Abstract
Objective
To identify links between altered gene imprinting in the placenta and infant neurobehavioral profiles.
Study design
We used qRT-PCR to examine the expression of 22 imprinted candidate genes in a series of 106 term human primary placenta tissues and associated that expression with summary scores from the NICU Network Neurobehavioral Scales performed on the corresponding infants. Clustering of the expression data was used to define distinct classes of expression.
Results
Significant associations were identified between classes of expression and the NICU Network Neurobehavioral Scales quality of movement (P=0.02) and handling (P=0.006) scores. Multivariable regression demonstrated an independent effect of imprinted gene expression profile on these neurobehavioral scores after controlling for confounders.
Conclusion
These results suggest that alterations in imprinted gene expression in the placenta are associated with infant neurodevelopmental outcomes. Our results suggest a role for the placenta and genomic imprinting in the placenta beyond intrauterine growth regulation.
Keywords: neurodevelopment, epigenetics, fetal programming, trophoblast, mental health
The paradigm of the developmental origins of health and disease posits that the period of intrauterine growth is critical in determining health outcomes throughout life. The focus is on outcomes associated with metabolic diseases and related disorders, such as diabetes, obesity, and coronary heart disease[1, 2]. The concept of fetal programming, and the importance of the intrauterine environment beyond metabolic outcomes has now been linked to behavioral and psychosocial outcomes[3, 4], including schizophrenia[5] and depression[6, 7]. Most studies have linked birth size with these health outcomes, but it is generally accepted that birth size is a proxy for a complex interplay of underlying etiologic mechanisms and that there are common factors that influence intrauterine growth as well as adult physiological systems [8]. Defining the molecular basis of fetal programming is critical to understanding the mechanistic basis of these observational findings. Such understanding can provide insight into early identification of individuals who might benefit from intervention and to design novel ways to prevent and treat health problems later in life.
Many of the adverse effects related to the intrauterine environment may result in epigenetic alterations [9–11]. A key mode of epigenetic regulation is that of genomic imprinting, which refers to the monoallelic expression of a subset of genes in a conserved parent-of-origin fashion. This mode of control of expression is orchestrated by the timely placement of epigenetic signals including DNA methylation and histone modification. Among the key functions of imprinted genes is the control of the allocation of maternal resources to the fetus, regulation of metabolism in the early postnatal period, and the determination of the metabolic states of developing metabolic organs such as the pancreas, muscle, adipocytes, and the hypothalamus [12]. It is likely that imprinted genes play a crucial role in neurodevelopment. A high proportion of imprinted genes are expressed in the central nervous system. Maternally expressed imprinted genes are thought to favor the development of larger brains[13]. Children with Beckwith-Wiedemann syndrome, which results in part from inappropriate imprinting of specific imprinted gene clusters, demonstrate greater than expected proportions of abnormal scores on emotional and behavioral scales[14]. Female mice engineered to be null for the paternal Peg3 gene, exhibit a reduced number of oxytocin-producing neurons in the hypothalamus[15].
The placenta serves as a central regulator of the intrauterine environment, and through its immune-endocrine role may be important in appropriate infant neurodevelopment[16, 17]. Utilizing a population-based birth cohort, we examined the expression of 22 imprinted genes in 106 human term placenta samples and associated the profiles of expression of these genes with measures of newborn neurodevelopment. Our hypothesis was that altered gene expression profiles of imprinted genes have a quantifiable impact on neurobehavioral development in an unbiased, population-based cohort.
METHODS
Subjects are part of the ongoing Rhode Island Child Health Study, which is enrolling mother-infant pairs following delivery at Women and Infants Hospital of Rhode Island. Term infants born small for gestational age (SGA, lowest 10th percentile), or large for gestational age (LGA, highest 10th percentile), based on birthweight and gestational age and calculated from the Fenton growth chart[18], were selected; infants appropriate for gestational age (AGA) matched on sex, gestational age (±3 days), and maternal age (±2 years) were also enrolled. Only singleton, viable infants were included in the study. Other exclusion criteria were maternal age <18 years or a life-threatening medical complication of the mother, and congenital or chromosomal abnormality of the infant. A structured chart review was used to collect information from the maternal inpatient medical record from delivery, and mothers had to participate in an interviewer-administered structured questionnaire for information on lifestyle, demographics, and exposure histories. Neurodevelopmental status at birth was assessed with the NICU Network Neurobehavioral Scales, administered by certified psychometrists, who were blinded to the study hypothesis. The test was administered during the newborn inpatient stay after the day of birth, but prior to discharge. Initially designed for the observation of high-risk term and preterm infants, the NICU Network Neurobehavioral Scales are a comprehensive evaluation of the neurobehavioral performance that includes neurological and behavioral measures and signs of stress [19, 20]. The NICU Network Neurobehavioral Scales have demonstrated predictive validity for medical outcomes such as cerebral palsy diagnosis, neurological abnormalities, and diseases with risks to the brain, as well as developmental outcomes such as mental and motor functioning, behavior problems, school readiness and IQ [21] and norms have been developed for this examination in healthy, full term infants [22]. Items on the NICU Network Neurobehavioral Scales were scored using previously established summary scores. We focused on the 9 summary scores (attention, arousal, excitability, hypertonicity, stress/abstinence, self-regulation, non-optimal reflexes, handling, and quality of movement) shown to be most sensitive based on previous studies[21]. 106 subjects, enrolled between September 2009 and May 2010, were examined. All subjects provided written informed consent as approved by the Institutional Review Boards for Women and Infants’ Hospital and Brown University.
