Summary
Neurite formation is a seminal event in the early development of neurons. However, little is known about the mechanisms by which neurons form neurites. F-BAR proteins function in sensing and inducing membrane curvature [1, 2]. Cdc42-interacting protein 4 (CIP4), a member of the F-BAR family, regulates endocytosis in a variety of cell types [3–9]. However, there is little data on how CIP4 functions in neurons [10, 11]. Here we show that CIP4 plays a novel role in neuronal development by inhibiting neurite formation. Remarkably, CIP4 exerts this effect not through endocytosis, but by producing lamellipodial protrusions. In primary cortical neurons CIP4 is concentrated specifically at the tips of extending lamellipodia and filopodia, instead of endosomes as in other cell types. Overexpression of CIP4 results in lamellipodial protrusions around the cell body, subsequently delaying neurite formation and enlarging growth cones. These effects depend on the F-BAR and SH3 domains of CIP4 and on its ability to multimerize. Conversely, cortical neurons from CIP4-null mice initiate neurites twice as fast as controls. This is the first study to demonstrate that an F-BAR protein functions differently in neuronal vs. non-neuronal cells and induces lamellipodial protrusions instead of invaginations or filopodia-like structures.
Results and Discussion
CIP4 is expressed in cortex only during early development
During brain development neurons migrate to their final destinations with little process outgrowth. As they approach their final location in the brain they extend a single axon, followed by multiple dendrites [12, 13]. In culture, cortical neurons undergo a stereotyped series of events whereby they first attach to the substrate and extend lamellipodia and filopodia (stage 1), which over time coalesce into several distinct neurites tipped by motile growth cones (stage 2). One of these neurites begins to extend rapidly to become the axon (stage 3), while the other neurites slowly develop into dendrites (stage 4) [14]. Several cytoskeletal proteins, including the actin-binding Ena/VASP family and microtubule-associated proteins (MAPs), have been shown to participate in filopodia and neurite formation [15–19]. In contrast, evidence for negative regulators of neurite formation remains scant. We hypothesized that there may be other proteins associated with the cytoskeleton that play opposing roles to Ena/VASP proteins and MAPs in neurite formation. Since filopodia are necessary for neurite formation [18] we reasoned that proteins that favor lamellipodia formation may inhibit neurite formation.
The F-BAR (Fer/CIP4 homology (FCH) domain and BAR (Bin, Amphiphysin, Rvs)) domain-containing proteins have emerged as important players in bridging the membrane and cytoskeleton [1, 20, 21]. The F-BAR domain forms a banana-shaped dimer that adopts a more gentle curvature than BAR domain proteins [22, 23]. The concave side of the F-BAR region plays an important role in endocytosis by sensing and altering membrane curvature [24]. However, recent reports indicate that some members of the F-BAR proteins are capable of inducing filopodia [25–29] or filopodia-like structures [28] and can localize to lamellipodia [30]. However, there are few studies of CIP4 in neurons [10, 11] and no studies that suggest a function for CIP4 in this cell type. To test whether CIP4 has a role in the neuronal development, we first determined its expression in the cortex and dissociated cortical neurons. We found that CIP4 is expressed in mouse cortex prenatally but is dramatically downregulated at birth (Figure 1A). Furthermore, in mouse cortical neurons cultured from E15.5 brains CIP4 protein levels decreased sharply within the first few days in culture, as neurons extend neurites (Figure 1B). These data are consistent with CIP4 functioning specifically in the early development of the cortex.
