Abstract
Many viruses introduce DNA into the host-cell nucleus, where they must either embrace or confront chromatin-factors as a support or obstacle to completion of its life cycle. Compared to the eukaryotic cell, viruses have compact and rapidly evolving genomes. Despite their smaller size, viruses have complex life-cycles that involve dynamic changes in DNA structure. Nuclear entry, transcription, replication, genome stabilization, and virion packaging involve complex changes in chromosome organization and structure. Chromatin dynamics and epigenetic modifications play major roles in viral and host chromosome biology. In some cases, viruses may use novel or viral-specific epigenetic modifying activities, which may reflect variant pathways that distinguish their behavior from the bulk of the cellular chromosome. This review examines several recent discoveries that highlight the role of chromatin dynamics in the life cycle of DNA viruses.
Keywords: chromatin, nucleosome, virus, latency, reactivation, epigenetic
Chromatin is a term that describes structures that organize DNA within the nucleus of of eukaryotic cells. In its simplest conception, chromatin is composed of histone proteins that form nucleosomes on DNA. However, the spacing and modifications of these histones is complex and non-random, and provide structural and signaling capacity to the protein scaffold surrounding genes, replicons, telomeres, and centromeres (Berger, 2007; Henikoff, 2008). The role of chromatin in virus biology depends largely on the life style of the virus, but for all viruses that transverse the nucleus, interactions with chromatin is unavoidable. A number of recent discoveries in virology underscore the importance of chromatin dynamics in the regulation of essential viral processes, including entry, gene expression, replication, and encapsidation. As in so many examples of cell biology, the virus model system can provide an instructive alternative view of mechanisms that can be captured and modified for the purposes of virus survival, but may also provide insight into the possibilities of mechanisms that occur within the cell.
Virus Entry and Chromatinization
Most viruses package their nucleic acids in protective nucleoprotein particles that can enter cells through a variety of receptor-mediated or endocytic mechanisms to gain access to the cellular machinery necessary to complete the reproductive life cycle of the virus. The transition from virion package to active nucleic acid (DNA or RNA) is a complex process that varies among the different classes of viruses. However, these early stages share some important mechanisms among the virus families, and with cellular processes where nucleic acids are mobilized from one compartment or from one activation state to another.
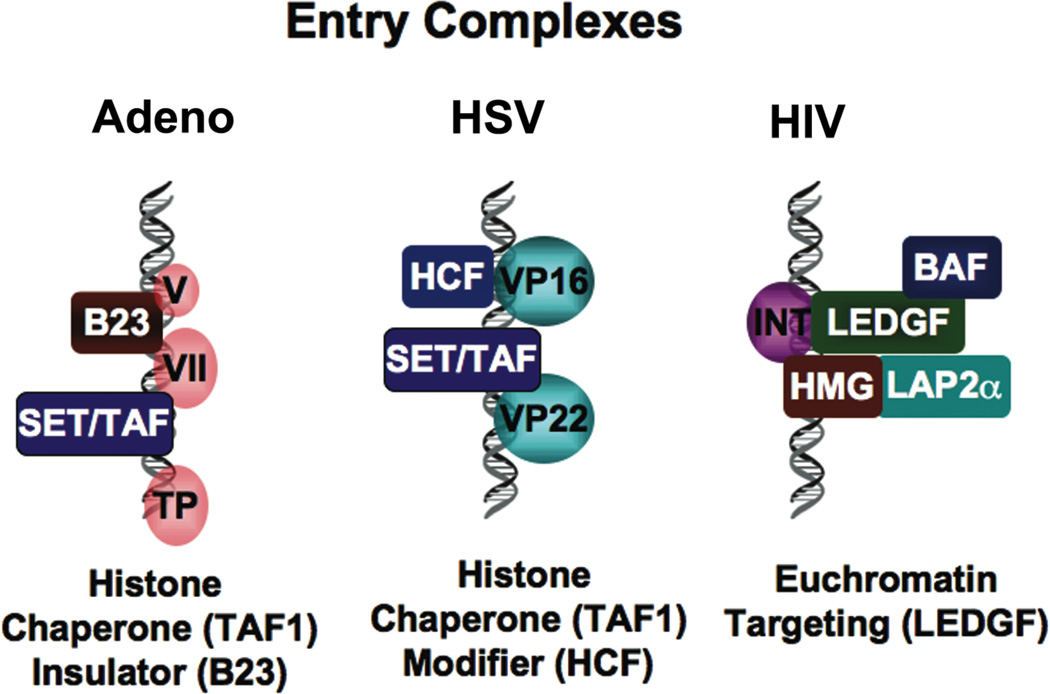
Most virus package their nucleic acid genomes at very high molecular density with specialized viral packaging proteins to form a capsid particle. Adenovirus genomes are packaged as linear double stranded DNA with viral core proteins that form a chromatin-like structure. The viral DNA is covalently linked with the viral encoded terminal binding protein (TP) and core proteins VII, V, and X (Haruki et al., 2006; Spector, 2007; Xue et al., 2005). Core protein VII has sequence similarity to histone H3 and basic sperm-specific protein. Transcription of the newly infecting genome utilizes a template that decondenses during entry into the nucleus. A cellular activity, referred to as the Template Activating Factor (TAF) is required for the replication competence of the viral DNA. Several cellular proteins contribute to TAF activity, including the SET/TAF1B and pp32 proteins. These proteins can interact with adenovirus core protein VII, and functions in remodeling of the incoming viral DNA (Haruki et al., 2006; Spector, 2007; Xue et al., 2005). The SET/TAF1B and pp32 proteins are also components of the INHAT complex that inhibits histone acetyltransferase (HAT) activity of cellular proteins p300 and CBP (Seo et al., 2002; Seo et al., 2001). SET/TAF1B has histone chaperone activity and can bind unmodified histone H3 tails (Schneider et al., 2004), which may account for its ability to inhibit histone H3 HAT activity. In addition to SET/TAF1B, the nucleolar protein nucleophosmin/B23 was isolated as a component of the template activating complex (Samad et al., 2007). Adenovirus protein V was responsible for relocalizing nucleophosmin from the nucleolus to the nucleoplasm (Hindley et al., 2007; Matthews, 2001). Nucleophosmin has been implicated in several chromatin functions including histone chaperone, nucleosome assembly, transcriptional co-activator/co-repressor, and chromatin boundary factor (Frehlick et al., 2007). One interesting feature of nucleophosmin is its ability to decondense sperm cell chromatin during early stage embryogenesis, suggesting that viral DNA may share some features with chromatinization of the paternal genome.
Herpe Simplex Virus (HSV), like adenovirus, enters cells as a linear double stranded DNA molecule. Several virion-associated proteins are essential for productive infection. The HSV VP22 protein binds to TAF1B, similar to that of Ad VII protein (van Leeuwen et al., 2003). The function of VP22 binding to TAF1B is not completely clear, but appears to contribute to the assembly of an active chromatin structure on viral genomes. VP16 is another HSV-encoded virion protein that plays a very active role in transcription activation of the viral immediate early genes. As I will discuss later, VP16 recruits histone modifying and nucleosome remodeling activities which indicate that nucleosomes are assembling during viral entry into the nucleus. The precise details of nucleosome assembly during the early stages of herpesvirus entry have not been completely elucidated.