Placenta Sample Collection and RNA Extraction
Within 2 hours of delivery, for each subject 12 biopsies of placenta tissue, 3 from each of 4 quadrants (totaling approximately 1 g of tissue) were excised, from the maternal side of the placenta 2 cm from the umbilical cord insertion site. All samples were from the placenta parenchyma and were homogenous, without areas of calcification, without hemorrhage or clots and free of maternal decidua. The samples were placed immediately in RNAlater and stored at 4°C. At least 72 hours later, placenta samples were removed from RNAlater, blotted dry, snap-frozen in liquid nitrogen, homogenized using a mortar and pestle, and stored in sample tubes at −80°C until examination. RNA was isolated from appr oximately 100 mg of placental sample using the RNeasy system (Qiagen) including the on-column DNAse digestion to assure removal of residual DNA contamination. Extracted RNA was quantified using a Nanodrop spectrophotometer and stored at −80°C.
Quantitative RT-PCR for Imprinted Genes
Twenty-two imprinted genes were chosen for this investigation including twenty that were found to be dysregulated in our previous gene expression experiments when comparing severe growth restricted with appropriate-for-gestational-age placentas[23, 24]. We also included two homeobox genes, HOXA11 and HOXD10, which are putatively imprinted genes fundamental for proper fetal development. The full gene list can be found in Table I (available at www.jpeds.com) that also provides the list of the primer sets used for the gene expression analysis.
Table 1.
(online only). List of the tested genes with their primer sets used for the expression test
| Gene | PoO(1)(2) | Primers
|
|
|---|---|---|---|
| Forward | Reverse | ||
| CCDC86 | ND | CAAGGACTTTGCCCGTCA | CGGCGTTTCAGGTTCTCA |
| CD44 | ND | CGACAGCACAGACAGAATCC | TCCAAATCTTCCACCAAACC |
| CDKAL1 | ND | TTAACCAATTTTACCCAAGACCA | TCTTTTGTCCTTTGCTTTTTCA |
| DHCR24 | ND | TTATGACAGGGGTCATGACAGA | AATTGCCAATGCTATTCAGCTT |
| DLK1 | P | AGCTGCACCCCCAACC | CTGCTGGCGCAGTTGGTC |
| EPS15 | ND | CTCCCTTGCATGATGGAAAC | CAGGATTCCCACTTGATAACTGT |
| H19 | M | ATTTGCACTAAGTCATTTGCACTG | CAGTCACCCGGCCCAGAT |
| IGF2 | P | TTTGTCCCTCTCCTCCTCCA | CAAGGCTCTCTGCCGAAACT |
| ILK | ND | AACACGGAGAACGACCTCAA | CATCTCAACCACAGCAGAGC |
| MEG3 | M | GTTTCTGGACTGTGGGCTGT | CAACAGCAACAAAACTCAGAACATTC A |
| MEST | P | TGACCACATTAGCCACTATCCA | CCTGCTGGCTTCTTCCTATACA |
| NNAT | P | GTACATCTTCCGCGTGCTG | CTTCTCGCAATGGGCTGT |
| PEG3 | P | ACATTTCTGGTGTTTGAGGAGTT | AGACCAGGTTCCGGTAATTCT |
| PEG10 | P | AAATTGCCTGACATGAAGAGGAGTCTA | AAGCCTAGTCACCACTTCAAAACACACTAAA |
| PHLDA2 | M | TCCATCCTCAAGGTGGACTG | ATCTCCTTGTGGTCGGTGGT |
| PLAGL1 | P | CATATTTGCATGTTAGAAGAATCAGC | TGAGTCAGTTAGGTCAGTGTAGAGAGA |
| SLC22A18 | M | CCTCAGCTTCACCTGCATCC | GTGTACCTGGCGGGGCTTAC |
| SNRPN | P | GAGGAGTTGGGGGACCAT | CAGCTGCTACAGTGCCTCTTC |
| TP73 | M | AGGCAGGTGGGCCAATG | TGGGAGATGTTAGTAGGGGAAGC |
| ZNF331 | P | AGGAGGAAGACGAATCGTTAAA | CTCGGGGATGCTTTTTCTG |
| HOXA11 | ND | AGGCAGCTGCAGTGGAGAAT | GGGACCACGCTCATCAAAAT |
| HOXD10 | ND | TGTCATGCTCCAGCTCAACC | CACGGGCTCGTTCATCTT |
Parent-of-Origin
P= paternally expressed/maternally imprinted; M = maternally expressed/paternally imprinted; ND = expressed/silenced allele not determine
Quantitative, real-time reverse transcription PCR (qRT-PCR) was performed at the Mount Sinai School of Medicine Real-Time PCR facility which uses automated fluid handling systems for running 384 well plates. Expression values were cascade normalized against the three housekeeping genes RPS11, ACTB and TUBB. The copy number for each imprinted gene for each sample was then calculated as:
where CN is the copy number, k is the equation constant, eff is the average replication efficiency and Ctcn and Ctgt are the Ct values for each specific sample for (1) the 3 housekeeping genes cascade normalized (cn), (2) each gene tested (gt), respectively.
