Abstract
Human mesenchymal stem cells (hMSC) have numerous potential advantages over terminally differentiated cells and embryonic stem cells for use in tissue engineering applications. The aims of this study were to develop methods to test the hypothesis that hMSC could be differentiated using cyclic compressive strain alone. hMSC were successfully isolated, purified using D7-FIB antibody, cloned, and characterized. The cells were subsequently analyzed using fluorescence-activated cell sorting using a panel of antibodies and differentiation into multiple cell lineages. D7FIB-positive cells were then seeded into collagen–alginate scaffolds and subjected to 10% or 15% cyclic compressive strain for 4 out of 24 hours for up to 21 days in a bespoke servo-assisted displacement-controlled device. Cells were analyzed using adenosine triphosphate assay to determine cell number, live–dead cell assay, and quantitative real-time polymerase chain reaction at 7 and 21 days. Cloned D7-FIB-positive hMSCs showed evidence of differentiation to an osteogenic lineage under 10% cyclic compressive strain alone (core binding factor alpha 1 (CBFA-1) was significantly upregulated at 7 and 21 days by a factor of 18.3 and 32.2, respectively) and to an osteo-chondrogenic lineage under 15% cyclic compressive strain alone (increased expression of CBFA-1, Sox9, and aggrecan). A combination of a composite viscoelastic scaffold and controlled cyclic compressive strain may be useful for study of the differentiation of MSC.
Introduction
In recent years, mesenchymal stem cells (MSCs) have shown considerable promise as an adaptable cell source for use in tissue engineering and other therapeutic applications. They have advantages over terminally differentiated cells in that the cells have been shown to be capable of differentiation in vitro into multiple cell lineages.1–3 MSC are straightforward to isolate and culture, and autologous cells can be harvested from the intended recipient and, therefore, not constrained by immunological problems. Nevertheless, although MSCs can be readily differentiated using specific biochemical supplements, little is known about the effects of physical stimulation alone on the differentiation of these cells.
Mechanical stimulation is an essential regulator of tissue homeostasis and is indispensable for the normal function of connective tissues. In 1980, Pauwels4 proposed that physical factors cause stress and deformation of mesenchymal cells and that these stimuli could determine cell differentiation pathways. Depending on the magnitude, direction, and distribution of mechanical forces, cells can respond in a variety of ways. For example, the stretching of cells attached to a substrate can alter cell motility and orientation.5 Moreover, the mechanical compression of cells, such as chondrocytes, has been shown to modulate proteoglycan synthesis.6 It has also been shown that mechanical loading stimulates bone formation by acting alone or in combination with hormones such as parathyroid hormone or estrogen on bone cells.7 Other reports have suggested that osteoblasts can be directly activated by loading, leading to an increase in proliferation and matrix protein synthesis, in addition to being activated by growth factors and by prostaglandins and nitric oxide released by osteocytes.7 Fluid shear acting on endothelial cells has been shown to activate hormone release and intracellular calcium signalling and to stiffen cells by inducing rearrangement of the cytoskeleton.8 Based on such evidence, it is now recognized that mechanical stimulation of cells plays an important role in blood pressure regulation, the response of the vasculature to shear stress, bone remodelling, and the maintenance of muscle and perception of touch and sound.9 Mechanical strain has also been shown to stimulate differentiation and structural alignment of MSC cultures.10 Specifically, there have been reports demonstrating that equiaxial strain promotes differentiation of MSC into osteoblastic cells in osteogenic media11 and that translational and rotational strain can increase the expression of collagen I and III and tenascin-C in bovine bone marrow cell cultures in collagen gels with ascorbate.12
The aim of this study was to test the hypothesis that cyclic compressive strain alone would influence the differentiation of human MSC (hMSC). Cloned hMSC were seeded into viscoelastic collagen–alginate scaffolds that were then cultured statically or dynamically without the use of additional biochemical cues to assess the effects of cyclic compressive strain on their functional differentiation over a 21-day period. Differentiation was assessed using quantitative real-time polymerase chain reaction (Q-PCR) for peroxisome proliferator-activated receptor gamma (PPAR-γ), core binding factor alpha-1 (CBFA-1), Sox9, and aggrecan.
Materials and Methods
Isolation and culture of hMSC
Human bone marrow was obtained as waste material from hip replacement surgeries at the Leeds Teaching Hospitals Trust, Leeds General Infirmary (Leeds, United Kingdom), after the acquisition of informed written consent of patients and approval from the local ethics committee. hMSC were separated using a Percoll gradient (22.05 mL of Percoll, 2.45 mL of 1.5M sodium chloride (NaCl), and 10.5 mL of Tyrode's balanced salt solution, all Sigma-Aldrich), centrifuged at 20,000 g for 15 minutes, and the cells were cultured in complete medium (Dulbecco's modified Eagle medium (DMEM) supplemented with 10 % (v/v) fetal calf serum (FCS), 100 U/mL of penicillin, 100 mg/mL of streptomycin, and 2mM L-glutamine; all Invitrogen) under standard conditions (37°C, 5 % carbon dioxide (CO2) (v/v) in air).
Selection of D7-FIB-positive hMSC
hMSC from two donors (73- and 55-year-old women) were allowed to grow for three passages. hMSC were magnetically labelled with antifibroblast microbeads (D7-FIB, Miltenyi Biotech) and separated on a magnetic activated cell sorting (MACS, Miltenyi Biotech) column according to the manufacturer instructions. The cells were then cultured under standard conditions. hMSC from the 73-year-old donor were analyzed by fluorescence activated cell sorting (FACS) at passages 5 and 11. hMSC from the 55-year-old donor were cloned and used in the differentiation assays (passage 10) and the mechanical loading experiments (passage 8).
Cloning of D7-FIB+hMSC
Once a near-confluent cell layer was obtained (∼70% confluent), approximately 30 cells were reseeded into each of twenty 60-mm tissue culture–treated dishes and cultured for 24 hours. Individual cells were isolated using a cloning ring then trypsinized and cultured in conditioned medium (waste medium removed while growing D7-FIB-positive hMSC from passages 0 to 3, mixed 50:50 with fresh complete culture medium). Culture medium was replaced three times per week, and once the clones were approximately 70% confluent, the cells were trypsinized and transferred to tissue culture plastic of ever-increasing surface area.