Retrovirus and lentivirus genomes also enter the nucleus as double stranded DNA, but this process appears to differ significantly from that of the constitutively double stranded nuclear DNA viruses, like adeno and herpesvirus. One major difference is that the double stranded DNA genome is synthesized by virion-associated reverse transcriptase in the cytoplasm. The newly synthesized cytoplasmic DNA forms a pre-integration complex (PIC) that consists of viral and cellular proteins. For MoMLV, the viral proteins include integrase (IN), nucleocapsid (NC), matrix (MA), reverse transcriptase (RT), and Vpr. Cellular proteins include the barrier-to-autointegration (BAF), HMG proteins, Ku, lamina-associated polypeptide 2α (LAP2α), and lens-epithelium-derived growth factor (LEDGF/p75) (Suzuki and Craigie, 2007). Entry into the nucleus is cell cycle dependent and utilizes the cellular nuclear import machinery, often through a nuclear localization signal on one or more of the PIC components. Once in the nucleus, PIC components like LEDGF have been implicating in anchoring to chromatin. HIV was found to integrate at chromatin sites enriched in euchromatic histone modifications, including H3 K4 methylation and histone H3 and H4 acetylation (Wang et al., 2007). LEDGF appears to be a decisive and essential factor in mediating the chromatin attachment of the PIC and viral integration (Llano et al., 2006). LEDGF was originally identified as a transcriptional coactivator and subsequently shown to have non-specific chromatin binding activity mediated by two AT-hook DNA binding domains, similar to those in HMGI/Y proteins (Turlure et al., 2006). In addition to LEDGF, the cellular polycomb protein EED was found to bind to viral integrase, Gag, and Nef (Violot et al., 2003). EED and other polycomb proteins are thought to establish epigenetically stable transcription repression through methylation of H3 K27. Overexpression of EED inhibits HIV replication, which could be reversed by cytoplasmic export mediated by viral encoded Nef (Rakotobe et al., 2007). Although the mechanism of EED inhibition of HIV is not clear, these findings suggest that cellular chromatin factors can restrict HIV gene expression, while viral factors, like Nef, can bypass this restriction (Fig. 1).
Figure 1. Chromatin and Viral Entry.
The establishment of chromatin structure may be directed by viral proteins that associate with viral DNA during entry into the nucleus. Adenovirus VII protein associates with SET/TAF1 histone chaperones and the B23/nucleophosmin protein implicated in chromatin insulators. Herpesvirus VP22 also binds SET/TAF1 and VP16 interacts with cellular histone methylation complex through HCF. HIV DNA forms a pre-integration complex with the HMG, BAF, LEDGF and LAP2α proteins that target proviral DNA to euchromatic domains in the host genome.
Chromatin and the Evasion of Host-Cell DNA Damage Response
After entry and delivery to the appropriate subcellular compartment, virus nucleic acid must adapt a structure that is competent for transcription, translation, and replication or establish a stable intermediate for latent infection or long-term integration. There are a variety of mechanisms and strategies to accomplish these fundamental requirements. For nuclear DNA viruses, there is the challenge of evading the host DNA damage recognition machinery, which may serve as an innate anti-viral defense mechanism, reminiscent of the prokaryotic restriction modification system that prevents phage infection. There have been several excellent reviews that describe the DNA damage response to viral infections (Davy and Doorbar, 2007; Lilley et al., 2007). DNA and chromatin structures formed during the infection process may evoke a DNA damage response that may inhibit or enhance the viral replication process. An exciting recent discovery described the phosphorylation of histone H2AX by a viral encoded kinase, ORF36 from murine gammaherpesvirus 68 (MHV68) (Tarakanova et al., 2007). ORF36 is not-essential in fibroblasts, but important for viral replication in macrophages. The ORF36 orthologue in EBV, BGLF4, is known to phosphorylate the EBV replication DNA polymerase processivity factor BMRF1, which is functionally related to cellular PCNA. BGLF4 can also phosphorylate H2AX, as might the KSHV orthologue ORF36. BGLF4 has been shown to localize to viral replication compartments (Wang et al., 2005a) and have cdc2-like activity, which when ectopically expressed induces unscheduled cellular chromosome condensation and topoisomerase II decatenation activity (Lee et al., 2007). One hypothesis for the BGL4 kinase function is that it reorganizes the nuclear structure to favor viral lytic replication, perhaps by increasing the available internuclear space for viral replication machinery to assemble. The function of virus-induced H2AX phosphorylation is not completely clear, especially since it is not known if ORF36 phosphorylation of H2AX leads to a productive or aberrant DNA damage response. Viral modulation of the cellular DNA damage response during the lytic phase of infection may be necessary to accommodate the complex DNA structures and double strand breaks formed during the viral replication process (Fig. 2).
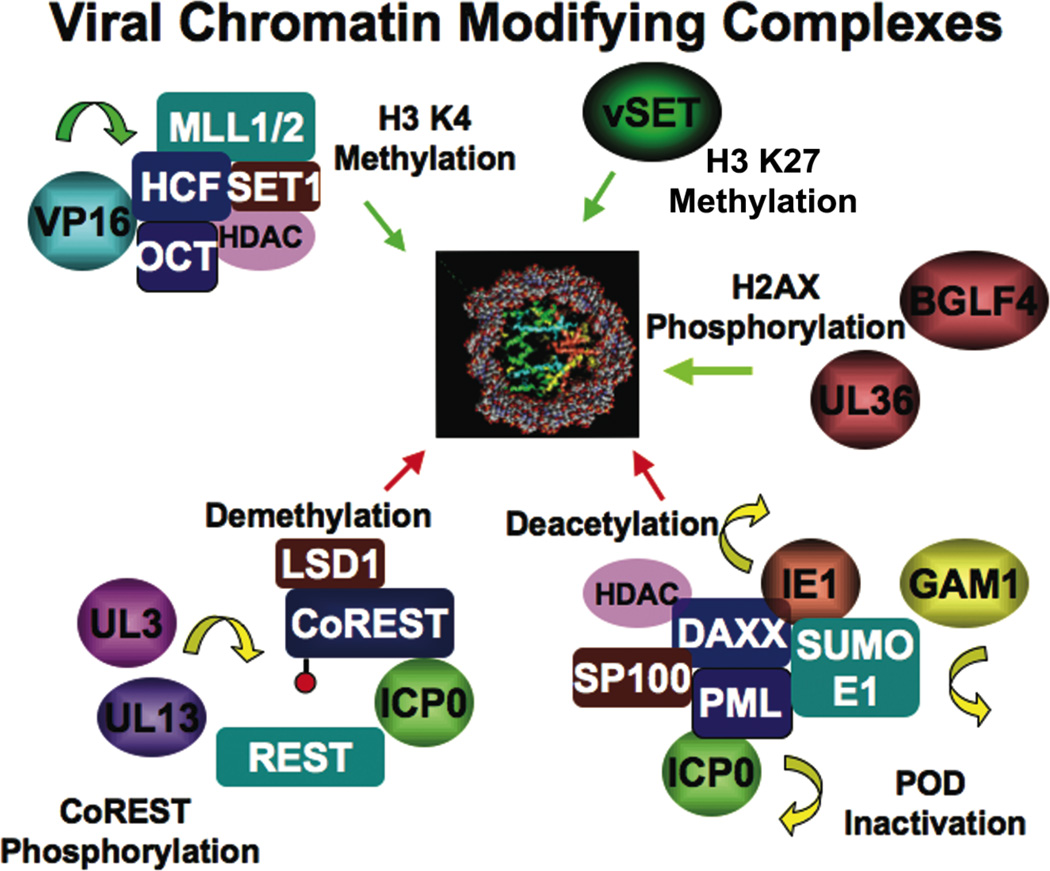
Figure 2. Viral Proteins that Effect Chromatin Modifications.