Statistical Analyses
Correlation of expression between imprinted genes was evaluated using the Spearman Rank Correlation. Due to the high degree of correlation in expression between imprinted genes and to allow for subsequent inference, subjects were clustered based on gene expression data using recursively partitioned mixture modeling[25] fit with a mixture of Gaussian distributions on log-transformed gene expression data. This modeling strategy allows samples to be clustered together into groups based on similarity in the gene expression profiles, and for the membership in those groups to be associated with variables of interest. The robustness of the recursively partitioned mixture modeling was assessed by comparison with a principle components analysis, using singular value decomposition, and which revealed similar class structure based on gene expression pattern. Class membership was obtained from the mixture model and subsequent bivariate associations were tested via permutation test with 10,000 permutations each. For continuous variables, the Kruskal-Wallis test statistic was used, and for categorical variables, the standard chi-square goodness-of-fit test was used. For those variables demonstrating a significant bivariate association, multinomial logistic regression was used to model expression class while controlling for potential confounders including infant birthweight, gestational age, sex, maternal age, delivery method, and maternal insurance as these have been previously linked to both altered placental gene expression and NICU Network Neurobehavioral Scales outcomes[20, 21, 23, 26, 27]. The functional network structure of the gene clusters resulting from the recursively partitioned mixture modeling were assessed using Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com).
RESULTS
The demographics of the population of infants, as well as the mean NICU Network Neurobehavioral Scales summary scores are provided in Table II. Based on the a priori design of the cohort, the population is over-represented by small and large for gestational age infants. All infants were near term with a mean gestational age of 38.9 weeks. Also included in Table II are the descriptive statistics of the summary scores for the 9 NICU Network Neurobehavioral Scales measures.
Table 2.
Population Demographic and Clinical Characteristics and NICU Network Neurobehavioral Scales Summary Scores
| Variable | N | % |
|---|---|---|
| Growth Status | ||
| SGA | 33 | 31 |
| AGA | 51 | 48 |
| LGA | 22 | 21 |
| Infant Sex | ||
| Female | 59 | 56 |
| Male | 47 | 44 |
| Delivery Method | ||
| Vaginal | 67 | 63 |
| Caesarean | 38 | 36 |
| Section | ||
| Unknown | 1 | 1 |
| Maternal Ethnicity | ||
| White | 79 | 75 |
| African American | 7 | 7 |
| Asian | 3 | 3 |
| Other | 14 | 12 |
| Unknown | 3 | 3 |
| Maternal Insurance | ||
| Public | 45 | 42 |
| Private | 61 | 58 |
| Variable | N | Mean | Std. Dev. | Min | Max |
|---|---|---|---|---|---|
| Birth weight (g) | 106 | 3310 | 744.9 | 1705 | 5080 |
| Gestational Age (weeks) | 106 | 38.9 | 1.1 | 37 | 41 |
| Maternal Age (yrs) | 106 | 29.4 | 6.0 | 18 | 40 |
| Arousal | 94 | 4.1 | 0.9 | 1.9 | 6.2 |
| Attention | 86 | 3.8 | 1.1 | 1.6 | 6.2 |
| Excitability | 94 | 4.6 | 3.0 | 0 | 13 |
| Hypertonicity | 94 | 0.34 | 0.82 | 0 | 5 |
| Stress Abstinence | 94 | 0.22 | 0.07 | 0.08 | 0.41 |
| Handling | 94 | 0.38 | 0.24 | 0 | 0.88 |
| Non-optimal Reflexes | 94 | 6.77 | 2.29 | 0 | 11 |
| Self-regulation | 93 | 4.68 | 0.91 | 2.3 | 6.5 |
| Quality of Movement | 94 | 4.0 | 0.69 | 2.2 | 5.5 |
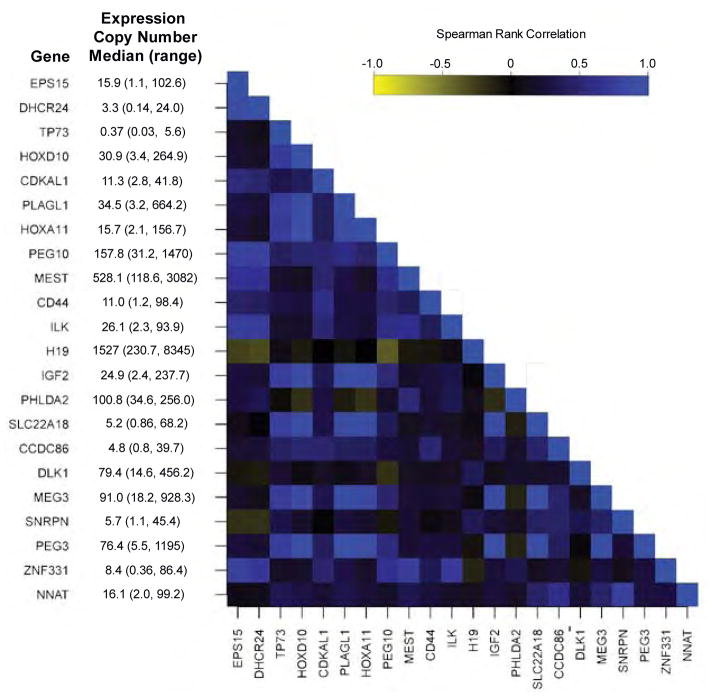
Using a quantitative reverse transcription RT-PCR normalized to RPS11, ACTB and TUBB, we calculated the copy number of transcripts for each of the 22 imprinted genes examined in the 106 placenta samples. The median and range of expression of these genes is given on Figure 1 (available at www.jpeds.com). As imprinted gene expression is tightly controlled, and as many of these imprinted genes are co-regulated in clusters, we examined the correlation in expression between each of the imprinted genes. The heatmap in Figure 1 depicts the Spearman Rank Correlation coefficient for the correlations in expression between each of the imprinted genes, ordered by chromosomal location. This correlation matrix suggests moderate to strong correlation between many of the imprinted genes, including those genes localized in the same chromosomal region, but also between those located on different chromosomes.