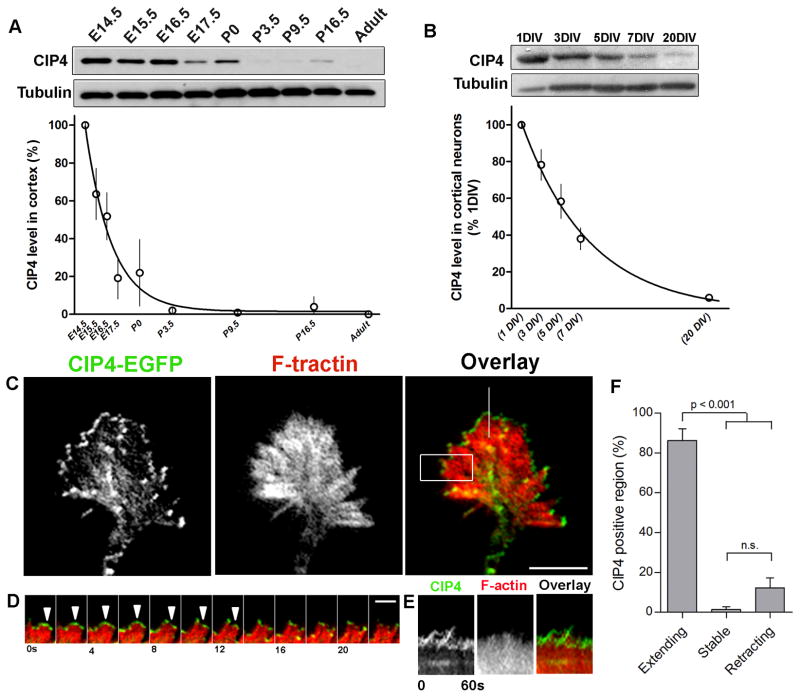
Figure 1. CIP4 is localized to the distal lamellipodia in the growth cones and is rapidly downregulated in developing cortex and cultured cortical neurons.
(A) Western blot and a non-linear regression plot of endogenous CIP4 expression in mouse cortex during development (n=3) and (B) in dissociated E15.5 mouse cortical neurons during development in culture (n=3). The CIP4 levels shown in the graphs were normalized to tubulin. (C) Images of a lamellipodial growth cone from a cortical neuron transfected with CIP4-EGFP and F-tractin (to label F-actin) showing that CIP4 is localized to the edge of the growth cone lamellipodia. (D) Selected frames from a time-lapse series (boxed region in C). White arrows indicate the extending edge of lamellipodia. (E) Kymographs of CIP4 and F-actin from the line shown in C. Note that CIP4 is only concentrated at the edge during extension. (F) Quantification showing that CIP4 is localized predominantly to extending lamellipodia (n=5 neurons, 40 regions, 3 independent experiments). Data are expressed as mean ± SEM. P values are indicated (One-way ANOVA with Bonferroni post-test comparison). Scale bars represent 10 μm in C and 3 μm in D.
CIP4 is localized to the distal lamellipodia and filopodia in cortical neurons
Since none of the antibodies that we have tested to date reliably label endogenous CIP4 in cultured mouse cortical neurons by immunocytochemistry (data not shown), we expressed CIP4-EGFP to determine its distribution and dynamics. Surprisingly, we found that CIP4-EGFP concentrated at the periphery of both stage 1 neurons (Figure 2A) and cortical neuron growth cones (Figures 1C and S1A; Movie 1 and 2). Rapid time-lapse imaging of living transfected neurons with total internal reflection fluorescence microscopy (TIRFM) confirmed that CIP4 concentrated at the periphery of extending lamellipodia (Figures 1D and 1F; Movie 1) and filopodia (Figures S1B and S1D; Movie 2), but disappeared as the lamellipodia (Figures 1E and 1F) or filopodia (Figures S1C and S1D) paused and retracted. This distribution of CIP4 in cortical neurons was quite surprising given that previous studies in non-neuronal cells showed CIP4 concentrated on tubulovesicular structures in the cytoplasm of cells [3–9, 31].
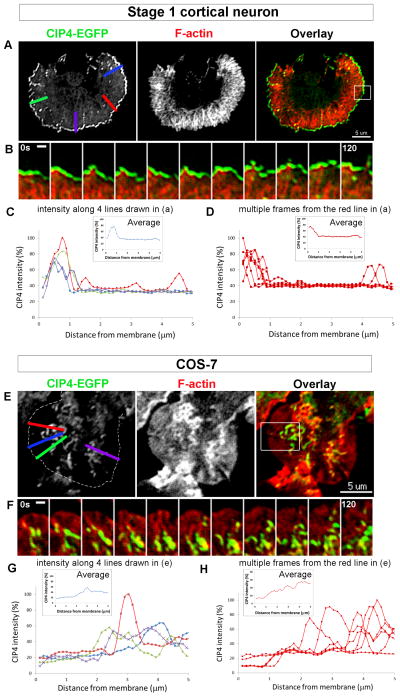
Figure 2. CIP4 localizes differently in cortical neurons and COS-7 cells.
