittner M, Poyser KA, Poyser JP, Silva M, Weldt E, Sammes PG. Phytochemistry. 1973;12:1427–1431. [Google Scholar]
- 7.Schenkel EP, Farias MR, Mayer R, Breitmaier E, Rücker G. Phytochemistry. 1992;31:1329–1333. [Google Scholar]
- 8.Achenbach H, Horn K, Dominguez XA, Rombold C, Gómez López EG. Phytochemistry. 1993;33:437–435. [Google Scholar]
- 9.Fang X, Phoebe CH, Jr, Pezzuto JM, Fong HHS, Farnsworth NR, Yellin B, Hecht SM. J. Nat. Prod. 1984;47:988–993. doi: 10.1021/np50036a013. [DOI] [PubMed] [Google Scholar]
- 10.Ito A, Chai H-B, Lee D, Kardono LBS, Riswan S, Farnsworth NR, Cordell GA, Pezzuto JM, Kinghorn AD. Phytochemistry. 2002;61:171–174. doi: 10.1016/s0031-9422(02)00232-7. [DOI] [PubMed] [Google Scholar]
- 11.Meng DH, Qiang SG, Lou LG, Zhao WM. Planta Med. 2008;74:1741–1744. doi: 10.1055/s-2008-1081356. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Tao Z-M, Zhang Y, Shen Z-W, Qin G-W. Chin. J. Nat. Med. 2010;8:21–24. [Google Scholar]
- 13.Chand L, Dasgupta S, Chattopadhyay SK, Ray AB. Planta Med. 1977;32:197–199. doi: 10.1055/s-0028-1097584. [DOI] [PubMed] [Google Scholar]
- 14.Ray AB, Dutta SC, Dasgupta S. Phytochemistry. 1976;15:1797–1798. [Google Scholar]
- 15.Carroll AR, Arumugan G, Quinn RJ, Redburn J, Guymer G, Grimshaw P. J. Org. Chem. 2005;70:1889–1892. doi: 10.1021/jo048525n. [DOI] [PubMed] [Google Scholar]
- 16.Katavic PL, Venables DA, Rali T, Carroll AR. J. Nat. Prod. 2007;70:866–868. doi: 10.1021/np060577f. [DOI] [PubMed] [Google Scholar]
- 17.Katavic PL, Venables DA, Rali T, Carroll AR. J. Nat. Prod. 2007;70:872–875. doi: 10.1021/np060607e. [DOI] [PubMed] [Google Scholar]
- 18.Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. Nat. Prod. Rep. 2005;22:794–795. doi: 10.1039/b418841c. [DOI] [PubMed] [Google Scholar]
- 19.Rios JL, Escandell JM, Recio MC. In: Studies in Natural Product Chemistry: Bioactive Natural Products (Part L) Atta-ur-Rahman Ed., editor. vol 32. Amsterdam: Elsevier B.V; 2005. pp. 429–469. [Google Scholar]
- 20.Saba BA, Oridupa AO. J. Med. Plant. Res. 2010;4:2821–2826. [Google Scholar]
- 21.Lee DH, Iwanski GB, Thoennissen NH. Sci. World. J. 2010;10:413–418. doi: 10.1100/tsw.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinghorn AD, Carcache-Blanco EJ, Chai H-B, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. In: Bioactive Compounds from Natural Sources, Second Edition. Natural Products as Lead Compounds in Drug Discovery. Tringali C, editor. London: Taylor & Francis; 2012. pp. 37–63. [Google Scholar]
- 24.Loub WD, Farnsworth NR, Soejarto DD, Quinn ML. J. Chem. Inf. Comput. Sci. 1985;25:99–103. doi: 10.1021/ci00046a009. [DOI] [PubMed] [Google Scholar]
- 25.Seger C, Sturm S, Haslinger E, Stuppner H. Monatsh. Chem. 2005;136:1645–1649. [Google Scholar]
- 26.Halaweish FT. J. Chem. Ecol. 1993;19:29–37. doi: 10.1007/BF00987468. [DOI] [PubMed] [Google Scholar]
- 27.Abd El-Fattah H. Phytochemistry. 1994;36:159–161. [Google Scholar]
- 28.Wu P-L, Lin F-W, Wu T-S, Kuoh C-S, Lee K-H, Lee S-J. Chem. Pharm. Bull. 2004;52:345–349. doi: 10.1248/cpb.52.345. [DOI] [PubMed] [Google Scholar]
- 29.Alley MC, Pacula-Cox CM, Hursey ML, Rubinstein LR, Boyd MR. Cancer Res. 1991;51:1247–1256. [PubMed] [Google Scholar]
- 30.Hollingshead MG, Alley MC, Camalier RF, Abbott BJ, Mayo JG, Malspeis L, Grever MR. Life Sci. 1995;57:131–141. doi: 10.1016/0024-3205(95)00254-4. [DOI] [PubMed] [Google Scholar]
- 31.Mi Q, Lantvit D, Reyes-Lim E, Chai H, Zhao W, Lee IS, Peraza-Sanchez S, Ngassapa O, Kardono LB, Riswan S, Hollingshead MG, Mayo JG, Farnsworth NR, Cordell GA, Kinghorn AD, Pezzuto JM. J. Nat. Prod. 2002;65:842–850. doi: 10.1021/np010322w. [DOI] [PubMed] [Google Scholar]
- 32.Mi Q, Pezzuto JM, Farnsworth NR, Wani MC, Kinghorn AD, Swanson SM. J. Nat. Prod. 2009;72:573–580. doi: 10.1021/np800767a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan L, Kardono LBS, Riswan S, Chai H-B, Carcache de Blanco EJ, Pannell CM, Soejarto DD, McCloud TG, Newman DJ, Kinghorn AD. J. Nat. Prod. 2010;73:1873–1878. doi: 10.1021/np100503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rieser MJ, Hui YH, Rupprecht JK, Kozlowski JF, Wood KV, McLaughlin JL, Hanson PR, Zhuang Z, Hoye TR. J. Am. Chem. Soc. 1992;114:10203–10213. [Google Scholar]
- 35.Su B-N, Park EJ, Mbwambo ZH, Santarsiero BD, Mesecar AD, Fong HHS, Pezzuto JM, Kinghorn AD. J. Nat. Prod. 2002;65:1278–1282. doi: 10.1021/np0202475. [DOI] [PubMed] [Google Scholar]
- 36.Chhabra BR, Hayano K, Ohtsuka T, Shirahama H, Matsumoto T. Chem. Lett. 1981;10:1703–1706. [Google Scholar]
- 37.Deng Y, Balunas MJ, Kim J-A, Lantvit DD, Chin Y-W, Chai H, Sugiarso S, Kardono LBS, Fong HHS, Pezzuto JM, Swanson SM, Carcache de Blanco EJ, Kinghorn AD. J. Nat. Prod. 2009;72:1165–1169. doi: 10.1021/np9001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan L, Matthew S, Lantvit DD, Zhang X, Ninh TN, Chai H-B, Carcache de Blanco EJ, Soejarto DD, Swanson SM, Kinghorn AD. J. Nat. Prod. 2011;74:2193–2199. doi: 10.1021/np200557e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan L, Lantvit DD, Riswan S, Kardono LBS, Chai H-B, Carcache de Blanco EJ, Farnsworth NR, Soejarto DD, Swanson SM, Kinghorn AD. Phytochemistry. 2010;71:635–640. doi: 10.1016/j.phytochem.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.