Figure 1.
Correlation matrix of gene expression between imprinted genes in human placenta samples (n=106). Gene names and median expression levels are given in rows, and the spearman rank correlation between the expression of these genes is provided as a heatmap.
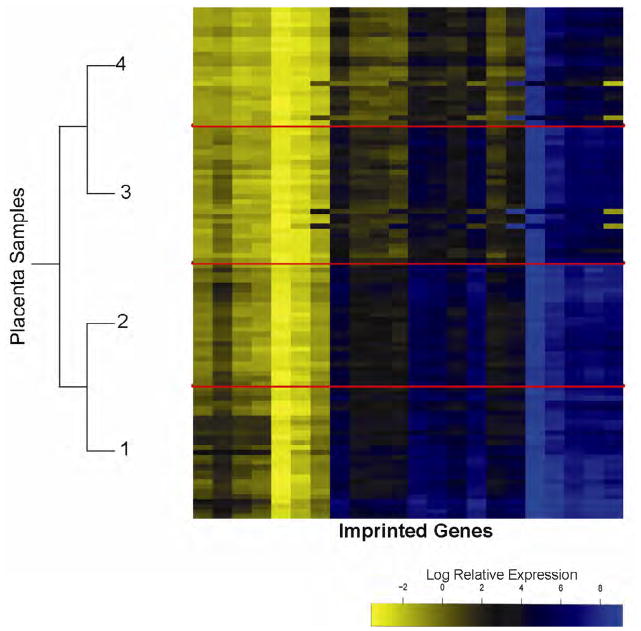
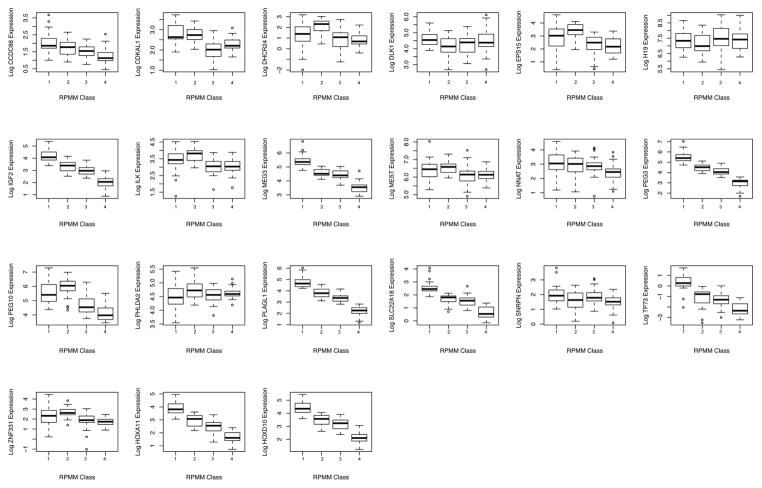
Due to the strong correlations in expression of these genes, we utilized a model-based hierarchical clustering strategy, Gaussian recursively partitioned mixture modeling, to group subjects into clusters based on the profiles of imprinted gene expression across all 22 genes examined. This modeling resulted in 4 classes of imprinted genes, depicted in the heatmap of Figure 2, where the color intensity represents the log relative expression of the genes in columns, and the classes of samples (rows) are separated by solid red lines which have been denoted by numbers 1 to 4. The log relative expression of each of the individual genes by class is shown in Figure 3 (available at www.jpeds.com). As expected the majority of genes show highly significant differential expression by class. A number of specific genes showed a trend of reduced expression from the class 1 to Class 4, including CCD86, IGF2, MEG3, PEG3, PLAGL1, SLC22A18, TP73, HOXA11, and HOXD10. Ingenuity network analysis suggests these genes all have either a function related to nervous system development and function or are associated with skeletal, muscular, or nervous system diseases. Alternatively, DHCR24, EPS15, ILK, PEG10, PHLDA2, and ZNF331 exhibited similar expression patterns with each having the highest expression occurring in class 2. Ingenuity pathway analysis on these genes places them in functional areas of cellular development, cell cycle control, and growth-associated diseases such as cancer.
Figure 2.