FACS analysis
Anticluster of differentiation-13 (CD13), anti-CD90, anti-CD105, anti-D7-FIB, anti-CD45, anti-human leukocyte antigen DR (HLA-DR), anti-CD34, and anti-CD117 antibodies were directly conjugated to phycoerythrin (PE). Anti-alkaline phosphatase and anti-osteocalcin, were not conjugated and were visualized using a polyclonal rabbit antimouse immunoglobulin (IG) control directly conjugated to PE. Anti-IgG1 and anti-IgG2a were used as controls. Anti-IgG1 and anti-IgG2a were obtained from DAKO, all other antibodies were obtained from Chemicon. Tris-buffered saline was used throughout as diluent and wash buffer (50mM Tris pH 7.6, 1.5M NaCl), and all incubations were performed at 4°C. Cells were incubated sequentially in primary antibody for 15 minutes and secondary antibody for a further 15 minutes, where required, with washing between each step. For each antibody, 10,000 cells were counted and analyzed for the presence of the respective markers on the F2 channel of a Beckman Coulter EPICS XL-MCL model FACS machine. The cells were gated to exclude cell clumps, fragments, and other artefacts.
MSC differentiation
hMSC were differentiated into adipogenic, myogenic, neurogenic, and chondrogenic lineages (n=4 cultures per assay). For adipogenic differentiation, confluent hMSC monolayers were fed with adipogenic medium (50 mL of adipogenic differentiation supplement added to 450 mL of human Mesencult medium; Stem Cell Technologies) for up to 4 weeks. Accumulation of lipid vacuoles was visualized using 0.5% (v/v) Oil Red O, as described previously.1 The method for myogenic differentiation was derived from that of Wakitani et al..13 Briefly, cells were fed with complete medium supplemented with 10μM 5-azacytidine for 24 hours, which was subsequently replaced with complete medium, and the cells were cultured for up to 4 weeks. Differentiation was assessed using immunofluorescent labelling using monoclonal antibodies to heavy-chain myosin–slow (HCMs) and desmin (Vector Laboratories). The method for neurogenic differentiation was derived from that of Woodbury et al.14 Briefly, cells were fed with induction medium (complete medium containing 20% (v/v) FCS supplemented with 1mM β-mercaptoethanol) for 24 hours followed by differentiation medium (serum-free DMEM supplemented with 10mM 2-mercaptoethanol) for up to 8 hours. Differentiation was assessed according to immunofluorescent evaluation using monoclonal antibodies to tau and neuron-specific enolase (NSE; both Vector Laboratories). Each marker is thought to be expressed at different time points during neurogenic differentiation, so positive labelling with either marker would indicate successful neurogenic differentiation. For chondrogenic differentiation,15 cells were cultured in chondrogenic medium consisting of serum-free medium supplemented with 100 μg/mL of sodium pyruvate, 40 μg/mL of L-proline, 50 μg/mL of L-ascorbic acid-2-phosphate, 1 mg/mL of bovine serum albumin, 1×insulin-transferrin-sodium selenite, 100nM dexamethasone (all Sigma-Aldrich) and 10 ng/mL of transforming growth factor beta-3 (R&D Systems) for up to 4 weeks in a polypropylene tube with a loosened cap to allow gaseous exchange. Cell pellets were harvested at 2 and 4 weeks and cryo-embedded for frozen sectioning, toluidine blue staining, and immunofluorescent evaluation using an antibody to collagen II (Sigma-Aldrich).
Immunofluorescent labelling of differentiated cells
Monoclonal antibodies to HCMs, tau, NSE (all Vector Laboratories), and collagen II (Sigma-Aldrich) were used to identify biochemical differentiation of hMSC as outlined above. Immunofluorescent labelling was performed using an indirect method. Tris-buffered saline was used throughout as diluent and wash buffer, and all incubations were performed at ambient temperature. Sections were incubated sequentially in primary antibody for 1 hour and fluorescein isothiocyanate –labelled goat antimouse secondary antibody (Molecular Probes) for 30 minutes, with washing between each step. Slides were air-dried and mounted using 1,4- diazobicyclo-(2,2,2)-octane (DABCO) –glycerol solution (9:1 solution glycerol (VWR International):2.5% (w/v) DABCO (Sigma-Aldrich) in 0.1M sodium bicarbonate, pH 9.0). Isotype control antibodies (IgG1 and IgG2a; Dako) were used to verify the specificity of the antibodies. Omission of the primary antibody served as a negative control.
Preparation of collagen sponges
Collagen I (2.8% w/v) was obtained from rat tails using acid-solubilized extraction. Briefly, rat tails were obtained from Lewis strain rats killed by schedule 1 as surplus to requirements in Central Biomedical Services, University of Leeds. The skin was removed under aseptic conditions and discarded. Tendon bundles were excised and finely minced under ethanol and transferred to a 500-mL glass bottle containing 17.4mM acetic acid and stirred at 4°C for 48 hours. The solution was centrifuged at 30,000 g at 4°C for 60 minutes before the supernatant was removed, the volume determined, and the solution neutralized with 0.1M sodium hydroxide. The neutralised solution was stirred at 4 °C for 90 min and further centrifuged at 15,000 g at 4°C for 30 minutes. The pellet was then resuspended in 17.4mM acetic acid. The sterility of the collagen solution was checked using standard microbiological plating. Ice-cold collagen solution; cold, buffered Hank's balanced salt solution (Invitrogen); and cold 0.1M NaOH were mixed together at a ratio of 3:5:1 to neutralize the acid and gel the collagen in a 24-well culture plate. The neutralized gels were incubated at 37°C for 30 minutes to allow fibrillogenesis to occur to form a firm gel. The gels were then washed with sterile pyrogen-free water to dilute and remove the salts and frozen at −50°C for 24 hours before being freeze-dried at −40°C for 24 hours and sterilized using gamma irradiation (Gamma Cell 1000 Elite, Nordion International I.W.C, Ontario, Canada) using 137Cs at a nominal dose of 30 Gy using two cycles of 11 minutes and 32 seconds.
Preparation of cell-seeded collagen–alginate scaffolds
A minimum volume (200 μL) of cell suspension containing the 70,000 cells was added to each 2.8% (w/v) collagen sponge and incubated at room temperature for 45 minutes to allow cell attachment. Each cell-seeded 2.8% (w/v) collagen sponge was then submerged in 400 μL of alginate solution (sodium alginate from Laminaria hyperborea (Pronova Biomedical, Norway) resuspended at 2.12% (w/v) in complete culture medium containing 20mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) in a 24-well plate. Once absorption of alginate solution was complete (22 minutes), the scaffolds were flooded with calcium chloride (80mM) in Hank's balanced salt solution for 1 hour at room temperature to allow cross-linking of the alginate. The cell-seeded collagen–alginate scaffolds produced were 12 mm in diameter and 3.6 mm thick. The cell-seeded collagen–alginate scaffolds were then transferred to a 24-well plate containing 1.0 mL/well of standard culture medium and incubated under standard conditions. The homogeneity of the alginate within the collagen sponges was evaluated using Alcian blue staining of histological sections.