(A and E) Images of a stage 1 neuron and a COS-7 cell transfected with CIP4-EGFP and mRuby-Lifeact. (B and F) Selected frames from the boxed region in A showing that CIP4 dynamically remains associated with the edge of lamellipodium in the neuron, and in E forms tubular structures in a COS-7 cell. Dashed line in E indicates the edge of the COS7 cell. (C and G) Graphs representing the intensity of CIP4 along the lines drawn in A and E. The inset shows the average intensity along those 4 lines. (D and H) Graphs of CIP4 intensity along the red line shown in A and E throughout the time-lapse (120 seconds). The inset shows the average intensity from multiple time points. Scale bars represent 5 μm in A and E and 2 μm in B and F.
CIP4 localizes to different structures in cortical neurons and COS7 cells
To determine if the distribution of CIP4 in cortical neurons was unique we transfected the same fusion construct (CIP4-EGFP) into COS7 cells, which have been used to study CIP4 previously [5, 31]. Remarkably, CIP4-EGFP segregated to entirely different regions of neurons and COS7 cells. In stage 1 cortical neurons (Figure 2A; Movie 3) CIP4 concentrated at the periphery of the lamellipodium and was dynamically associated with extending lamellipodia (Figure 2B; Movie 3). Line scans perpendicular to the lamellipodium revealed that most of the CIP4 was concentrated within one micron of the periphery (Figures 2C and 2D). Conversely, CIP4 strongly labeled tubulovesicular structures in the interior of COS7 cells (Figures 2E and 2F; Movie 4), consistent with previous experiments in these cells [5]. CIP4 also dynamically associated with these structures, but they rarely extended into the periphery of the cell (Figures 2G and 2H). Thus, CIP4 localizes to distinct structures in neurons and COS7 cells. In neurons CIP4 is associated with peripheral protrusions, while in COS7 cells CIP4 is localized to tubulovesicular structures in the cytoplasm.
We also discovered that CIP4 was capable of concentrating at either convex or concave protrusions (Figures S2A-S2C) in stage 1 neurons and stage 3 axonal growth cones (data not shown). Quantification showed that CIP4-EGFP-positive protrusions included approximately 40% convex and 60% concave protrusions, which is consistent with a previous study of growth cone protrusion shape [32]. These data indicate that although CIP4 usually associates with concave membrane protrusions it can also associate with convex protrusions in cortical neurons.
CIP4 does not localize to endocytic structures in cortical neurons
To our knowledge no study has demonstrated endocytosis at the tips of extending lamellipodia and filopodia. However, we do note that CIP4 puncta near the cell periphery can be transported retrogradely (Figures S2D and S2E). The vast majority of these CIP4 puncta (94.4±1.4%, n=205 puncta, 3 neurons) move at the same rate as actin retrograde flow, indicating they are likely associated with actin filaments that continuously undergo flow from peripheral to central regions of cells.
To determine if CIP4 was associated with endocytic vesicles we transfected COS-7 cells and cortical neurons with CIP4-EGFP and either DsRed-clathrin or mCherry-Rab5, two proteins known to associate with endocytic vesicles [33, 34], and imaged them with TIRFM. In COS-7 cells, CIP4 colocalizes with clathrin (23.5% fluorescent overlap) and Rab5 (27.4% fluorescent overlap), consistent with a previous study [5]. In contrast, in stage 1 cortical neurons, we observed very little overlap of CIP4 and clathrin (2.7%) or Rab5 (2.1%) (Figures S2F-S2P; Movie 5). Therefore, in cortical neurons, it appears that CIP4 does not have a prominent role in endocytosis, since we do not detect colocalization of CIP4 with either clathrin or Rab5 in space or time. Rather, CIP4 appears to be functioning primarily in protrusive activity at the plasma membrane.
CIP4 inhibits neurite formation by inducing lamellipodial protrusions
To determine the function of CIP4 in neuronal development we cultured neurons from homozygous CIP4 knock-out mice [3]. Western blots from cortical lysates confirmed that the deletion of CIP4 protein did not change the levels of the other CIP4 family members, FBP17 and Toca-1 (data not shown), indicating that any defects were due to CIP4 specifically. Strikingly, CIP4-null cortical neurons formed neurites precociously compared to neurons from wild-type littermates (Figure 3A). Indeed, CIP4-null neurons advanced to stage 2 (Figure 3B) twice as fast as controls (Figure 3C). Because CIP4-null neurons were specifically precocious in forming neurites (stage 1–2 transition) but not polarization (stage 2–3 transition) (Figure 3D), CIP4 appears to act distinctly at the stage when neurons form neurites, but not during the transition from a neurite to an axon. However, in contrast to Ena/VASP proteins which are necessary for neurite formation [18, 19], CIP4 expression inhibits neuritogenesis. These data are also consistent with our data showing rapid down-regulation of CIP4 during prenatal development (Figure 1A), a period of rapid axon outgrowth.