Gaussian recursively partitioned mixture models, a hierarchical clustering, based on imprinted gene expression identifying 4 distinct classes of expression signature. Samples are in rows and the expression of the 22 imprinted genes examined is shown by the heatmap in column. The dendrogram depicts the splitting of the samples into 4 classes, which are separated on the heatmap by red lines and are labeled as Classes 1 to 4.
Figure 3.
Boxplots of the expression of each of the 22 imprinted genes in each of the recursively partitioned mixture model classes. Boxes represent 25th and 75th percentile, the dark horizontal line the median value, and the whiskers the 5th and 95th percentile.
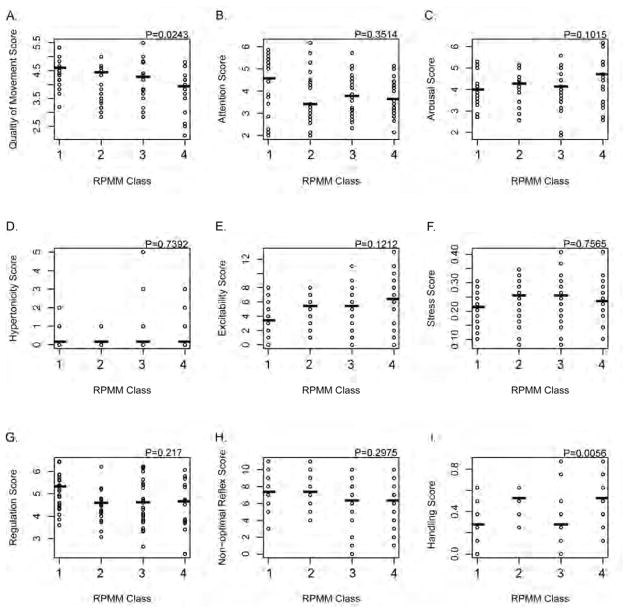
Utilizing the classes resulting from the Gaussian recursively partitioned mixture model, we then examined the associations between imprinted gene expression class and the NICU Network Neurobehavioral Scales summary scores for the individual newborns (Figure 4). A permutation-based Kruskal-Wallis test identified significant differences in the NNNS summary scores for Quality of Movement (P=0.02) and Handling (P=0.006) by imprinted gene expression class. For quality of movement, there was a trend for poorer quality of movement moving from class 1 to 4 of imprinted gene expression, and the poorest handling scores were observed for gene expression classes 1 and 3. Although not statistically significant, scores for arousal and excitability demonstrated an opposite trend than quality of movement, with greater arousal and excitability identified in infants whose imprinted gene expression was classified in the right (3 and 4) classes.
Figure 4.
Scatterplots of NICU Network Neurobehavioral Scales scores by gene-expression based recursively partitioned mixture models. The distribution of scores for (A) quality of movement, (B) attention, (C) arousal, (D) hypertonicity, (E) excitability, (F) stress/abstinence, (G) self-regulation, (H) non-optimal reflexes, and (I) handling are examined across each class, and the P-values provided are based on a non-parametric permutation-based Kruskal-Wallis test of differences across groups.
To model the magnitude of difference in NICU Network Neurobehavioral Scales score by imprinted gene expression class, and to control for potential confounders, we performed multivariable generalized linear modeling of the NICU Network Neurobehavioral Scales scores found to be significantly associated with imprinted gene expression class in bivariate analyses (Table III). In a model controlling for infant birthweight, gestational age, sex, maternal age, delivery method, and maternal insurance, membership in class 3 compared with 1 resulted in a significant reduction in quality of movement summary score of 0.4. By contrast, membership in class 4 compared with 1 resulted in a significant reduction by 0.6. Thus, having a gene expression profile which assigns an infant to the class 4 is associated with a reduction in the quality of movement score of nearly a full standard deviation compared with infants assigned to the class 1. In an additional model where we controlled for the same confounders, infants whose expression profile was a member of class 2 compared with 1 had a significantly increased handling score of 0.1, and those in class 4 compared with 1 significantly increased by 0.2, which represents nearly a full standard deviation greater score in infants assigned to class 4 compared with 1.
Table 3.
Multivariable Generalized Linear Models for NICU Network Neurobehavioral Scales Quality of Movement and Handling Scores Associated with Imprinted Gene Expression Classes Controlled for Confounders
| Imprinted Gene Expression Cluster | NNNS Quality of Movement | NNNS Handling | ||
|---|---|---|---|---|
| Estimate (95% CI) | P | Estimate (95% CI) | P | |
| 1 | Referent | Referent | ||
| 2 | −0.4 (−0.8, 0.04) | 0.08 | 0.1 (0.001, 0.3) | 0.05 |
| 3 | −0.4 (−0.8, −0.03) | 0.03 | 0.1 (−0.1, 0.2) | 0.31 |
| 4 | −0.6 (−1.0, −0.2) | 0.004 | 0.2 (0.05, 0.3) | 0.01 |
Models are controlled for infant birthweight, gestational age, sex, maternal age, delivery method, and maternal insurance.