Alcian blue staining
Samples were fixed in 30% Lillie formalin (100 mL of 30% formalin, 1 g of calcium acetate, pH 7.4) and dehydrated through increasing concentrations of alcohol followed by xylene and embedded in paraffin wax. Sections (6 μm) were cut and, after dewaxing and rehydration, were rinsed in 100 mL of 2% (v/v) acetic acid (pH 2.5) for 2 minutes before they were immersed in l00 mL of Alcian blue stain (1 g in 2% (v/v) acetic acid) for 5 minutes and blotted dry. The slides were counterstained using eosin, mounted using DPX (VWR), and viewed under light microscopy.
Biomechanical testing of scaffolds
The purpose of mechanical testing was to investigate the stress strain behavior of the collagen–alginate scaffolds. For these tests, five collagen–alginate scaffolds discs 12 mm in diameter were prepared as described above, but without cells, and subjected to uniaxial compression. Before testing, the thickness of the samples was measured using a gauge with a resolution of 0.01 mm (Mitutoyo, Andover, UK), and their average thickness was recorded. The samples were then placed on the base part of a Shimadzu testing machine operated using a 50-N load cell and compressed to failure in a quasistatic test at a rate of 5 mm/min. All testing was conducted at room temperature (20°C), and during testing, the samples were continuously sprayed with phosphate buffered saline (PBS). During testing, load data from the load cell and specimen compression data from the stroke of the cross-head of the testing machine were acquired at a rate of 20 Hz. To obtain an accurate measure of the tissue gauge length, the testing machine was set to produce a specimen preloading of 0.01 N before the operating program started to acquire any data. Therefore, zero compression was taken at the point where a load of 0.01 N was detected. The final gauge length (Lo) of the specimen was calculated as the initial gauge length minus the compression required to produce the specified preloading. Failure was taken to occur when the first decrease in load was detected during compression.
The recorded load (F, in Newtons) and specimen compression (ΔL, in mm) data of each specimen were converted to engineering stress and strain. Stress (σ, in MPa) was defined as F/Ao, with Ao (in mm2) representing the original cross-sectional area of the undeformed specimen, which was assumed to be circular. The axial compressive strain (ɛ) was defined as (ΔL/Lo)×100. The calculated stress–strain behavior of each specimen was analyzed according to six parameters. These have been described elsewhere16 and included the initial elastic modulus (El-E), collagen phase slope (Col-E), transition stress (σtrans) and strain (ɛtrans), ultimate compressive strength (σucs), and failure strain (ɛucs). The biomechanical parameters were averaged over the number of samples (n=5) tested.
Application of cyclic compressive strain to hMSC-seeded collagen–alginate scaffolds
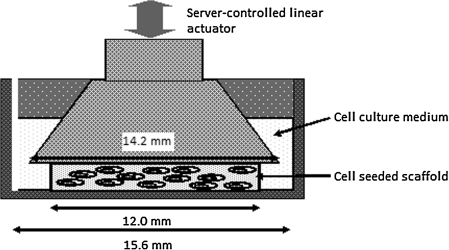
D7-FIB-positive cloned hMSC (7×104 cells) were seeded into each of 27 collagen–alginate scaffolds. Three cell-seeded scaffolds were used immediately to extract the RNA for gene expression analysis at the start of the experiments (controls). The cell-seeded collagen–alginate scaffolds (n=24) were then transferred into individual culture wells of a 24-well plate containing 1 mL/well of standard culture medium. The 24-well plate was then transferred to the ComCell,17 a servo-assisted displacement controlled device for applying cyclic compressive strain to cell-seeded collagen–alginate scaffolds in 24-well plates (Fig. 1). Using this functional simulator, strain could be simultaneously applied to up to 12 wells of the culture plate, leaving the remaining wells static. The simulator was housed within a chamber, which allowed maintenance of high humidity and temperature of between 35°C and 37°C and an atmosphere of 5% (v/v) CO2 in air. Cell-seeded scaffolds were then subjected to cyclic compressive strain of 10% or 15% (in separate experiments) using a sinusoidal waveform with a frequency of 1 Hz for 4 hours per day for 21 days. The strain amplitude was controlled to within 0.5% across all stations, and the 0 position was controlled to within 1% across all wells. After 7 and 21 days of culture, six statically cultured and six dynamically cultured scaffolds were harvested. Three of each were used to extract the RNA for gene expression analysis. The remaining three were divided, and half were used for ATP assay and the other half for live–dead cell staining.
FIG. 1.
Schematic diagram illustrating a single tissue culture well in the ComCell cyclic compressive strain device.
ATPLite assay
All reagents and solutions were allowed to reach room temperature and prepared according to the manufacturer's instructions. Seeded scaffolds were transferred to the wells of 96-well plates, and finely minced. Cell lysis solution (50 μL) was added to each well, and the plate was placed on an orbital plate shaker (700 rpm) for 5 minutes. Substrate solution (50 μL) was then added to each lysate, followed by a further 5 minutes of shaking. Luminescence was measured using a TopCount (NXT) luminometer for 5 seconds after a 10-minute pause to dark-adapt the plate, and data were recorded as counts per second (CPS). The raw data (CPS) for each experiment (10% and 15% compressive strain) was analyzed using one-way analysis of variance (ANOVA) followed by calculation of the minimum significant difference (P<0.05) between group means using the T-method. Data were converted to equivalent cell numbers according to interpolation from a standard curve (MSC number vs CPS) and are presented as the mean (n=3) cell number with 95% confidence intervals.
Live–dead cell staining
Seeded collagen–alginate composites were rinsed in PBS and then stained with 1mM ethidium homodimer and 1mM calcein acetoxymethyl (AM) for 30 minutes at 37°C. The samples were then rinsed, cryo-embedded, sectioned (6 μm), wet mounted, and viewed using a confocal microscope (40× magnification, Zeiss LSM 510 Meta Axioplan 2). Live cells appeared green, and dead cells fluoresced red. Relative numbers of live and dead cells were estimated from digital images.
Quantitative real-time polymerase chain reaction
The relative expression of a number of genes of interest was determined using a Stratagene MX-3000P Real-Time PCR System and SYBR Green I dye.
Isolation of RNA
Total cellular RNA was extracted using the GenElute Mammalian Total RNA Kit RTN10. Briefly, cell-seeded scaffolds were finely minced and transferred to a 5-mL sterile test tube containing 1 mL of lysis solution. A further 500 μL of lysis solution containing 5 μL of 2-mercaptoethanol was added to each sample, and the sample was homogenized. The resulting solution was added to a Genelute filtration column and centrifuged at 12,000 g for 2 minutes. Ethanol (70% v/v, 500 μL) was then added to the filtered lysate, and the mixture was vortexed. The solution (500 μL) was added to a Genelute binding column and centrifuged at 12,000 g for 15 seconds. Wash solution 1 (500 μL) was then added to the column and centrifuged at 12,000 g for 15 seconds. Wash solution 2 (500 μL) was added to the column and centrifuged at 12,000 g for 15 seconds. This step was then repeated, extending the centrifuging to 3 minutes. Elution solution (50 μL) was added to the binding column and centrifuged at 12,000 g for 1 minute. Purified RNA was then ready for immediate use or storage at −70°C.
cDNA synthesis reaction
cDNA was synthesized from the extracted RNA using the Stratascript Q-PCR cDNA synthesis kit (Stratagene, CA). The first-strand cDNA synthesis reaction was prepared in a microcentrifuge tube by adding 5 μL of RNAse-free water, 10 μL of first-strand master mix, 2.0 μL of oligo (DT) primer, 1.0 mL of Stratascript RT-RNAse block enzyme mixture, and 2 μL of extracted RNA. The solution was vortexed and annealed in a thermal cycler (Thermo Electron Corporation) at 25°C for 5 minutes. The solution was then incubated at 42°C for 15 minutes to allow cDNA synthesis, followed by 95°C for 5 minutes to terminate the cDNA synthesis reaction. The reactions were placed on ice for immediate use in Q-PCR or stored at −20°C.