HSV VP16 assembles a histone H3 K4 methylation complex through its association with HCF. HSV UL3 and UL13 kinases have been implicated in phosphorylation of CoREST, a co-repressor of REST that can also associate with the histone demethylase LSD1. HSV ICP0 can also dissociate CoREST from REST to disrupt transcription reprepression. PBCV-1 virus encoded methyltransferase vSET1 can methylate histone H3 K27. Gammaherpesvirus kinases UL36 and BGLF4 can phosphorylate H2AX. Murine CMV IE1 can interact with DAXX and disrupt transcription repression through SUMO1 modification. Similarly, HSV ICP0 can interfere with PML SUMO1 modification, and avian adenovirus GAM1 can cause degradation of the SUMO1 E1 and E2 ligases.
PODS, Heterochromatin, and Innate Anti-Viral Defense
Once in the nucleus, many DNA and retroviruses localize to subnuclear structures, referred to as PODS or ND10s, because of they contain the promyeolocytic leukemia-associated (PML) protein (Maul, 1998). The PODs consist of a complex mixture of proteins, but are defined by the invariant presence of the PML and SP100. Many viruses encode proteins that remodel POD structure and interact with POD proteins, but the precise function of the PODS is not clear (Everett and Chelbi-Alix, 2007). PML has tumor suppressor activity and its translocation leads to several cancers. PML is a member of the TRIM family and contains a RING domain with E3 SUMO (small ubiquitin-like modifier) ligase activity. PML interacts with several viral proteins, including HSV ICP0, an E3 ubiquitin ligase, which facilitates the degradation of PML and the disruption of PODs. SP100 can function as a transcription repressor, and at least one isoform contains an HMG domain indicating that it has a non-specific DNA binding activity. Another major components of the PODs is the DAXX protein, which has transcriptional repression activity in the nucleus as a result of its interactions with numerous chromatin inhibitory complexes, including HDACS 1 and 2, and the heterochromatin associated protein ATRX (Lin et al., 2006). DAXX can also function in the cytoplasm as a FAS-associated death domain interacting protein that promotes apoptosis (Salomoni and Khelifi, 2006). DAXX transcription repression function is associated with its ability to mediate and bind SUMO (Small Ubiquitin-Related Modifier) modifications (Lin et al., 2006). The SUMO interacting domain (SIM) of DAXX was required for localization at PODs and at regions of condensed heterochromatin. SUMO modification has been detected on numerous transcription factors, and typically correlates with transcription repression functions. DAXX is targeted for degradation by the human cytomegalovirus (hCMV) pp71 protein (Everett and Chelbi-Alix, 2007; Hwang and Kalejta, 2007). Viruses lacking pp71 are repressed by histone deacetylation and condensed chromatin structure. Conversely, cells depleted in DAXX enhance CMV IE gene expression, especially in pp71-deleted strains. Thus, DAXX may provide an important mechanism for the formation of repressive chromatin structure on newly infecting or early replicating virus DNA. Like DAXX, other components of the POD, including PML and SP100 isoforms contribute to transcription repression and a more general antiviral response. Disruption of the POD structure by viral prorteins may be necessary to escape transcription repression and heterochromatinization.
The importance of SUMO1 modification in transcription repression is underscored by the activities of the avian adenovirus GAM1 protein. GAM1 is a viral encoded transcriptional activator that was found to promiscuously activate transcription by a general inhibition of HDAC activity (Chiocca et al., 2002). Subsequent characterization of GAM1 revealed that it also inhibits SUMO modification of PML and other substrates. The inactivation of SUMO modification was a consequence of the proteosome dependent-degradation of the SAE1/SAE2 (E1) and Ubc9 (E2) SUMO-conjugating enzymes. GAM1 was shown to form an E3-ubiquitin ligase complex with cellular proteins and target this complex to SAE1 for ubiquitin-mediated degradation (Boggio et al., 2007; Chiocca, 2007). A similar mechanism of transcription activation may be utilized by other promiscuous transactivators, especially the HSV ICP0, which also possesses E3 ubiquitin-ligase activity and causes PML degradation.
Viral Gene Expression and Chromatin Modification
Transcription activation of virus genes may have unique features. Episomal genomes may have different topology than cellular chromosomal DNA, and therefore may utilize different mechanisms for enhancer-promoter communication and chromatin-structure. Lytic DNA viruses, like herpes simplex and adenoviruses, may have genomes that are only partially assembled into chromatin or assembled into a chromatin in the absence of cellular DNA replication. Replication-independent chromatin assembly may involve variant histones and alternative histone chaperones from those used for replication-associated chromatin assembly. For these reasons, atypical chromatin-patterns may assemble on viral genomes as they enter a newly infected cell nucleus. The precise composition of the DNA templates of these lytic virus has been explored in a few model systems, and each virus may have different strategies to accomplish the goal of setting up a transciptionally active template. For adenovirus, the initial transcription events occur at the E1A promoter region, which is activated by multiple cellular transcription factors that may bind DNA that is not completely chromatinized. The chromatin structure of the E1A promoter region has not been investigated in detail for histone modifications or nucleosome organization. One possibility is that this region of the genome remains unchromatinized due to its location at the termini, and that viral tegument proteins prevent repressive chromatin structure from forming. As noted above, INHAT interacts with Ad protein VII on the viral genome during the early stages of transcription initiation. Production of E1A protein resets the cellular milieu in multiple ways to facilitate viral transcription and replication, including interactions with host histone acetylases CBP and p300. The fact that some of the viral genome is delayed for expression and requires E1A mediated histone activation and de-repression, suggests that these regions of the genome have been chromatinized, to a greater extent than the E1A promoter region. Interestingly, the major late promoter, which can be activated by E1A, associates with the MLTF/USF transcription factor, which has been implicated in chromatin boundary functions at the chicken beta-globin insulator locus (West et al., 2004). In this context, USF prevented the spread of repressive H3 mK9 into the euchromatic H3 mK4 region. It will be interesting to determine if USF provides this function to the MLP during adenovirus infection.