DISCUSSION
There is growing interest in the molecular mechanisms which underlie the epidemiologic observations of the developmental origins of health and disease, as well as a focus of study on the critical role of the placenta in mediating these effects[16, 28, 29]. Most studies to date have emphasized gross placental morphology and its relationship to near-term adverse birth outcomes or later life cardiovascular disease and metabolic syndrome related phenotypes[28–31]. We systematically quantify imprinted gene expression using a targeted qRT-PCR strategy in the placenta and to relate variability in that expression to another important phenotype, infant neurobehavioral profiles. These results support a critical role for the function of the placenta and for genomic imprinting of genes in the placenta in neurodevelopmental outcome. The neurobehavioral profiles captured through the NICU Network Neurobehavioral Scales assessment have robust, prospective predictive potential for later behavior problems, school readiness, and IQ through 4.5 years of age[21]. Thus the associations we have identified between the placenta imprinted gene expression profiles and NICU Network Neurobehavioral Scales informs the molecular mechanism through which the intrauterine environment plays a role in mental health and behavior beyond birth.
Variability in expression of imprinted genes in this population is consistent with our previous work, where we observed up to 2–4 fold variation in expression in human placentas[23]. This would indicate that mechanisms other than classical loss of imprinting may be affecting gene expression, as loss of imprinting has a maximum effect of doubling the expression level. Our observation of a strong correlation in the expression between many of the imprinted genes, and most particularly amongst genes located on the same chromosomal regions suggest coordinated alterations in expression potentially results from a common stress or exposure encountered during intrauterine development and/or disruption of a single imprinting control region. This also suggests that the association of complicated phenotypes, such as neurobehavioral profiles, with changes in gene expression may not result from a single altered gene, but from coordinated alterations in a number of genes. Multiple gene expression signatures are now being recognized as important predictors of outcome in oncologic diagnostics[32]. This observation should extend to other complex outcomes and diseases. Our statistical modeling allows for detailed but unbiased examination of imprinted gene expression as expression profiles, thereby improving our power and taking into account this potentially important coordinated variation.
The placenta plays a key role in regulating the intrauterine environment and in the appropriate development of the fetus. Once developed, the placenta serves as the source of fetal nutrients, water and gas exchange, excretion of toxic waste products, and serves many immune-endocrine interactions at the maternal fetal interface. These effects are modulated by simultaneous production of many pregnancy related hormones, proteins and growth factors thereby fulfilling a critical role in proper intrauterine development. The placenta produces an array of neuropeptide hormones that are analogous to those produced by the hypothalamus and pituitary of the brain, including GnRH, TRH, GHRH, CRH, and oxytocin[33]. Rapid advancements in understanding the integrated regulation of neuropeptide/neuroendocrine homeostasis within and outside the brain as well as placenta[34–36] has led to the formulation of a new concept that placenta is the “third brain” that links the developed (maternal) and developing (fetal) brains [35], and have expanded the role of the placenta in the pathophysiology of intrauterine insults[16]. Thus, altered placental function, through altered expression of imprinted genes may play a critical role in mediating various health outcomes related to the intrauterine environment[37].
We identified that infants’ whose placentas exhibited an expression profile of class 4 had significantly reduced quality of movement in the NNNS assessment. There were a number of infants in this category who exhibited scores considered in the lowest 10th percentile from normative scales of performance[22]. These measures examine motor control including smoothness, maturity, and lack of startles and tremors. Reduced quality of movement has been observed among infants exposed to prenatal methamphetamine[27], infants born small for gestational age infants to adolescent mothers[38], and infants requiring pharmaceutical treatment for neonatal abstinence syndrome[39]. Our findings, in non-drug exposed infants suggest that there may be other exposures or stressors during the intrauterine developmental period that can, through alterations in the expression of imprinted genes, and thus the function of the placenta, lead to similar poorer quality of movement phenotypes. Imprinted genes such as MEG3, HOXA11, and HOXD10 demonstrated greatly reduced expression in infants with this expression profile compared with those in class 1, and network analysis suggests these genes have functional overlap in nervous system, skeletal, and muscular development. HOXA11 plays a key role in joint and skeletal development[40], HOXD10 is important in motorneuron development[41], and MEG3 alterations have been linked with Prader-Willi syndrome like phenotypes[42]. These genes encode non-coding RNAs, which may play a critical role in modifying placental gene expression, and may have resulted in the observed phenotypes.
Both classes 2 and 4 showed greater handling scores, suggesting more handling strategies were needed to maintain the infant in an alert state during the NICU Network Neurobehavioral Scales orientation examination. The observed relationship between handling score and imprinted gene expression class was most similar for the genes CDKAL1, ILK, MEST, and PHLDA2. ILK is highly expressed in first trimester placentas, regulating migration, and signaling through ILK may be involved in trophoblast syncytialization[43, 44]. MEST is related to placental angiogenesis[45], and increased expression of placental maternally expressed PHLDA2 has been associated with reduced infant birthweight[46]. An Ingenuity network analysis suggested that these genes are involved in cell cycle control and cell growth, and it may be possible that it is through these functions and their role in placental development that these genes can influence neurodevelopment.
A limitation of this study is the moderate sample size in examining the large number of neurobehavioral outcomes available through the NICU Network Neurobehavioral Scales assessment. The length of follow-up is also limited for these infants, so definitive relationships between imprinting signatures and long-term neurobehavioral outcomes await validation. Finally, our samples are limited to term placentas, which may not accurately reflect the expression pattern throughout pregnancy. We cannot make definitive statements regarding the mechanistic link between these profiles, neurobehavior, and birth at earlier gestational ages. Nonetheless, we evaluated a representative collection of samples from the single, large, high-risk delivery service which serves an entire geographic region.