Q-PCR procedure
After optimization of the reaction conditions, the reaction vessel contained 30 μL of 2× SYBR Green I QPCR Master Mix, 0.9 μL of passive reference dye (diluted 1:200), 6.5 μL of deionized water, and 1 μL template DNA. The relevant forward primer (3.6 μL of 5μM stock) and reverse primer (3.6 μL of 5μM stock) combined in 14.4 μL of deionized water were then added to give a final volume of 60 μL. This solution was then split into 2×25 μL samples and placed in the thermal block of the Stratagene MX-3000P Real-Time PCR System. All reactions were performed in optical tube strips in triplicate under the conditions found in Table 1. A dissociation (melt) curve was produced by gradual heating of the end PCR product to allow identification of nonspecific PCR products. No-template controls and reverse transcription–negative controls were included to ensure that there was no DNA contamination in the reverse transcription or PCR reactions. The level of expression of each target gene (aggrecan, CBFA-1, Sox9, and PPAR-γ) was normalized to the reference gene 18s rRNA, and the resulting data were expressed as a ratio of the levels of expression to the untreated D7-FIB-positive hMSC control group. Genomic DNA (gDNA, 0.2 ng/μL) was used as a positive control. A passive reference dye was included to normalize levels of fluorescence between wells. Primers were designed using OligoPerfect (Invitrogen) primer design software (Table 2) to target the sequence of the gene of interest within an exon, allowing amplification of gDNA in positive controls and to produce standard curves for Q-PCR, and giving PCR products of 100 to 400 bp. All reagents were obtained from Stratagene, except for the primers, which were synthesized by Sigma-Aldrich, and the genomic DNA, obtained from Promega.
Table 1.
Reaction Conditions of Each Step of Quantitative Real-Time Polymerase Chain Reaction (PCR)
| Step | Duration, seconds | Temperature, °C | Cycles, n |
|---|---|---|---|
| Initial denaturation | 600 | 95 | 1 |
| Denaturation | 30 | 95 | 40 |
| Annealing | 60 | 1–5 below Tm | 40 |
| Extension | 60 | 72 | 40 |
Tm, melting temperature.
Table 2.
Primer Pairs and PCR Conditions
| Control Gene | Forward Sequence | Reverse Sequence | Tm (°C) | Base Product |
|---|---|---|---|---|
| CBFA-1 | GTTCAACGATCTGAGATTTG | GGGGTCTGTAATCTGACTCT | 66.7 | 322 bp |
| PPAR-γ | AGTTTGAGTTTGCTGTGAAG | TGTCTGAGGTCTGTCATTTT | 61.1 | 232 bp |
| Sox9 | ATAACCGAAGAAAGAGAGGA | CAAACAGGCAGAGAGATTTA | 67.9 | 308 bp |
| Aggrecan | GCCTTGAGCAGTTCACCTTC | ACTTGGGTCACGATCCACTC | 64.0 | 355 bp |
CBFA-1, core binding factor alpha 1; PPAR-γ, peroxisome proliferator-activated receptor gamma; bp, base pair.
Data acquisition and analysis
Fluorescent data was acquired using the FAM filter set for the SYBR I Green dye and the ROX filter set for the reference dye during the annealing and extension step, as well as throughout the dissociation program. The software produced amplification plots of the reaction using fluorescence (normalized to reference dye) versus cycle number. The Ct values for each sample, which refer to the threshold cycle at which exponential amplification of target is occurring significantly above background fluorescence levels, were automatically calculated. From this, the standard curves of the Ct values versus the relative quantity of target were plotted for the gene of interest and the reference gene (18s rRNA). Gene expression levels in cells cultured statically and dynamically for 7 and 21 days (n=3 for each condition) were compared with gene expression levels in the controls using the pair-wise fixed reallocation randomization test.
Results
Phenotype of hMSCs
Control hMSC (not purified using the D7-FIB antibody) and D7-FIB-positive cells were negative for rabbit antimouse Ig, IgG1, IgG2a, and more than 69% positive for CD-13, CD-90, CD-105, and D7-FIB. Control cells were 2.5% to 15% positive for alkaline phosphatase (ALP), osteocalcin (OC), CD-45, HLA-DR, CD-34, and CD-117, whereas D7-FIB-positive cells were negative for these markers, indicating no detectable hematopoietic or osteoblastic cell differentiation (Table 3).
Table 3.
Fluorescence Activated Cell-Sorting of Control and D7-FIB Purified Human Mesenchymal Stem Cells
| |
Control |
D7-FIB-Positive |
|---|---|---|
| Marker | % | |
| Secondary antibody alone | 0.00 | 0.00 |
| IgG1 | 0.00 | 0.00 |
| IgG2a | 0.00 | 0.00 |
| CD-13 | 90.38 | 93.12 |
| CD-90 | 79.25 | 73.94 |
| CD-105 | 69.32 | 72.33 |
| D7-FIB | 72.63 | 91.55 |
| Alkaline phosphatase | 3.55 | 0.00 |
| Osteocalcin | 2.73 | 0.00 |
| CD-45 | 14.80 | 0.16 |
| Human leukocyte antigen DR | 3.32 | 0.32 |
| CD-34 | 3.64 | 0.00 |
| CD-117 | 2.87 | 0.00 |
Differentiation of hMSC
Adipogenic differentiation
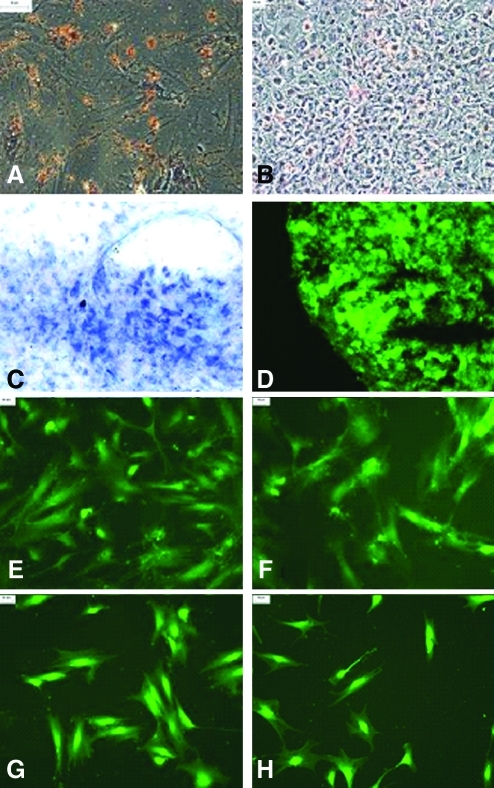
After 2 and 4 weeks of culture, Oil Red O dye was used to detect possible adipogenic differentiation. Approximately 8% to 10% of cloned D7-FIB-positive hMSC had differentiated (Fig. 2A, B).