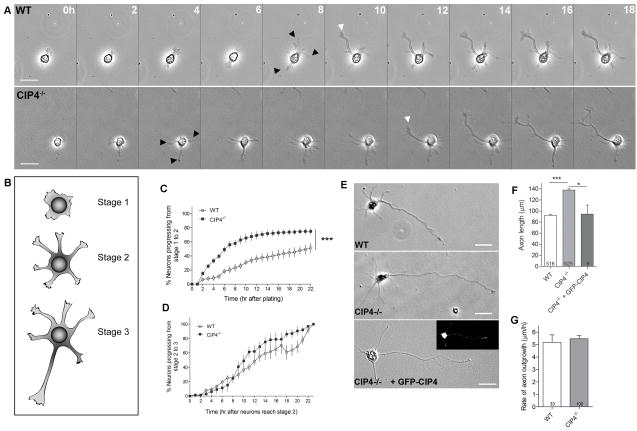
Figure 3. CIP4 negatively regulates neurite formation.
(A) Frames from a time-lapse series showing the extent of outgrowth of living neurons from wild-type and CIP4-null neurons after 1DIV. Black arrowheads indicate extension of neurites (stage 1 → stage 2) and white arrowheads indicate extension of axons (stage 2 → stage 3). (B) Model of stage 1, 2 and 3 neurons. (C) Cumulative histogram comparing the rate at which WT and CIP4-null neurons progress from stage 1 to 2 (n=100 WT and 211 CIP4-null neurons) or (D) from stage 2 to 3 (n=62 WT and 64 CIP4-null neurons). ***, P <0.001 (Two-way repeated measures ANOVA). (E) Images of a wild type neuron, a CIP4-null neuron and a GFP-CIP4-transfected CIP4-null neuron after 24 hours in culture. The fluorescence image of the GFP-CIP4-transfected CIP4-null neuron is shown in the inset. (F) Bar graphs of the axon length of wild type, CIP4-null and GFP-CIP4 transfected CIP4-null neurons at 1DIV. *, P <0.05 and ***, P <0.001 (One-way ANOVA with Bonferroni post-test comparison). (G) The rate of axon outgrowth of wild type and CIP4-null neurons. P >0.05 (two-tailed unpaired T-test with Welch’s correction). All error bars represent ± SEM. Scale bars represent 25 μm.
If CIP4 is a negative regulator of neuritogenesis, CIP4-null neurons should extend longer axons than wild-type littermate controls. This was indeed the case, with CIP4-null cortical neurons extending axons 1.5 times longer than controls at one day in vitro (1DIV) (Figure 3E and 3F). Importantly, the increase in axon length can be rescued by expressing GFP-CIP4 in CIP4-null neurons (Figure 3E and 3F). However, when we measured the instantaneous axon extension rate of stage 3 CIP4-null neurons over shorter time courses, we discovered they extended axons at the same rate as wild-type controls (Figure 3G). Therefore, CIP4 deletion does not change the instantaneous speed of axon outgrowth. Rather, CIP4-null neurons form neurites and therefore axons more rapidly than controls, giving them a “head start”, resulting in significantly longer axons at early times in culture.
Based on these results, overexpression of CIP4 should inhibit neurite formation and subsequently decrease axon length. Consistent with the results obtained by knockout of CIP4, GFP-CIP4 expressing neurons developed shorter axons (77.7 ± 5.1 μm, n= 38 neurons) compared to GFP-transfected controls after 2 DIV (117.9 ± 12.8 μm, n= 30 neurons), (p<0.01, Student t-test with Welch’s correction). However, the instantaneous rate of axon outgrowth did not differ between GFP- (5.3 ± 0.9 μm/hr, n= 6 neurons) and GFP-CIP4-transfected (4.5 ± 0.9 μm/hr, n=5 neurons) neurons. These data indicate that CIP4 overexpression inhibits neurite formation, resulting in shorter axons without affecting instantaneous axon outgrowth rates. Thus, neither removal nor overexpression of CIP4 had any effect on instantaneous axon outgrowth rate, but resulted in longer or shorter axons, respectively, at early times in culture because of its effects on neurite formation.