The chromatin structure of HSV DNA during the early stages of lytic infection has been investigated (Herrera and Triezenberg, 2004; Huang et al., 2006; Kent et al., 2004). Like adenovirus, the initial genome may associate only partially with nucleosomes, some of which may be regulated by the action of VP22 interactions with the INHAT complex. Transcription of the immediate early genes is largely dependent on the action of the VP16 virion component. Virus engineered with mutant VP16 were incapable of forming transcription complexes and recruiting histone modifying enzymes (Herrera and Triezenberg, 2004). During early stages of productive wt virus infection, the immediate early regions of HSV were enriched with trimethylated H3 lysine 4 (H3mK4). Small interfering RNA was used to demonstrate that this H3mK4 was Set1, but not Set7/9 dependent (Huang et al., 2006). Set1 is one of many proteins that can be recruited to the HSV genome by VP16. VP16 binds directly to the Host Cell Factor (HCF) which serves as a scaffold for recruiting a large histone modifying complex that includes, Set1, MLL1/2, and HDACS 1 and 2 (Julien and Herr, 2004; Narayanan et al., 2007; Yokoyama et al., 2004). MLL and SET1 have histone H3 K4 methylase activity, but it is not yet known whether these proteins have redundant or complementary activities. VP16 can also recruit transcription factors and coactivators, including histone acetyltransferases CBP and p300, and chromatin-remodeling factors, Swi/Snf. Thus, for HSV the establishment of euchromatin at the IE genes appears to be an essential step in the activation of immediate early genes (Fig. 3).
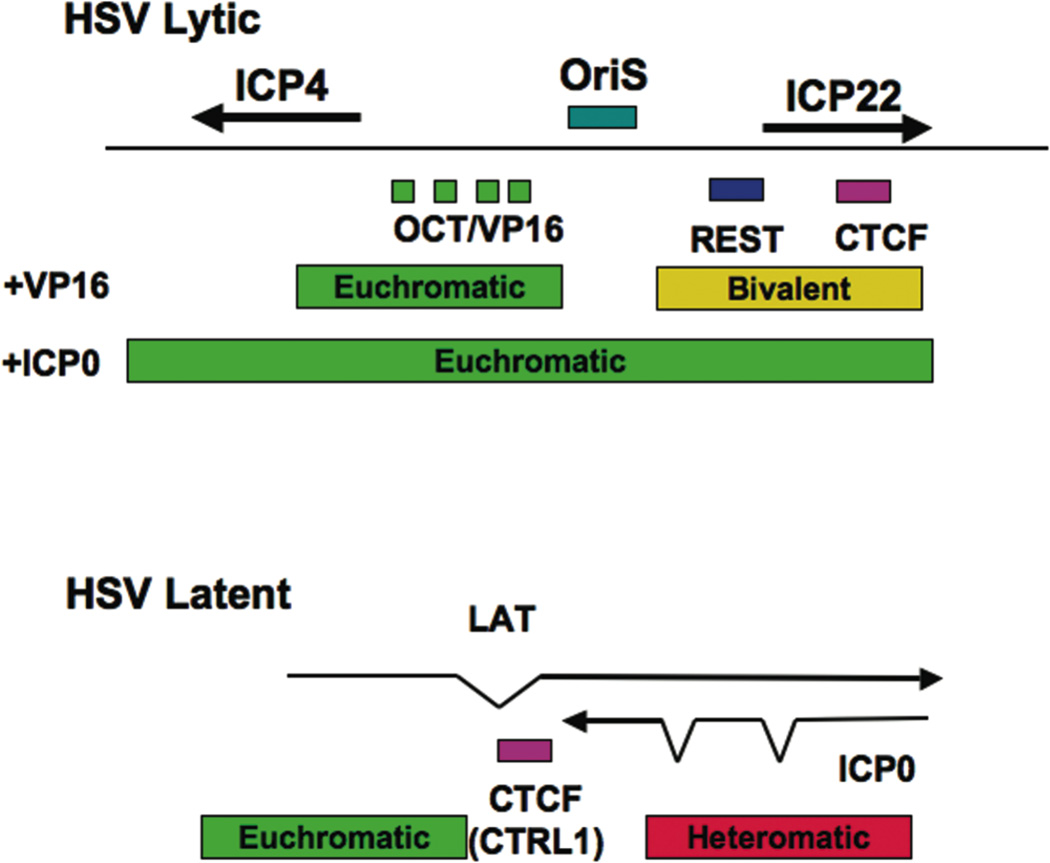
Figure 3. HSV Chromatin Organization.
Octamer/VP16 binding sites determine the gene expression pattern during lytic infection. The localization of REST and CTCF sites at the ICP22 location may help to limit the spread of euchromatin during the earliest stages of lytic cycle gene expression. During latent infection, CTCF binding sites within the LAT exon may help to control euchromatin formation at the ICP0 promoter to limit lytic reactivation.
Sequence specific-DNA binding complexes that can function as activators and repressors are also critical for modulating chromatin during viral infection. There are numerous examples of this level of gene regulation, which is in many ways similar to cellular gene regulation. The list of these activities among the DNA viruses is too extensive to review here, but there are a few new developments that are particularly notable because of their implications for chromatin organization. For example, the Repressor Element Silencing Transcription and Corepressor complex (REST-CoREST) was found to bind to the HSV1 immediate early promoter region located between the divergent promoter for ICP4 and ICP22 (Pinnoji et al., 2007). REST/CoREST bound to a specific site upstream of ICP4 and inhibited ICP4 transcription. This repression could be reversed by HDAC inhibitors. A more recent study revealed that HSV ICP0 can bind CoRest and displace it from its endogenous partner REST(Gu and Roizman, 2007). Additionally, HSV protein kinases UL3 and UL13 phosphorylate CoREST which facilitates its export from the nucleus (Gu et al., 2005). CoREST has been shown to associate with an HDAC and histone H3 K4 demethylase, LSD1/BCC110 (Lee et al., 2005). In the absence of ICP0, CoREST is predicted to prevent the euchromatinization of the ICP22 promoter, and of OriS, which is located adjacent to the REST binding site. The chromatin organizing potential of this region is further supported by the identification of a cluster of CTCF consensus sites located immediately downstream of the REST binding site. CTCF may insulate this region from other regions of the genome, but may also provide additional organizing functions including a DNA loop that allows long-distance communication with other regions of the genome, such as other immediate early control regions and the LAT promoter.
Reversible Epigenetic Silencing of Viral Genomes
Deferring gene expression and replication is a strategy that benefits many of the persistent and latent viruses. In this respect, they suppress or postpone viral gene expression until an opportune later occasion, perhaps during host-immune suppression or age-associated changes when the host environment is less hostile to virus infection. One of the best studies examples of this is the latent infections of HIV. A recent study has shown that HIV genomes integrate at locations in the cellular genome than are typically associated with euchromatin (Wang et al., 2007). This may represent the fact that integration events prefer sites where DNA is accessible. However, in the rare events where HIV establishes latent infection, the regions surrounding the genome are associated with heterochromatin-generating repressor proteins. An example of this has been studied in the murine leukemia virus (M-MLV), which has a restricted replication in embryonic carcinoma and embryonic stem cells due to a block at the level of viral mRNA transcription (Wolf and Goff, 2007). The cellular TRIM28 protein was found to bind to M-MLV LTR that is required for viral transcription control and assemble a transcription repressor and heterochromatin-forming complex (Wolf and Goff, 2007). TRIM28, the mouse orthologue of KAP1, is a transcriptional co-repressor that mediates the interactions between the KRAB zinc finger DNA binding proteins and several repressor complexes, including the NURD deacetylase complex, the histone H3 K9 methyltransferase ESET, and heterochromatin protein 1 (HP1). A similar role for TRIM28 in HIV transcription regulation has been reported (Eberhardy et al., 2006; Pengue et al., 1995).