FIG. 2.
Differentiation of cloned D7FIBpositive human mesenchymal stem cells (hMSC). Adipogenic differentiation (A) After 2 weeks (×200), (B) after 4 weeks (×100) as indicated by lipid vesicles stained with Oil Red O. Chondrogenic differentiation after 4 weeks, (C) toluidine blue staining for glycosaminoglycan production (×400), (D), immunolabeling with collagen II antibody (×400). Myogenic differentiation after 4 weeks, (E) immunolabeling with desmin (×100), (F) immunolabeling with myosin heavy chain (×100). Neurogenic differentiation after8 hours, (G) immunolabeling with tau (×100), (H) immunolabeling with neuron-specific enolase (NSE) (×100). Color images available online at www.liebertonline.com/tec
Chondrogenic differentiation
Toluidine blue staining of hMSC after 4 weeks in culture demonstrated blue-stained regions, indicative of extracellular matrix production by the cells (Fig. 2C). Immunofluorescent labelling of the pellet using anti-collagen II indicated extensive collagen II production (Fig. 2D).
Myogenic differentiation
After 4 weeks, the hMSC were fixed and labelled with antibodies to desmin and HCMs. Approximately 60% to 70% of the cells had differentiated (Fig. 2E, F).
Neurogenic differentiation
After 2, 4, 6, and 8 hours in culture, cells were fixed and labelled with fluorescent antibodies to tau and NSE. Approximately 15% to 20% of cloned D7-FIB-positive hMSC had differentiated after 8 hours (Fig. 2G, H).
Biomechanical testing of collagen–alginate scaffold
Collagen–alginate scaffolds were manufactured, and their overall histoarchitectural morphology was assessed using Alcian blue staining (Fig. 3). The biomechanical properties of the scaffold, obtained under compression testing and averaged over the number of specimens tested (n=5), are presented in Table 4.
FIG. 3.
(A) Collagen–alginate scaffold, pink color due to alginate being prepared in culture medium. (B) Alcian blue staining of scaffold section. Blue areas show alginate filling pores in collagen sponge, shown by pink eosin staining, to give the overall collagen–alginate scaffold structure (scale bar 50 μm). Color images available online at www.liebertonline.com/tec
Table 4.
Biomechanical Properties of the Collagen–Alginate Scaffold Under Low Compressive Strain Rate Failure Testing (N=5)
| Young's elastic modulus, MPa | Collagen phase slope. MPa | Transition stress, MPa | Strain, % | Ultimate compressive strength, MPa | Failure strain, % | Thickness, mm |
|---|---|---|---|---|---|---|
| Mean±95% Confidence Limits | ||||||
| 3.8×10−2±4.4×10−3 | 0.52±5.1×10−5 | 0.09±2.7×10−3 | 25.5±1.0 | 0.2±0.01 | 81.8±1.7 | 3.6±7.9×10−3 |
Application of cyclic compressive strain to hMSC seeded on collagen–alginate scaffolds
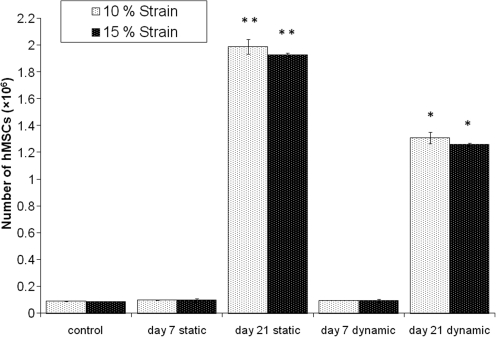
The proliferation of the hMSC seeded on the collagen–alginate scaffolds and cultured statically or under 10% or 15% cyclic compression for 21 days was determined by measuring the levels of ATP (Fig. 4). Under 10% cyclic compressive strain, the number of hMSC increased from 7×104 cells at day 0 to 1.35×106 cells at day 21, whereas the number of cells in the static controls increased from 7×104 to 1.93×106 cells during the same interval. The number of cells in the scaffolds cultured under static conditions was significantly greater than in scaffolds cultured under 10% cyclic compressive strain at 21 days (P<0.05; ANOVA). Under 15% cyclic compressive strain, the number of hMSC increased from 7×104 at day 0 to 1.22×106 cells at day 21, whereas the number of cells in the static controls increased from 7×104 to 1.99×106 cells during the same interval. Again, the number of cells in the scaffolds cultured under static conditions was significantly greater than in scaffolds cultured under 15% cyclic compressive strain at 21 days (P<0.05; ANOVA).
FIG. 4.
Proliferation of hMSC seeded on composites and cultured statically or subjected to 10% or 15% cyclic compressive strain for 21 days determined according to adenosine triphosphate assay. Data are expressed as the mean (n=3) number of cells with 95% confidence limits. The raw data (counts per second) was analyzed using one-way analysis of variance for each experiment (10% and 15% cyclic compressive strain) followed by determination of the minimum significant difference between group means using the T-method (P<0.05). **Significantly higher (P<0.05) cell numbers in the static cultures at 21 days than in all other groups. *Significantly higher (P<0.05) cell numbers in the dynamic cultures at 21 days than in the static and dynamic cultures at 7 days.