In contrast to CIP4 depletion results, overexpression of CIP4 markedly reduced the number of neurons progressing from stage 1 to stage 2, with half still in stage 1 after 48 hours in culture (Figure 4A and B). Note that cortical neurons overexpressing myc-CIP4 displayed the same distribution of protein and phenotype as CIP4-EGFP overexpressing neurons (Figure 4A, E(FL-CIP4) and Figure 2A), indicating the fidelity of the CIP4 localization and dynamics described above. In addition, the CIP4-EGFP construct was as efficient at inducing this phenotype as the myc-CIP4 construct (Figure 4B), suggesting that the addition of EGFP does not disrupt CIP4 function.
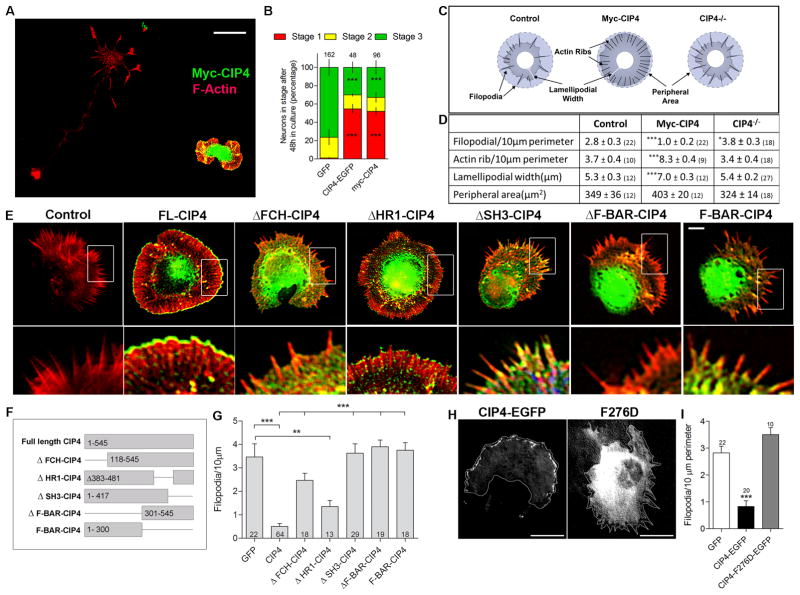
Figure 4. Overexpression of full length CIP4 alters the morphology and development of cortical neurons.
(A) Images of a myc-CIP4 positive and negative neuron fixed and stained with a myc-antibody and phalloidin. (B) Stacked bar graph comparing the number of control neurons in stage 1, 2 and 3 in either CIP4-EGFP or myc-CIP4 expressing neurons after 2 DIV. ***, P <0.001 compared with GFP transfected neurons (Two-way ANOVA with Bonferroni post-test comparison). (C) Diagram showing models of control, myc-CIP4 overexpressing and CIP4-/- stage 1 neurons. (D) Table comparing morphology of stage1 control, myc-CIP4 overexpressing and stage 1 CIP4-null neurons. The numbers in parentheses indicate the number of neurons examined in 3 independent experiments. (E) Images of stage1 CIP4−/− neurons transfected with full length (FL)-mycCIP4, ΔFCH-mycCIP4, ΔHR1-mycCIP4, ΔSH3-mycCIP4, ΔF-BAR-CIP4-EGFP and F-BAR-CIP4-EGFP. FL-CIP4 (full-length) overexpression induces lamellar structures around cell bodies of stage 1 neurons. ΔHR1-CIP4 has a similar effect as FL-CIP4, Δwhile FCH-CIP4, ΔF-BAR-CIP4, F-BAR alone and ΔSH3-CIP4 fail to induce the lamellar structures, resulting in more filopodia. (F) Schematic representations of full-length CIP4 and mutations. (G) Bar graph of filopodia number per 10 μm of perimeter showing that CIP4 and ΔHR1-CIP4 decrease filopodia number of stage 1 neurons. The numbers on the bars indicate the number of neurons examined. (H) Images of CIP4-EGFP and CIP4-F276D-EGFP transfected stage1 neurons. (I) Bar graph comparing filopodia number in GFP, CIP4-EGFP and CIP4-F276D-EGFP transfected control neurons. All error bars represent ± SEM. ***, P <0.001, **, P <0.01 and *, P <0.05 (One-way ANOVA with Bonferroni post-test comparison). Scale bars represent 25 μm in A and 10 μm in E and H.