Establishment of HIV latent infection has been an area of intense investigation. In model systems of LTR integration and long-term transcription repression, the heterochromatin proteins HP1γ and the histone H3 K9 methylase SUV39H1 were found to associate and functionally contribute to the stable repression of the LTR(du Chene et al., 2007). Earlier studies showed that YY1 and NF-kBp50 homodimers recruit HDAC1 to the LTR to repress transcription(Williams and Greene, 2005). Another study found c-myc and Sp1 recruited HDAC to the HIV LTR (Jiang et al., 2007). C-myc may also interact with YY1 to form a larger order repression complex at the HIV LTR(Stojanova et al., 2004). Other HDACs may also be involved in HIV latency, including the class III NAD-dependent deacetylase SIRT1. SIRT1 can bind and deacetylate HIV TAT protein, and this may reduce TAT interaction with TAR during the reactivation process (Pagans et al., 2005). It is not known whether the interaction of TAT with SIRT1 contributes to the formation of stable heterochromatin at the HIV LTR.
Lentiviral gene expression is of great interest in gene therapy. However, recent studies indicate that many lentivirus vectors, like those derived from HIV-1, suffer from a similar problem of transcription repression as has been observed with retrovirus genomes(Hofmann et al., 2006). In one third of the viral integrants, transcription was not detected and correlated with a high degree of CpG methylation. This finding is in contrast to a previous study that found that histone deacetylation was the primary mechanism of lentivirus repression (Pannell et al., 2000). The mechanism directing CpG methylation to the HIV LTR has not been investigated in detail, but there is evidence indicating that DNA methylation prevents transcription factor binding to the HIV LTR and promotes latent infection (Bednarik et al., 1990; Yedavalli and Jeang, 2007). Whether histone deacetylation and heterochromatin formation drives DNA methylation remains an important area of investigation.
The contribution of cellular and viral microRNAs in regulating viral replication and chromatin is also under intense investigation. A number of cellular microRNAs have been shown to restrict HIV replication in CD4 T-cells (Han and Siliciano, 2007). One model suggests that HIV-I TAR element is processed to yield a viral micro-RNA which functions in the chromatin-mediated repression of the viral LTR (Klase et al., 2007). In support of this model is the finding that the TAR binding protein (TRBP) recruits the Dicer complex for microRNA processing and for gene silencing (Chendrimada et al., 2005). The prototype for this mechanism of RNAi mediated heterochromatin formation comes from plant and S. pombe model systems (Buhler and Moazed, 2007). A similar model has been proposed for the role of a viral encoded microRNA in the formation of heterochromatin on the latent HSV genome (Wang et al., 2005b). Despite the many intriguing hints and early studies, there has been no clear demonstration that the viral RNA (microRNA, RNAi, or ncRNA) or their processing leads to heterochromatin formation in these viral model systems.
Chromatin Organization of Viral Genomes
For the larger DNA viruses that establish latent infection, it is imperative that they repress the majority of lytic replication, and express a limited set of latency-associated genes. The contribution of chromatin to the latent state has been studied in some detail among the herpesviruses, which have complex transcription patterns during latency. The chromatin regulation of these latent viruses may be most similar to the cellular chromosomes. However, the compact, episomal nature of the viral genome suggests that it may have unique features not commonly found at most cellular chromosomal regulatory elements. For EBV, histone modifications patterns share many properties with that of the cellular chromosome. Actively transcribed viral genes have euchromatic marks, including histone H3 and H4 acetylation, and H3 K4 trimethylation (Day et al., 2007). However, the EBV genomes were not enriched for stable repressive chromatin, like that associated with constitutive heterochromatin, and the formation of H3 mK9 modifications and HP1 binding was limited. H3 mK9 modifications were detected at some of the repetitive elements in the EBV genome (at IR1 or the W repeats) and at the terminal repeats of the KSHV genome (Sakakibara et al., 2004). As of yet, there is no evidence for histone modifications associated with polycomb group silencing through histone H3 K27 that is commonly observed at sights of variegation in developmental cell systems. To date, the methylases responsible for the K4 methylation on the EBV genome have not been identified (Fig. 4).
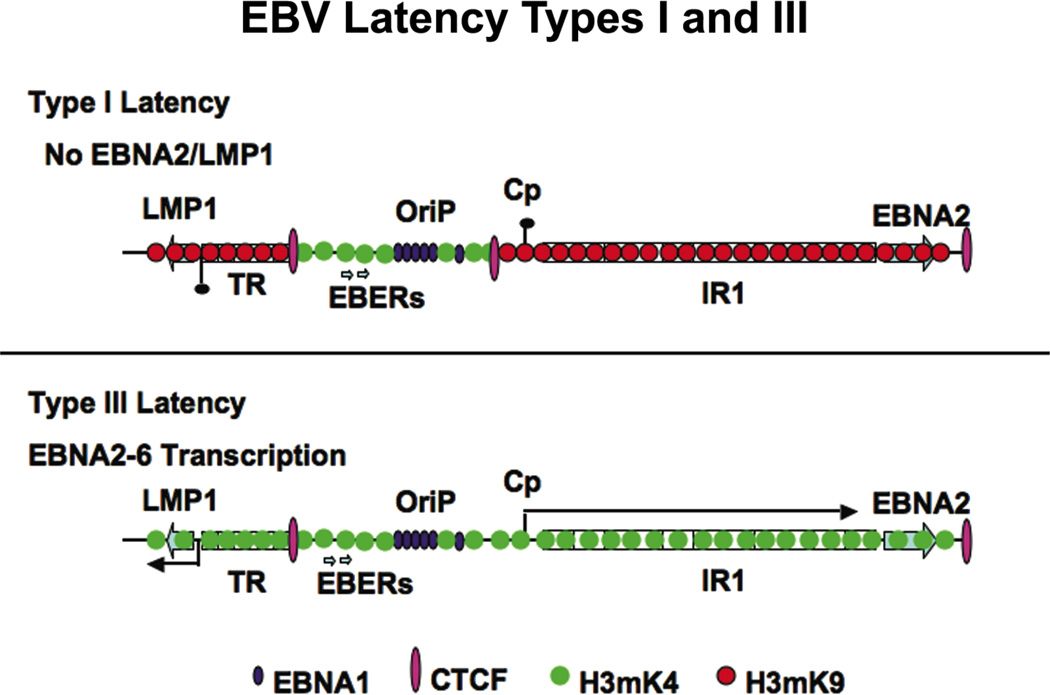
Figure 4. Chromatin Organization of EBV Latency Programs.
EBV type I latency is restricted by CTCF binding to the region between the OriP enhancer and the Cp control region for the multicistronic gene encoding EBNA-LP,-2, 3A,B,C. CTCF binding is reduced in type III latency and euchromatic marks of H3 K4 methylation are expanded through the Cp and LMP1 control regions.