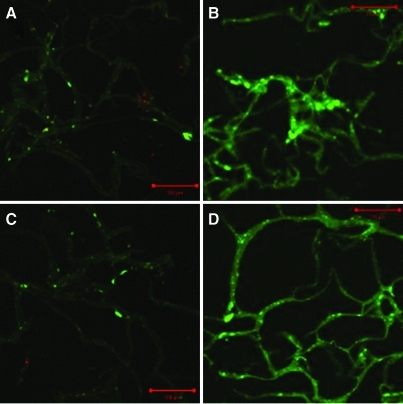
The live–dead cell assay was used to estimate the proportion of cells that were alive and dead in the scaffolds at different depths. The hMSC cultured dynamically at 10% cyclic compressive strain appeared viable at all depths of the scaffolds at days 7 and 21. In the statically cultured control scaffolds, approximately 50% to 55% of the cells appeared dead at depths of 1.6 mm and 2.4 mm at days 7 and 21, respectively. In the 15% strain study, hMSC cultured dynamically were viable at all depths of the scaffolds at days 7 and 21. In the respective statically cultured control scaffolds, 10% to 25% of hMSC were dead at depths of 1.6 mm and 2.4 mm at 7 and 21 days, respectively. Representative images of live–dead staining at a depth of 2.4 mm are shown in Fig. 5. Because dead cells were seen only in sections taken at depths of 1.6 to 2.4 mm from the surface of all of the statically cultured scaffolds, this gave an indication of the limits of nutrient diffusion in the scaffolds in the absence of cyclic compression. The large difference between the percentage of dead cells in the scaffolds cultured statically between the two experiments is most likely explained by sampling error because of uneven dispersion of the dead cells.
FIG. 5.
Live–dead cell assay of hMSC seeded on collagen–alginate scaffolds and cultured for up to 21 days (images taken at a depth of 2.4 mm through scaffold): (A) static culture for 21 days, (B) 10% cyclic compressive strain for 7 days, (C) static culture for 21 days, (D) 15% cyclic compressive strain for 21 days. Original magnification ×100. Color images available online at www.liebertonline.com/tec
Q-PCR of hMSC seeded on collagen–alginate scaffold and cultured statically or subjected to cyclic compressive strain for up to 21 days
The results of the Q-PCR analysis are shown in Table 5. All data were normalized to the reference gene (18s rRNA) and the resulting data expressed as a ratio of the levels of expression to the untreated D7FIB-positive cells at 7 and 21 days. For the 10% strain study, under static control conditions, PPAR-γ was upregulated significantly after 21 days by a factor of 1.8 but not significantly at 7 days. Under dynamic conditions, PPAR-γ was significantly upregulated at 7 days by a factor of 2.0 but not significantly at 21 days. CBFA-1 was significantly upregulated after 21 days by a factor 1.8 under static culture but not significantly after 7 days. Under dynamic culture, CBFA-1 was significantly upregulated at 7 and 21 days by factors of 18.3 and 32.2, respectively. Sox9 was significantly upregulated at 7 and 21 days by factors of 1.3 and 1.6, respectively, under static culture. Under dynamic conditions, Sox9 at 7 and 21 days was significantly upregulated by factors of 2.6 and 1.9, respectively. Aggrecan was significantly upregulated at 7 and 21 days by factors of 1.4 and 1.6, respectively, under static culture. Under dynamic conditions, aggrecan was significantly upregulated at 7 and 21 days by factors of 1.3 and 2.0, respectively.
Table 5.
Expression of PPAR-γ, CBFA-1, Sox9, and Aggrecan by Human Mesenchymal Stem Cells Seeded into Collagen–Alginate Scaffolds and Subjected to 10% and 15% Cyclic Compressive Strain
| |
7 Days |
21 Days |
||
|---|---|---|---|---|
| |
Static |
Compressive Strain |
Static |
Compressive Strain |
| Compressive | % | |||
| Strain 10% | ||||
| Control group | 1.0 | 1.0 | 1.0 | 1.0 |
| PPAR-γ | 1.6 | 2.0* | 1.8* | 1.1 |
| CBFA-1 | 1.9 | 18.3* | 1.8* | 32.2* |
| Sox9 | 1.3* | 2.6* | 1.6* | 1.9* |
| Aggrecan | 1.4* | 1.3* | 1.6* | 2.0* |
| 15% | ||||
| Control group | 1.0 | 1.0 | 1.0 | 1.0 |
| PPAR- γ | 1.0 | 3.3 | 1.0 | 2.3 |
| CBFA-1 | 3.1* | 6.8* | 2.8* | 2.9* |
| Sox9 | 2.5 | 3.7* | 1.6 | 3.4* |
| Aggrecan | 1.4* | 1.3 | 1.0 | 3.5* |
Significant difference from control group determined using the pair-wise fixed reallocation randomization test (p<0.05).
For the 15% strain study, PPAR-γ showed no significant upregulation at 7 or 21 days under static or dynamic culture. CBFA-1 was significantly upregulated at 7 and 21 days by factors of 3.1 and 2.8, respectively, under static culture and by factors of 6.8 and 2.9, respectively, under dynamic culture. Sox9 showed no significant upregulation at 7 or 21 days under static culture, although under dynamic conditions, there was significant upregulation by factors of 3.7 and 3.4, respectively. Aggrecan was significantly upregulated only after 7 days by a factor of 1.4 under static conditions but not significantly after 21 days, whereas under dynamic culture, no change was seen at 7 days, but at 21 days, there was a significant increase by a factor of 3.5.
Discussion
hMSC have been widely shown to be capable of differentiating into multiple cell lineages in vitro using biochemical stimulation,1–3 although it is likely that mechanical stimulation will also prove to be a significant factor in inducing and controlling the differentiation of these cells once implanted in vivo. In this study, hMSC were isolated, separated into D7-FIB-positive cells, and characterized as MSC according to FACS analysis. One population of hMSC was then cloned and the differentiation potential determined before seeding into collagen–alginate scaffolds, which were loaded for 4 hours per day for 21 days under 10% or 15% cyclic compressive strain, together with unstrained static controls. At 7 and 21 days of culture, composites were harvested and analyzed for gene expression using Q-PCR, cell proliferation, and cell viability.
hMSC were routinely isolated from the bone marrow of patients undergoing hip replacement operations. Cells were initially phenotyped using a panel of 10 different antibodies used by several researchers.1,15,18 These included CD-13, CD-34, CD-45, CD-90, CD-105, CD-117, D7-FIB, HLA-DR, OC, and ALP. CD-34, CD-45, and HLA-DR are commonly expressed by hematopoietic cells, D7-FIB and CD-90 by human fibroblast cells, CD-105 by endothelial cells, and CD-13 and CD-117 by myeloid and epithelial cells. OC and ALP were used as bone differentiation markers to exclude osteogenic differentiation. The D7-FIB antibody was used to purify the cells to determine whether this gave rise to cell populations with better growth and MSC characteristics. Workers in the field have shown that hMSC express a nonhematopoietic phenotype,1,19 which is usually identified by STRO-120 and D7-FIB antibody,15 although it has been shown that the STRO-1 antibody is not sufficient to obtain the purity of bone marrow stem cells required to study their properties properly, owing to the presence of a contaminating population of glycophorin-A-positive nucleated red cells and a small subset of B-lymphocytes.21 In the present study, the D7-FIB antibody was found to identify putative hMSC among adherent human bone marrow cells. Flow cytometric analysis indicated that D7-FIB-positive cells were negative for markers such as ALP, OC, CD-45, HLA-DR, CD-34, and CD-117, indicating no detectable differentiation into hematopoietic or osteoblastic cells, and showed phenotypic characteristics of the undifferentiated state of hMSC comparable with those that others have isolated and characterized.1,15,18 This was in contrast to nonpurified cells, which showed positive labelling for these markers.