In summary, this study provides support for relationships between imprinted gene expression and critical, prospectively predictive early neurobehavioral measures. Replication and expansion of these findings in additional cohorts is warranted, with additional examination of the important confounding clinical factors during pregnancy which mediate the variation in imprinted gene expression and the relationship to developmental outcome. This research also raises critical questions on the mechanisms through which imprinted genes become deregulated, if and for how long their alteration may persist beyond birth, and the potential for additional neurodevelopmental and other health outcomes to be associated with this altered expression.
Acknowledgments
Funded by National Institutes of Health (NIH) grants from the NIMH [R01 MH094609] and the NCRR [P20 RR018728] and the Venture Capital Research Funding Program of the Mount Sinai Children’s Environmental Health Center and Mount Sinai Child Health and Development Institute. The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barker DJ. Fetal programming of coronary heart disease. Trends Endocrinol Metab. 2002;13:364–8. doi: 10.1016/s1043-2760(02)00689-6. [DOI] [PubMed] [Google Scholar]
- 2.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 3.Allin M, Rooney M, Cuddy M, Wyatt J, Walshe M, Rifkin L, et al. Personality in young adults who are born preterm. Pediatrics. 2006;117:309–16. doi: 10.1542/peds.2005-0539. [DOI] [PubMed] [Google Scholar]
- 4.Wals M, Reichart CG, Hillegers MH, Van Os J, Verhulst FC, Nolen WA, et al. Impact of birth weight and genetic liability on psychopathology in children of bipolar parents. J Am Acad Child Adolesc Psychiatry. 2003;42:1116–21. doi: 10.1097/01.CHI.0000070242.24125.78. [DOI] [PubMed] [Google Scholar]
- 5.Cannon TD, Rosso IM. Levels of analysis in etiological research on schizophrenia. Dev Psychopathol. 2002;14:653–66. doi: 10.1017/s0954579402003139. [DOI] [PubMed] [Google Scholar]
- 6.Gale CR, Martyn CN. Birth weight and later risk of depression in a national birth cohort. Br J Psychiatry. 2004;184:28–33. doi: 10.1192/bjp.184.1.28. [DOI] [PubMed] [Google Scholar]
- 7.Thompson C, Syddall H, Rodin I, Osmond C, Barker D. Birth weight and the risk of depressive disorder in late life. Br J Psychiatry. 2001;179:450–5. doi: 10.1192/bjp.179.5.450. [DOI] [PubMed] [Google Scholar]
- 8.Welberg LA, Seckl JR. Prenatal stress, glucocorticoids and the programming of the brain. J Neuroendocrinol. 2001;13:113–28. doi: 10.1046/j.1365-2826.2001.00601.x. [DOI] [PubMed] [Google Scholar]
- 9.Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, et al. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011:6. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myatt L. Placental adaptive responses and fetal programming. J Physiol. 2006;572:25–30. doi: 10.1113/jphysiol.2006.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robins JC, Marsit CJ, Padbury JF, Sharma SS. Endocrine disruptors, environmental oxygen, epigenetics and pregnancy. Front Biosci (Elite Ed) 2011;3:690–700. doi: 10.2741/e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 13.Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–30. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Kent L, Bowdin S, Kirby GA, Cooper WN, Maher ER. Beckwith Weidemann syndrome: a behavioral phenotype-genotype study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1295–7. doi: 10.1002/ajmg.b.30729. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–3. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 16.Lester BM, Padbury JF. Third pathophysiology of prenatal cocaine exposure. Dev Neurosci. 2009;31:23–35. doi: 10.1159/000207491. [DOI] [PubMed] [Google Scholar]
- 17.Lester BM, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa CW, et al. Behavioral epigenetics. Ann N Y Acad Sci. 2011;1226:14–33. doi: 10.1111/j.1749-6632.2011.06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenton TR. A new growth chart for preterm babies: Babson and Benda’s chart updated with recent data and a new format. BMC Pediatr. 2003;3:13. doi: 10.1186/1471-2431-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lester B, Tronick E. The Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113:631–95. [PubMed] [Google Scholar]
- 20.Lester BM, Tronick EZ. History and description of the Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113:634–40. [PubMed] [Google Scholar]
- 21.Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, et al. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125:e90–8. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tronick EZ, Olson K, Rosenberg R, Bohne L, Lu J, Lester BM. Normative neurobehavioral performance of healthy infants on the Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113:676–8. [PubMed] [Google Scholar]
- 23.Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4:235–40. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