Chromatin boundary factor CTCF binding sites were identified at several interesting positions on the EBV genome (Chau et al., 2006). One noteable position was the region upstream of the major latency promoter, Cp, which controls the multicistronic transcript for EBNA-LP, EBNA-2, EBNA-3A, -3B, -3C, and EBNA-1. Cp is regulated by several sequence specific factors, including the EBNA2 and Notch-responsive CBF1 protein. EBNA-1 bound to OriP is also required for the proper gene regulation of Cp (Puglielli et al., 1996), and viruses engineered with EBNA-1 mutations that disrupt transcription activity fail to immortalize primary B-lymphocytes (Altmann et al., 2006). Evidence suggests that CTCF regulates the interactions between the EBNA1/OriP enhancer and Cp, but it was not clear that this regulation was strictly inhibitory(Chau et al., 2006). In type I latency, where Cp transcription is repressed, the CTCF binding was 3–4 fold greater than that detected in type III latency, where Cp is actively transcribed. The CTCF binding correlated with a decreased histone H3 K4 methylation pattern. A correlation between CTCF binding and elevated H3 K4 methylation was noted in a mammalian genome-wide analysis of CTCF and H3 K4 methylation sites, suggesting that CTCF may regulate the pattern of histone H3 K4 methylation(Barski et al., 2007).
For HSV, it is known that the latency transcript, LAT, is expressed in latently infected neurons, but that surrounding lytic regulatory proteins, including the transcription activators IE110/ICP0 and IE174/ICP4, are strictly repressed. The LAT transcript is now known to encode a viral microRNA which provides a cellular survival function by inhibiting the pro-apoptotic TGF-β protein expression (Gupta et al., 2006). It is not known if LAT microRNA or other non-coding regions contribute to the establishment or maintenance of viral heterochromatin that may be envisioned to repress lytic cycle activating proteins (e.g ICP0). It is known that the LAT promoter and enhancers are insulated from the ICP0 and other immediate early gene promoters through the cellular chromatin boundary factor, CTCF (Amelio et al., 2006). The CTCF site is positioned between the downstream enhancer and the control locus for immediate early gene activation, and has been shown to provide insulator activity in a reconstituted assay in Drosophila Sf9 cells (Chen et al., 2007). It is possible that CTCF provides additional functions to the latent genome, including organization of the chromosome to maintain heterochromatin-like histone modifications throughout the immediate early locus.
Genome Maintenance and Chromosomal Passengers
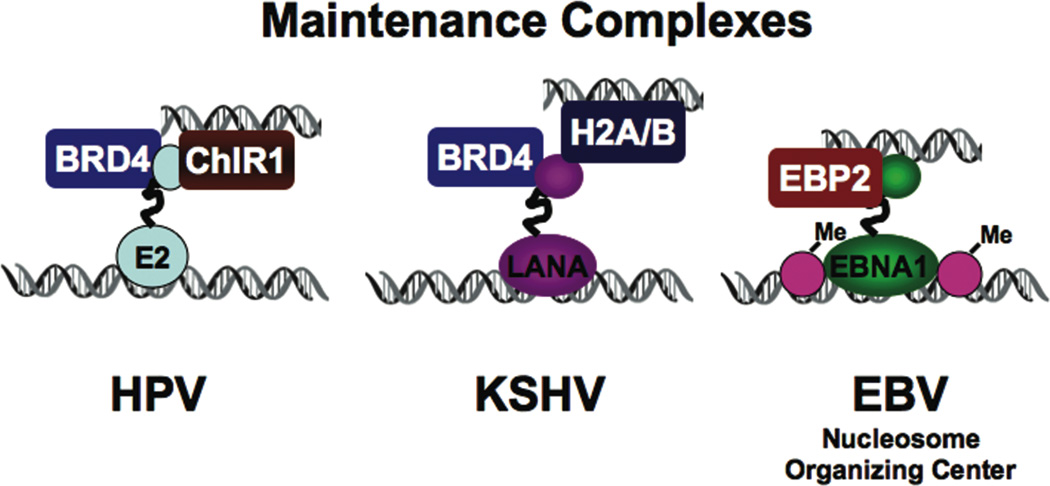
Episomal DNA viruses, like the latent forms of EBV, KSHV, and HPV, may require a chromatin organization that is compatible with passage through cellular mitosis. The viral episomes form non-integrated mini-chromosomes that tether non-covalently to metaphase chromosomes. Superficially, these episomal viruses use a very similar mechanism and viral encoded protein that tethers the viral genome to host-chromatin. The HPV E2, EBV EBNA1, and KSHV LANA proteins share both structural and functional features in their DNA binding and chromatin-attachment activities. However, the precise mechanism for chromosome tethering remains controversial since several different candidate targets have been identified for each virus. BRD4 was identified as an E2-associated protein using biochemical purification of affinity tagged E2 (Wu et al., 2006; You et al., 2004). A BRD4 peptide bound to E2 metaphase chromosome attachment domain was solved by X-ray crystallography (Abbate et al., 2006). Moreover, the BRD4 peptide was capable of blocking E2 attachment to metaphase chromosomes, indicating that it is either the metaphase chromosome receptor for E2 or binds to an E2 domain that overlaps the metaphase chromosome receptor. However, BRD4 may not be the whole story for E2 chromosome attachment. Another cellular protein, ChlR1, has been implicated in the chromosome attachment and segregation process (Parish et al., 2006a). ChlR1 has DNA helicase motifs and was found to function in cellular sister-chromatid cohesion (Parish et al., 2006b). Point mutations in E2 that distinguish segregation from transcription function, suggest that BRD4 binding correlates better with transcription function, more than segregation (McPhillips et al., 2006; Senechal et al., 2007). Consistent with this, BRD4 inhibits E2 transcription activity in vitro and in vivo (Wu et al., 2006). BRD4 may have a common chromatin-based function that contributes to the E2 transcription activation, repression, and chromosome segregation functions, depending on the particular the assay system. BRD4 and its related paralogue, BRD2 (Ring3), bind to the KSHV LANA protein, which has similarly complex functions as does E2, in transcription activation, repression, and chromosome segregation (Mattsson et al., 2002; Ottinger et al., 2006; Viejo-Borbolla et al., 2005; You et al., 2006). LANA can also interact with histones H2A/B (Barbera et al., 2006), histone H1 (Cotter and Robertson, 1999), and heterochromatin protein HP1 (Lim et al., 2003). The EBV functional paralogue, EBNA1, does not interact with BRD2 or BRD4. Rather, EBNA1 interacts with the nucleolar protein EBP2 which mediates its metaphase chromosome attachment and segregation function in mammalian and yeast model systems (Kapoor et al., 2005). The metaphase chromosome attachment domain of EBNA1 also possesses AT-hook DNA binding activity suggesting an alternative model where EBAN1 can bind AT-rich DNA on the metaphase chromosome without an accessory cellular protein receptor (Sears et al., 2004). Since the attachment to the metaphase chromosome may involve multiple steps, it is possible that all of these mechanisms have physiological relevance (Fig. 5).
Figure 5. Chromatin Targets of Episome Maintenance Factors.
HPV E2, EBV EBNA1, and KSHV LANA share functional and structural features necessary for episomal maintenance during latent infections. E2 and LANA target cellular BRD2/4, as well as ChLR1 and histone H2A/B, respectively, while EBNA1 interacts with EBP2 and binds AT-rich DNA directly. EBNA1 has been implicated as a chromatin organizing factor at OriP where nucleosomes are positioned and methylated at H3 K4.