Interestingly, morphological analysis of stage1 cortical neurons showed that overexpression of CIP4 resulted in almost three times fewer filopodia, but many more actin “ribs”, defined as bundled actin structures within the lamellipodium that do not protrude more than one micron past the cell periphery (Figure 4C and 4D). CIP4 overexpressing neurons also showed large lamellipodial protrusions around the cell body. However, the total peripheral area of CIP4 overexpressing neurons was not significantly different from control neurons (Figure 4C and 4D). CIP4-null neurons, on the other hand, developed significantly more filopodia compared to control neurons. Thus, CIP4 promoted polymerization of actin filaments between filopodia (veils), which resulted in a fewer filopodia, larger lamellipodial area, and more actin ribs. Although we have specifically defined actin ribs for purposes of quantification it is important to point out that actin ribs and filopodial actin bundles are inter-convertible structures that depend on how far the actin-based veils extend between filopodia or conversely, how far filopodia protrude (see Figure S3G, left box).
Since the coordination of actin filament and microtubule dynamics is crucial for neuritogenesis [18], we examined whether CIP4 overexpression affects microtubules in stage 1 neurons. We found that dynamic (tyrosinated) microtubules were excluded from the periphery of CIP4-overexpressing stage 1 neurons as measured by the distance of dynamic microtubules from the peripheral membrane (Control 0.7±0.1μm (N=6 cells) and CIP4-overexpressing neurons 2.9±0.4μm (N=11 cells)). However, the relative number and length of dynamic microtubules per unit area was not diminished significantly (data not shown). These results suggest that the increased actin network in the veils and ribs inhibits microtubule extension into the periphery and subsequent actin/microtubule interactions required for neurite formation [18].
CIP4 overexpression induces veil extension in growth cones
Because a portion of CIP4-overexpessing neurons did progress to stage 3, we investigated the effect of CIP4 expression on growth cones morphology. We found that stage 3 neurons overexpressing CIP4 possessed larger axonal growth cones (Figure S3A and S3B), which contained fewer filopodia (Figure S3C), excessive actin ribs (Figure S3D) and lamellipodial width (Figure S3E), but a similar peripheral area (Figure S3F). These findings are entirely consistent with our results in CIP4-overexpressing stage 1 neurons (Figure 4). Thus, overexpression of CIP4 appears to increase veil protrusion in growth cones as well, resulting in larger lamellipodia with more actin ribs, but fewer filopodia.
The F-BAR and SH3 domains and multimerization of CIP4 are required for targeting to the protruding membrane and inducing lamellipodia
If CIP4 is functioning at the plasma membrane to induce protrusion in neurons we would predict that removing it from the membrane would abolish the increase in lamellipodial veil extension (and therefore the decrease in filopodia) seen in CIP4-overexpressing neurons. The FCH and F-BAR (FCH with Coil-Coil domain) domains of CIP4 are required for membrane tubulation in non-neuronal cells [1]. Interestingly, deletion of either FCH or F-BAR domain prevented CIP4 from concentrating at the periphery of stage 1 neurons and markedly inhibited the effect of CIP4 to induce veil-like lamellipodial protrusions (measured as number of filopodia per unit perimeter) (Figure 4E-G). In addition to membrane binding, the ability of CIP4 to induce lamellipodia also depended on its ability to induce actin polymerization. The SH3 domain of F-BAR proteins has been shown to interact with actin associated proteins (e.g. N-WASP and DAAM1) and induce actin polymerization [35, 36]. Interestingly, transfection of the F-BAR alone or CIP4 lacking the SH3 domain also failed to concentrate CIP4 at the plasma membrane and induce lamellipodial protrusions in neurons (Figure 4E-G). In contrast, after deletion of the HR1 domain CIP4 still concentrated at the plasma membrane and induced lamellipodial protrusions although to a lesser extent than the full length CIP4. (Figure 4E-G). These data indicate that CIP4 requires both the F-BAR membrane-binding domain and the SH3 actin-associated protein-binding domain to concentrate at the peripheral plasma membrane and induce veil-like lamellipodial protrusions. Moreover, these two domains are also required for the effect of CIP4 on neurite formation since expression of ΔF-BAR and F-BAR alone fail to rescue the longer axon phenotype of neurons from CIP4 knockout mice as the full length CIP4 construct does (WT 92.2± 1.9, CIP4(−/−) 137.9 ± 2.3 μm, CIP4(−/−) + FL-CIP4 94.5 ± 16.2 μm, CIP4(−/−) + ΔF-BAR 120.1± 4.0μm and CIP4(−/−) + F-BAR 125 ± 6.0μm (P>0.5, One-way ANOVA with Bonferroni post-test comparison)).