- 24.Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3:261–9. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houseman EA, Christensen BC, Yeh RF, Marsit CJ, Karagas MR, Wrensch M, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boukydis CF, Lester BM. The NICU Network Neurobehavioral Scale. Clinical use with drug exposed infants and their mothers. Clin Perinatol. 1999;26:213–30. [PubMed] [Google Scholar]
- 27.LaGasse LL, Wouldes T, Newman E, Smith LM, Shah RZ, Derauf C, et al. Prenatal methamphetamine exposure and neonatal neurobehavioral outcome in the USA and New Zealand. Neurotoxicol Teratol. 2011;33:166–75. doi: 10.1016/j.ntt.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker DJ, Thornburg KL, Osmond C, Kajantie E, Eriksson JG. The surface area of the placenta and hypertension in the offspring in later life. Int J Dev Biol. 2010;54:525–30. doi: 10.1387/ijdb.082760db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roseboom TJ, Painter RC, de Rooij SR, van Abeelen AF, Veenendaal MV, Osmond C, et al. Effects of famine on placental size and efficiency. Placenta. 2011 doi: 10.1016/j.placenta.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Eriksson JG, Kajantie E, Osmond C, Thornburg K, Barker DJ. Boys live dangerously in the womb. Am J Hum Biol. 2010;22:330–5. doi: 10.1002/ajhb.20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajantie E, Thornburg KL, Eriksson JG, Osmond C, Barker DJ. In preeclampsia, the placenta grows slowly along its minor axis. Int J Dev Biol. 2010;54:469–73. doi: 10.1387/ijdb.082833ek. [DOI] [PubMed] [Google Scholar]
- 32.Conlin AK, Seidman AD. Use of the Oncotype DX 21-gene assay to guide adjuvant decision making in early-stage breast cancer. Mol Diagn Ther. 2007;11:355–60. doi: 10.1007/BF03256259. [DOI] [PubMed] [Google Scholar]
- 33.Liu JH. Endocrinology of Pregnancy. In: Resnik Ca., editor. Maternal-Fetal Medicine, Principles and Practice. 6. Philadelphia: Saunders Elsevier; 2009. pp. 111–24. [Google Scholar]
- 34.Petraglia F, Coukos G, Volpe A, Genazzani AR, Vale W. Involvement of placental neurohormones in human parturition. Ann N Y Acad Sci. 1991;622:331–40. doi: 10.1111/j.1749-6632.1991.tb37878.x. [DOI] [PubMed] [Google Scholar]
- 35.Yen SS. The placenta as the third brain. J Reprod Med. 1994;39:277–80. [PubMed] [Google Scholar]
- 36.Yen SS. Endocrine-metabolic adaptions in pregnancy. In: Yen SS, Jaffe RB, editors. Reproductive Endocrinology. 3. Philadelphia: WB Saunders; 1991. pp. 936–70. [Google Scholar]
- 37.Reik W, Constancia M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, et al. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol. 2003;547:35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Moraes Barros MC, Guinsburg R, Mitsuhiro SS, Chalem E, Laranjeira RR. Neurobehavior of full-term small for gestational age newborn infants of adolescent mothers. J Pediatr (Rio J) 2008;84:217–23. doi: 10.2223/JPED.1796. [DOI] [PubMed] [Google Scholar]
- 39.Jones HE, O’Grady KE, Johnson RE, Velez M, Jansson LM. Infant neurobehavior following prenatal exposure to methadone or buprenorphine: results from the neonatal intensive care unit network neurobehavioral scale. Subst Use Misuse. 2010;45:2244–57. doi: 10.3109/10826084.2010.484474. [DOI] [PubMed] [Google Scholar]
- 40.Castillo-Caro P, Dhanraj S, Haut P, Robertson K, Dror Y, Sharathkumar AA. Proximal radio-ulnar synostosis with bone marrow failure syndrome in an infant without a HOXA11 mutation. J Pediatr Hematol Oncol. 2010;32:479–85. doi: 10.1097/MPH.0b013e3181e5129d. [DOI] [PubMed] [Google Scholar]
- 41.Lin AW, Carpenter EM. Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J Neurobiol. 2003;56:328–37. doi: 10.1002/neu.10239. [DOI] [PubMed] [Google Scholar]
- 42.Hosoki K, Kagami M, Tanaka T, Kubota M, Kurosawa K, Kato M, et al. Maternal uniparental disomy 14 syndrome demonstrates prader-willi syndrome-like phenotype. J Pediatr. 2009;155:900–3. e1. doi: 10.1016/j.jpeds.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 43.Butler TM, Elustondo PA, Hannigan GE, MacPhee DJ. Integrin-linked kinase can facilitate syncytialization and hormonal differentiation of the human trophoblast-derived BeWo cell line. Reprod Biol Endocrinol. 2009;7:51. doi: 10.1186/1477-7827-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elustondo PA, Hannigan GE, Caniggia I, MacPhee DJ. Integrin-linked kinase (ILK) is highly expressed in first trimester human chorionic villi and regulates migration of a human cytotrophoblast-derived cell line. Biol Reprod. 2006;74:959–68. doi: 10.1095/biolreprod.105.050419. [DOI] [PubMed] [Google Scholar]
- 45.Mayer W, Hemberger M, Frank HG, Grummer R, Winterhager E, Kaufmann P, et al. Expression of the imprinted genes MEST/Mest in human and murine placenta suggests a role in angiogenesis. Dev Dyn. 2000;217:1–10. doi: 10.1002/(SICI)1097-0177(200001)217:1<1::AID-DVDY1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 46.Apostolidou S, Abu-Amero S, O’Donoghue K, Frost J, Olafsdottir O, Chavele KM, et al. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med. 2007;85:379–87. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]