Another feature of these episome maintenance proteins is their ability to organize viral chromatin. EBNA1 is known to displace histone binding at the family of repeats in EBV OriP (Avolio-Hunter and Frappier, 2003). Furthermore, the nucleosomes positioned adjacent to the EBNA1 binding sites in the Dyad Symmetry region undergo cell cycle acetylation and deacetylation corresponding to the activation of the DNA replication origin (Zhou et al., 2005). The region surrounding OriP is enriched in histone H3 K4 methylation, suggesting that EBNA1 may contribute to the euchromatinization of OriP (Day et al., 2007). In contrast, LANA binds to the terminal repeats (TR) of KSHV and recruits heterochromatic protein HP1 to the TR. Despite that enrichment of H3 K9 methylation at the flanks of the TR, the region immediately surrounding the LANA binding sites remain elevated in histone H3 K4 methhylation and K3 K9 acetylation (Stedman et al., 2004). How LANA maintains the origin initiation sites as a euchromatic region, and the surrounding TR as heterochromatic is not known, and may be complicated by heterogeneity among the different 800 bp repeats at the KSHV termini.
Viral Packaging and Epigenetic Memory
Are any epigenetic marks or memories transmitted to the next viral generation? Among the latent viral genomes (e.g. EBV or HIV), which do not produce infectious virus particles, DNA methylation and metaphase chromosome attachment proteins, which remain bound throughout multiple cell generations, may function as epigenetic memories that maintain a stable chromatin state and gene expression pattern. This process is most similar to that observed at many cellular chromosomal sites. In contrast, productive infections would require an epigenetic memory maintained within the newly assembly infectious virus particle. It has been well-established that histones and other chromatin-associated proteins assemble with SV40 genomes during virion packaging, and the newly infected virus comes pre-packaged in chromatin. Numerous cellular proteins associated with viral DNA or RNA have been identified in virus particles. Histone H2A was found in mCMV virions, although no histones were identified in proteomic analysis of most other herpesvirus virions, including EBV, KSHV, HSV, and CMV(Maxwell and Frappier, 2007). Retroviral genomes were also found to contain cellular proteins typically associated with template protection or preparation. For the RNA retrovirus genomes, these included the RNA binding proteins polyadenylate-binding protein-1 (PABP) and Poly (rc) binding protein (hnRNP-E1), as well as the DNA binding protein histone H4 (Segura et al., 2007). Other types of epigenetic marks may be transmitted in the viral genomes, such as sites of DNA methylation or modified bases. In CMV, there is evidence that an RNA-DNA hybrid exists in the virion DNA that marks the origin of DNA replication (Prichard et al., 1998). Structural RNAs and RNA editing enzymes are also known to be incorporated into the HIV virion, and these molecules may also transmit epigenetic content essential for establishing successful gene expression programs and productive infections of new host cells.
Concluding Remarks and Future Directions
A great deal of interest has arisen over the nature of epigenetic marks, and their contribution to genome organization and control of gene expression. Viruses establish stable genetic programs and can modulate cellular genetic programs to accommodate infection. To what extent viruses employ epigenetic marks similar or distinct from the cellular host remains an important avenue of future investigation. Recent data clearly indicates that viruses encode enzymes with capacity to modify histones and other chromatin associated proteins. These include the PBCV-1 virus encoded methyltransferase vSET1, which can methylate histone H3 K27, and the MHV68 virus encoded ORF35 kinase capable of phosphorylating H2AX. And it is likely that future investigation of these viral genes will reveal novel epigenetic marks, including those involving nucleotide editing and modifying enzymes, and non-coding RNAs and microRNAs.
Another important area of future investigation will be understanding the role of chromatin and DNA damage response proteins in restricting viral infection. PODs and other chromatin organizing machines may play a significant role in preventing infectious DNA and RNA from being transcribed and replicated. The mechanisms through which anti-viral heterochromatin is established and by which viruses inactivate this process will reveal important insights into the host-innate immune mechanisms against viruses, and may provide new strategies for the next generation of viral-mediated gene-therapy.
The role of chromatin in the organization of latent viruses may help to provide insight into genome stability in general. Viruses, although smaller than cellular chromosomes, confront many of the same challenges as the cellular chromosome. How viruses manage these tasks in the absence of canonical centromeres and telomeres, and sometimes lacking dedicated origins of DNA replication, will be important to define in the near future. The chromatin boundary factor CTCF has emerged as a central organizer of gene expression, but its precise biochemical function, if other than structural DNA binding protein, remains largely unexplored. A recent study suggests that CTCF may colocalize with cohesins, suggesting that chromatin boundaries are linked to regions of the genome involved in sister-chromatid or other inter-chromosomal interactions (Stedman et al., EMBO J., in press). How these larger order structures of chromosomes establish stable gene expression patterns and contribute to genome stability will be important to elucidate in the near future. Finally, the mechanisms of viral DNA condensation and decondensation during the virion packaging and unpackaging may provide important insights into the basic cellular mechanisms of chromosome dynamics and genome stability.
Acknowledgements
I apologize to my colleagues for the many relevant studies that were not sited due to space limitations. PML is funded by grants from the NIH (CA085678 and CA93606)
References
- Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol Cell. 2006;24(6):877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Altmann M, Pich D, Ruiss R, Wang J, Sugden B, Hammerschmidt W. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc Natl Acad Sci U S A. 2006;103(38):14188–14193. doi: 10.1073/pnas.0605985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio AL, McAnany PK, Bloom DC. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol. 2006;80(5):2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avolio-Hunter TM, Frappier L. EBNA1 efficiently assembles on chromatin containing the Epstein-Barr virus latent origin of replication. Virology. 2003;315(2):398–408. doi: 10.1016/s0042-6822(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bednarik DP, Cook JA, Pitha PM. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. Embo J. 1990;9(4):1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282(21):15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14(11):1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Chau CM, Zhang XY, McMahon SB, Lieberman PM. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol. 2006;80(12):5723–5732. doi: 10.1128/JVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Huang J, Berger SL, Zhou J. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol. 2007;81(10):5192–5201. doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocca S. Viral control of the SUMO pathway: Gam1, a model system. Biochem Soc Trans. 2007;35(Pt 6):1419–1421. doi: 10.1042/BST0351419. [DOI] [PubMed] [Google Scholar]
- Chiocca S, Kurtev V, Colombo R, Boggio R, Sciurpi MT, Brosch G, Seiser C, Draetta GF, Cotten M. Histone deacetylase 1 inactivation by an adenovirus early gene product. Curr Biol. 2002;12(7):594–598. doi: 10.1016/s0960-9822(02)00720-0. [DOI] [PubMed] [Google Scholar]
- Cotter MA, 2nd, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi's sarcoma- associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells [In Process Citation] Virology. 1999;264(2):254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368(2):219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day L, Chau CM, Nebozhyn M, Rennenkamp AJ, Showe M, Lieberman PM. Chromatin Profiling Of Epstein-Barr Virus Latency Control Region. J Virol. 2007 doi: 10.1128/JVI.02172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. Embo J. 2007;26(2):424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Goncalves J, Coelho S, Segal DJ, Berkhout B, Barbas CF., 3rd Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol. 2006;80(6):2873–2883. doi: 10.1128/JVI.80.6.2873-2883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89(6–7):819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29(1):49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci U S A. 2005;102(21):7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci U S A. 2007;104(43):17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442(7098):82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- Han Y, Siliciano RF. Keeping quiet: microRNAs in HIV-1 latency. Nat Med. 2007;13(10):1138–1140. doi: 10.1038/nm1007-1138. [DOI] [PubMed] [Google Scholar]
- Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J Virol. 2006;80(2):794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9(1):15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78(18):9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley CE, Davidson AD, Matthews DA. Relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J Gen Virol. 2007;88(Pt 12):3244–3248. doi: 10.1099/vir.0.83196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, Pfeifer A. Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther. 2006;13(1):59–66. doi: 10.1016/j.ymthe.2005.07.685. [DOI] [PubMed] [Google Scholar]
- Huang J, Kent JR, Placek B, Whelan KA, Hollow CM, Zeng PY, Fraser NW, Berger SL. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol. 2006;80(12):5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kalejta RF. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology. 2007;367(2):334–338. doi: 10.1016/j.virol.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81(20):10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14(6):713–725. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Kapoor P, Lavoie BD, Frappier L. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol Cell Biol. 2005;25(12):4934–4945. doi: 10.1128/MCB.25.12.4934-4945.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JR, Zeng PY, Atanasiu D, Gardner J, Fraser NW, Berger SL. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J Virol. 2004;78(18):10178–10186. doi: 10.1128/JVI.78.18.10178-10186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC molecular biology. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Chen JY, Wang JT, Kimura K, Takemoto A, Lu CC, Chen MR. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J Virol. 2007;81(10):5166–5180. doi: 10.1128/JVI.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Lim C, Lee D, Seo T, Choi C, Choe J. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J Biol Chem. 2003;278(9):7397–7405. doi: 10.1074/jbc.M211912200. [DOI] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24(3):341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Matthews DA. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J Virol. 2001;75(2):1031–1038. doi: 10.1128/JVI.75.2.1031-1038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson K, Kiss C, Platt GM, Simpson GR, Kashuba E, Klein G, Schulz TF, Szekely L. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J Gen Virol. 2002;83(Pt 1):179–188. doi: 10.1099/0022-1317-83-1-179. [DOI] [PubMed] [Google Scholar]
- Maul GG. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Maxwell KL, Frappier L. Viral proteomics. Microbiol Mol Biol Rev. 2007;71(2):398–411. doi: 10.1128/MMBR.00042-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol. 2006;80(19):9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104(26):10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol. 2006;80(21):10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, Hruby H, Jung M, Verdin E, Ott M. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS biology. 2005;3(2):e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz HD, Ellis J. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. Embo J. 2000;19(21):5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish JL, Bean AM, Park RB, Androphy EJ. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol Cell. 2006a;24(6):867–876. doi: 10.1016/j.molcel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci. 2006b;119(Pt 23):4857–4865. doi: 10.1242/jcs.03262. [DOI] [PubMed] [Google Scholar]
- Pengue G, Caputo A, Rossi C, Barbanti-Brodano G, Lania L. Transcriptional silencing of human immunodeficiency virus type 1 long terminal repeat-driven gene expression by the Kruppel-associated box repressor domain targeted to the transactivating response element. J Virol. 1995;69(10):6577–6580. doi: 10.1128/jvi.69.10.6577-6580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnoji RC, Bedadala GR, George B, Holland TC, Hill JM, Hsia SC. Repressor element-1 silencing transcription factor/neuronal restrictive silencer factor (REST/NRSF) can regulate HSV-1 immediate-early transcription via histone modification. Virology journal. 2007;4:56. doi: 10.1186/1743-422X-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Jairath S, Penfold ME, St Jeor S, Bohlman MC, Pari GS. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J Virol. 1998;72(9):6997–7004. doi: 10.1128/jvi.72.9.6997-7004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglielli MT, Woisetschlaeger M, Speck SH. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70(9):5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotobe D, Tardy JC, Andre P, Hong SS, Darlix JL, Boulanger P. Human Polycomb group EED protein negatively affects HIV-1 assembly and release. Retrovirology. 2007;4:37. doi: 10.1186/1742-4690-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S, Ueda K, Nishimura K, Do E, Ohsaki E, Okuno T, Yamanishi K. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J Virol. 2004;78(14):7299–7310. doi: 10.1128/JVI.78.14.7299-7310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Khelifi AF. Daxx: death or survival protein? Trends in cell biology. 2006;16(2):97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Samad MA, Okuwaki M, Haruki H, Nagata K. Physical and functional interaction between a nucleolar protein nucleophosmin/B23 and adenovirus basic core proteins. FEBS Lett. 2007;581(17):3283–3288. doi: 10.1016/j.febslet.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279(23):23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78(21):11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MM, Garnier A, Di Falco MR, Whissell G, Meneses-Acosta A, Arcand N, Kamen A. Identification of host proteins associated to retroviral vector particles by proteomic analysis of highly purified vector preparations. J Virol. 2007 doi: 10.1128/JVI.01909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology. 2007;358(1):10–17. doi: 10.1016/j.virol.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277(16):14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Spector DJ. Default assembly of early adenovirus chromatin. Virology. 2007;359(1):116–125. doi: 10.1016/j.virol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Stedman W, Deng Z, Lu F, Lieberman PM. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol. 2004;78(22):12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova A, Caro C, Jarjour RJ, Oster SK, Penn LZ, Germinario RJ. Repression of the human immunodeficiency virus type-1 long terminal repeat by the c-Myc oncoprotein. Journal of cellular biochemistry. 2004;92(2):400–413. doi: 10.1002/jcb.20065. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nature reviews. 2007;5(3):187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- Tarakanova VL, Leung-Pineda V, Hwang S, Yang CW, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HWt. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell host & microbe. 2007;1(4):275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34(5):1653–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen H, Okuwaki M, Hong R, Chakravarti D, Nagata K, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J Gen Virol. 2003;84(Pt 9):2501–2510. doi: 10.1099/vir.0.19326-0. [DOI] [PubMed] [Google Scholar]
- Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J Virol. 2005;79(21):13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violot S, Hong SS, Rakotobe D, Petit C, Gay B, Moreau K, Billaud G, Priet S, Sire J, Schwartz O, Mouscadet JF, Boulanger P. The human polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J Virol. 2003;77(23):12507–12522. doi: 10.1128/JVI.77.23.12507-12522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Yang PW, Lee CP, Han CH, Tsai CH, Chen MR. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J Gen Virol. 2005a;86(Pt 12):3215–3225. doi: 10.1099/vir.0.81313-0. [DOI] [PubMed] [Google Scholar]
- Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005b;102(44):16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16(3):453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Williams SA, Greene WC. Host factors regulating post-integration latency of HIV. Trends Microbiol. 2005;13(4):137–139. doi: 10.1016/j.tim.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131(1):46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20(17):2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Johnson JS, Ornelles DA, Lieberman J, Engel DA. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J Virol. 2005;79(4):2474–2483. doi: 10.1128/JVI.79.4.2474-2483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VR, Jeang KT. Methylation: a regulator of HIV-1 replication? Retrovirology. 2007;4:9. doi: 10.1186/1742-4690-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117(3):349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, Howley PM. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol. 2006;80(18):8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman PM. Cell cycle regulation of chromatin at an origin of DNA replication. Embo J. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]