Multimerization of F-BAR dimers has been shown to be important for tubule formation in non-neuronal cells as well [22, 23]. Replacing F276 in the F-BAR domain of CIP4 with a negatively charged residue (F276D) disrupted CIP4 multimerization and potently inhibited tubule formation in COS7 cells [22]. Strikingly, this same point mutation also abolished localization of CIP4 to the leading edge of stage 1 neurons (Figure 4H) and increased the number of filopodia to control levels (Figure 4I). These results suggest that the F-BAR/SH3 domains and multimerization are required for CIP4 to function in both tubule formation in COS7 cells and lamellar protrusion in neurons, indicating some conservation of function in both processes.
Conclusions
Together these data are the first to suggest a function for the F-BAR protein CIP4 in neuronal development. We show that CIP4 plays a unique role as a negative regulator of neurite formation by inducing lamellipodial and veil-like protrusions. This is also the first study to document that an F-BAR protein localizes to both convex and concave lamellipodial protrusions. Although many recent studies suggest that CIP4 is primarily involved in endocytosis, our results demonstrate strikingly that CIP4 plays an unconventional role in cortical neurons by inducing lamellipodial protrusions through alteration of actin filament structures. One possibility is that CIP4 is differentially regulated in cortical neurons and COS7 cells, possibly through phosphorylation [37]. It is also likely that cortical neurons and COS7 cells express unique proteins and/or lipids that could differentially target CIP4 to tubulovesiclular structures vs. protruding plasma membrane. Further studies will be required to test these hypotheses.
Supplementary Material
Highlights.
CIP4 localizes to protruding lamellipodia in neurons
CIP4-induced lamellipodia inhibit neurite formation
The F-BAR/SH3 domains and multimerization are required for CIP4 function in neurons
Acknowledgments
We thank Jon Audya, Bill Bement, Frank Gertler, Tim Gomez, Mary Halloran, Anna Huttenlocher, Katherine Kalil, Kate O’Connor Giles and all the members of the Dent lab for helpful discussions and critical comments on the manuscript. We thank Katherine Kalil for the use of the electroporator and the developmental neurobiology group at UW for critical advice during the project. This work was supported by grants from NIH (R01-NS064014), Dana Foundation and Whitehall Foundation to E.W.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aspenstrom P. Roles of F-BAR/PCH proteins in the regulation of membrane dynamics and actin reorganization. Int Rev Cell Mol Biol. 2009;272:1–31. doi: 10.1016/S1937-6448(08)01601-8. [DOI] [PubMed] [Google Scholar]
- 2.Frost A, De Camilli P, Unger VM. F-BAR proteins join the BAR family fold. Structure. 2007;15:751–753. doi: 10.1016/j.str.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, Hartig SM, Bechill JE, Blanchard EG, Caudell E, Corey SJ. The Cdc42-interacting protein-4 (CIP4) gene knock-out mouse reveals delayed and decreased endocytosis. J Biol Chem. 2010;285:4348–4354. doi: 10.1074/jbc.M109.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartig SM, Ishikura S, Hicklen RS, Feng Y, Blanchard EG, Voelker KA, Pichot CS, Grange RW, Raphael RM, Klip A, et al. The F-BAR protein CIP4 promotes GLUT4 endocytosis through bidirectional interactions with N-WASp and Dynamin-2. J Cell Sci. 2009;122:2283–2291. doi: 10.1242/jcs.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Troglio F, Mukhopadhyay A, Everingham S, Kwok E, Scita G, Craig AW. F-BAR-containing adaptor CIP4 localizes to early endosomes and regulates Epidermal Growth Factor Receptor trafficking and downregulation. Cell Signal. 2009;21:1686–1697. doi: 10.1016/j.cellsig.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Koduru S, Kumar L, Massaad MJ, Ramesh N, Le Bras S, Ozcan E, Oyoshi MK, Kaku M, Fujiwara Y, Kremer L, et al. Cdc42 interacting protein 4 (CIP4) is essential for integrin-dependent T-cell trafficking. Proc Natl Acad Sci U S A. 2010;107:16252–16256. doi: 10.1073/pnas.1002747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr Biol. 2008;18:1639–1648. doi: 10.1016/j.cub.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 8.Toguchi M, Richnau N, Ruusala A, Aspenstrom P. Members of the CIP4 family of proteins participate in the regulation of platelet-derived growth factor receptor-beta-dependent actin reorganization and migration. Biol Cell. 2010;102:215–230. doi: 10.1042/BC20090033. [DOI] [PubMed] [Google Scholar]