Cloned hMSC were then successfully differentiated into adipogenic, chondrogenic, neurogenic, and myogenic lineages using appropriately supplemented culture medium over 4 weeks or less, showing them to possess a multipotential differentiation capacity and therefore be stem cells. These studies did not result in 100% differentiation of the cells into the four cell lineages. Because the cells were cloned from a single cell before the differentiation assays were undertaken, the results reflected the multipotential of a single cell population because there was no possibility of contaminating precursors in the differentiated cell population. The finding that not all of the cells differentiated in response to the biochemical cues indicates that the differentiation conditions were probably suboptimal and require further refinement.
Before undertaking the experiments with hMSC, a great deal of effort was focused on producing a scaffold that would retain its biomechanical and elastic properties under repeated compressive strain regimens over a prolonged culture period. Collagen was chosen to provide cell adhesion motifs. Scaffolds made of collagen I have been used for tissue engineering of bone, muscle, and skin.22 Benefits include low antigenicity and ease of handling, but limitations include initial poor mechanical strength and difficulty obtaining large quantities of sterile collagen I.22, 23 Alginate was included in the scaffold to improve the elastic properties and enable the scaffold to recover during the relaxed unloading phase of the cyclic compressive culture. The macroscale strain was determined for the scaffold as a whole, but no attempt was made to determine the local mesoscale strain acting at a local level on the cell within the biological composite scaffold. The average stress–strain behavior of the scaffolds was similar to that of other biological tissues described elsewhere.16 The average transition strain of the scaffolds under compression was found to be 25.5±1.04%, indicating that the chosen 10% and 15% compression strains used during the conditioning experiments were well within the initial linear elastic region of the stress–strain behavior of the scaffolds.
When hMSC were seeded on the scaffolds and subjected to 10% or 15% cyclic compressive strain over a 21-day period, they did not proliferate to the same extent as hMSC cultured in scaffolds under static conditions, suggesting that the hMSC were losing viability or were slowing their rate of cell division and perhaps differentiating into a synthetic phenotype, although the live–dead cell assay showed that there were more dead hMSC at different depths of the scaffolds cultured statically than dynamically. In the dynamic cultures, mechanical stimulation may have helped to alleviate any diffusion limitations present in the statically cultured samples by creating an additional convective transport mechanism due to the compression-induced interstitial fluid flow within the scaffolds in combination with a secondary mixing effect on the surrounding medium, ensuring that the hMSC remained viable at all depths of the scaffold. Therefore, it was speculated that the hMSC cultured under dynamic conditions were differentiating and slowing their rate of division.
In analyzing the effects of cyclic compressive strain, Q-PCR was used to investigate changes in expression of CBFA-1, Sox9, and aggrecan, which are all associated with bone and cartilage cell lineages, and PPAR-γ, indicative of adipogenesis.12,25–28. CBFA-1 has been demonstrated to play an important role in bone formation and is reported to control the expression of osteoblast-specific genes that synthesize and deposit the bone extracellular matrix. Levels are higher in immature bone. Studies indicate that CBFA-1 has a distinct, albeit complementary, role in osteogenesis.26 PPAR-γ is a member of the PPAR subfamily, is expressed predominantly in adipose tissue, and plays a central role in the control of adipocyte differentiation. PPAR-γ agonists and overexpression of PPAR-γ2 have been reported to induce differentiation of bone marrow stromal cells into the adipocyte lineage and inhibit CBFA-1 expression and osteoblast differentiation. PPAR-γ insufficiency has been related to the regulation of bone mass in vivo by increasing osteoblastogenesis from bone marrow progenitors.27 Sox9 is a transcriptional activator that binds to the promoters and activates transcription of collagen type II and aggrecan genes and is usually expressed in chondrogenesis.28
Previous studies have shown a positive influence of compression on differentiation of the chondrogenic lineage and the upregulation of PPAR-γ and Sox9 genes. Takahashi et al. demonstrated that differences exist between undifferentiated MSC and differentiated chondrocytes in terms of their responses to compressive forces and demonstrated enhanced chondrogenesis of mouse embryonic limb bud mesenchymal cells seeded in a type I collagen gel after a static compressive force of up to 2 kPa.29 A study conducted with 2% agarose seeded with rabbit MSC demonstrated that 10% compressive loading for 4 hours per day in the absence of any growth factor could induce chondrogenic differentiation.30 Furthermore, a recent study on hMSC seeded in 6% alginate gel and subjected to 15% compression with repeated cycles of loading and recovery of 1.5 hours and 4.5 hours, respectively, indicated that dynamic compressive strain enhanced chondrogenic differentiation.31 Although all these studies demonstrated that cells were differentiated along the chondrogenic lineage under compression, the expression of any osteogenic differentiation markers was not examined.
In the present study, under 10% cyclic compressive strain, there were minor changes in the expression of genes indicative of an adipogenic and chondrogenic lineage, but the most significant result was observed for CBFA-1, which showed 18 and 32 times greater expression at 7 and 21 days, respectively. To some extent, the same pattern was observed at 15% cyclic compressive strain, with Sox9 and aggrecan markers also upregulated. This indicated that cells subjected to 10% cyclic compressive strain, in the absence of external chemical cues, were directed toward osteogenic differentiation, whereas under 15% cyclic compressive strain, chondrogenic markers were also upregulated, which may have indicated differentiation along an osteochondrogenic pathway. Future research should further examine the functional differentiation of hMSC in this system through determination of extracellular matrix deposition within the cell seeded constructs.
The question arises regarding the mechanobiological pathways that may be operating at the two magnitudes of strain used in this study. In addition to substrate deformation under load, the compression system applied in this study would also induce hydrostatic pressure and fluid flow. hMSC will have the capacity to respond to mechanical cues through integrin-mediated focal adhesions and mechanosensitive ion channels, which are sensitive to stretch and fluid flow. Substrate strain and fluid flow will cause cell deformation, and the type of deformation is likely to be different for substrate strain and fluid flow and may activate different cell signalling pathways. It may be speculated that, during the application of higher levels of compressive strain, substrate strain may play a more-dominant role than fluid flow, with the reverse being true at lower levels of cyclic compressive strain. It has been shown that application of low to moderate uniaxial tensile strain (10–12%) to cultures of hMSC in three-dimensional collagen–agarose matrices was sufficient to induce osteogenic differentiation in the absence of external chemical cues as determined by the expression of bone morphogenetic protein-2.32 It could be speculated that low to moderate substrate strain–induced fluid flow is an important factor in osteogenic differentiation of hMSC and that higher magnitudes of substrate strain favor chondrogenic differentiation, but unravelling the mechanobiological pathways that may be operating will require a much more detailed understanding of the cell-signalling pathways, the fluid dynamics of the system, and measurements of cell deformation.