- 9.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holbert S, Dedeoglu A, Humbert S, Saudou F, Ferrante RJ, Neri C. Cdc42-interacting protein 4 binds to huntingtin: neuropathologic and biological evidence for a role in Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2712–2717. doi: 10.1073/pnas.0437967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahm M, Kim S, Paik SK, Lee M, Lee S, Lee ZH, Kim J, Lee D, Bae YC. dCIP4 (Drosophila Cdc42-interacting protein 4) restrains synaptic growth by inhibiting the secretion of the retrograde Glass bottom boat signal. J Neurosci. 2010;30:8138–8150. doi: 10.1523/JNEUROSCI.0256-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 13.Polleux F, Snider W. Initiating and growing an axon. Cold Spring Harb Perspect Biol. 2010;2:a001925. doi: 10.1101/cshperspect.a001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lima AD, Merten MD, Voigt T. Neuritic differentiation and synaptogenesis in serum-free neuronal cultures of the rat cerebral cortex. J Comp Neurol. 1997;382:230–246. doi: 10.1002/(sici)1096-9861(19970602)382:2<230::aid-cne7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Caceres A, Mautino J, Kosik KS. Suppression of MAP2 in cultured cerebellar macroneurons inhibits minor neurite formation. Neuron. 1992;9:607–618. doi: 10.1016/0896-6273(92)90025-9. [DOI] [PubMed] [Google Scholar]
- 16.Chuang JZ, Yeh TY, Bollati F, Conde C, Canavosio F, Caceres A, Sung CH. The dynein light chain Tctex-1 has a dynein-independent role in actin remodeling during neurite outgrowth. Dev Cell. 2005;9:75–86. doi: 10.1016/j.devcel.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehmelt L, Smart FM, Ozer RS, Halpain S. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J Neurosci. 2003;23:9479–9490. doi: 10.1523/JNEUROSCI.23-29-09479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J, et al. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 19.Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, et al. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Dawson JC, Legg JA, Machesky LM. Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Itoh T, Takenawa T. Mechanisms of membrane deformation by lipid-binding domains. Prog Lipid Res. 2009;48:298–305. doi: 10.1016/j.plipres.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 24.Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Letters. 2010;584:1111–1118. doi: 10.1016/j.febslet.2010.02.058. [DOI] [PubMed] [Google Scholar]
- 25.Bu W, Lim KB, Yu YH, Chou AM, Sudhaharan T, Ahmed S. Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation and vesicle motility: implications for endocytosis. PLoS ONE. 2010;5:e12153. doi: 10.1371/journal.pone.0012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci. 2011;31:2447–2460. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaidel-Bar R, Joyce MJ, Lynch AM, Witte K, Audhya A, Hardin J. The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J Cell Biol. 2010;191:761–769. doi: 10.1083/jcb.201005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J, Mukhopadhyay A, Truesdell P, Chander H, Mukhopadhyay UK, Mak AS, Craig AW. Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis. J Cell Sci. 2011;124:1739–1751. doi: 10.1242/jcs.078014. [DOI] [PubMed] [Google Scholar]
- 31.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Mongiu AK, Weitzke EL, Chaga OY, Borisy GG. Kinetic-structural analysis of neuronal growth cone veil motility. J Cell Sci. 2007;120:1113–1125. doi: 10.1242/jcs.03384. [DOI] [PubMed] [Google Scholar]
- 33.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 34.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 35.Aspenstrom P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res. 2006;312:2180–2194. doi: 10.1016/j.yexcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Bu W, Chou AM, Lim KB, Sudhaharan T, Ahmed S. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J Biol Chem. 2009;284:11622–11636. doi: 10.1074/jbc.M805940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.