Further investigations into the specific mechanisms of how differentiation is induced could lead to the development of novel methods for enhancing and accelerating tissue repair and new directions for the tissue engineering of replacement bone and cartilage through the preconditioning and implantation of patient-derived MSC with or without an appropriate scaffold. This will lead to general advancement of our understanding of how cells perceive and respond to the application of different types of physical force.
Conclusion
Tissue mechanics depend not only on the features of individual cells and matrix molecules, but also on the complex organization and reciprocal interaction that occur between groups of cells and their surrounding matrix.33 We successfully isolated, purified, and characterized a clonal population of hMSC. Cloned D7-FIB-positive cells retained their ability to differentiate into four lineages (adipogenic, chondrogenic, myogenic, and neurogenic) after 10 passages. A novel 2.8% (w/v) collagen–alginate scaffold with nonlinear viscoelastic stress–strain behavior and reproducible structure and pore size was developed and shown to support the three-dimensional growth of cloned D7-FIB-positive hMSC. The cell-seeded collagen–alginate scaffolds were subjected to cyclic compressive strain on a servo-assisted displacement-controlled rig that was developed and manufactured in house. Cloned D7-FIB-positive hMSC showed evidence of differentiation to an osteogenic lineage under 10% cyclic compressive strain alone and to an osteochondrogenic lineage under 15% cyclic compressive strain alone.
Further investigations into the specific mechanisms of how differentiation is induced could lead to the development of novel methods for enhancing and accelerating tissue repair and new directions for the tissue engineering of replacement bone and cartilage through the preconditioning and implantation of patient-derived mesenchymal stem cells with or without an appropriate scaffold.
Acknowledgments
This work was funded jointly by the EPSRC and NHS BT (NHS Blood and Transplant) through a Case studentship to Efstathios Michalopoulos and through WELMEC, a Centre of Excellence in Medical Engineering funded by the Wellcome Trust and EPSRC, under grant WT 088908/Z/09/Z. John Fisher is an NIHR senior investigator.
Disclosure Statement
No competing financial interests exist.
References
- 1.Pittenger M.F. Mackay A.M. Beck S.C., et al. Multi-lineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Y. Jahagirdar B.N. Reinhardt R.L., et al. Pluripotency of mesenchymal stem cells derived from adult marrows. Nature. 2002;418:41. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 3.Pasini A. Krampera M. Rigo A., et al. HB-EGF/HER-1 signalling in bone marrow mesenchymal stem cells: inducing cell expansion and reversibly preventing multi-lineage differentiation. Blood. 2005;106:59. doi: 10.1182/blood-2004-09-3645. [DOI] [PubMed] [Google Scholar]
- 4.Pauwels F. A new theory concerning the influence of mechanical stimuli in the differentiation of supporting tissues. Biomech Locomotor Apparatus. 1980;375:458. [Google Scholar]
- 5.Liu S.Q. Influence of tensile strain on smooth muscle cell orientation in rat blood vessels. J Biomech Eng. 1998;120:313. doi: 10.1115/1.2797996. [DOI] [PubMed] [Google Scholar]
- 6.Buschmann M.D. Gluzband Y.A. Grodzinsky A.J., et al. Mechanical compression modulates matrix biosynthesis in chondrocyte/agarose culture. J Cell Sc. 1995;108:1497. doi: 10.1242/jcs.108.4.1497. [DOI] [PubMed] [Google Scholar]
- 7.Takagaki Y.M. Mechanical responses and signal transduction pathways in stretched osteocytes. J Bone Miner Metab. 1999;17:57. doi: 10.1007/s007740050065. [DOI] [PubMed] [Google Scholar]
- 8.Jen C.J. Jhiang S.J. Chen H.I. Cellular responses to mechanical stress: effects of flow on vascular endothelial intracellular calcium signaling of rat aortas ex vivo. J Appl Physiol. 2000;89:1657. doi: 10.1152/jappl.2000.89.4.1657. [DOI] [PubMed] [Google Scholar]
- 9.Katsumi A. Orr A.W. Tzima E., et al. Integrins in mechanotransduction. J Biolog Chem. 2004;279:12001. doi: 10.1074/jbc.R300038200. [DOI] [PubMed] [Google Scholar]
- 10.Neidlinger-Wilke C. Grood E.S. Wang J.H.C., et al. Cell alignment is induced by cyclic changes in cell length: studies of cells grown in cyclically stretched substrates. J Orthop Res. 2001;19:286. doi: 10.1016/S0736-0266(00)00029-2. [DOI] [PubMed] [Google Scholar]
- 11.Simmons C.A. Matlis S. Thornton A.J., et al. Cyclic strain enhances matrix mineralization by adult human mesenchymal stem cells via the extracellular signal-regulated kinase (ERK1/2) signaling pathway. J Biomechan. 2003;36:1087. doi: 10.1016/s0021-9290(03)00110-6. [DOI] [PubMed] [Google Scholar]
- 12.Altman G. Horan R. Martin I., et al. Cell differentiation by mechanical stress. FASEB J. 2002;16:270. doi: 10.1096/fj.01-0656fje. [DOI] [PubMed] [Google Scholar]
- 13.Wakitani S. Saito T. Caplan A.I. Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine. Muscle Nerve. 1995;18:1417. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- 14.Woodbury D. Schwarz E.J. Prockop D.J., et al. Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res. 2000;61:364. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Jones E.A. Kinsey S.E. English A., et al. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheum. 2002;46:3349. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 16.Korossis S. Booth C. Wilcox H.E. Ingham E. Kearney J.N. Watterson K.G. Fisher J. Tissue engineering a cardiac valve prosthesis II: biomechanical characterisation of decellularised porcine heart valves. J Heart Valve Dis. 2002;11:463. [PubMed] [Google Scholar]
- 17.Matthews J.B. Mitchell W. Stone M.H. Fisher J. Ingham E. A novel three-dimensional tissue equivalent model to study the combined effects of cyclic mechanical strain and wear particles on the osteolytic potential of primary human macrophages in vitro. Proc Inst Mech Eng H. 2001;215H:479. doi: 10.1243/0954411011536073. [DOI] [PubMed] [Google Scholar]
- 18.Lodie T.A. Blickarz C.E. Devarakonda T.J., et al. Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue Eng. 2002;8:739. doi: 10.1089/10763270260424105. [DOI] [PubMed] [Google Scholar]
- 19.De Boer J. Siddappa R. Gaspar C., et al. Wnt signalling inhibits osteogenic differentiation of human mesenchymal stem cells. Bone. 2004;34:818. doi: 10.1016/j.bone.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Colter D.C. Sekiya I. Prockop D.J. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci. 2001;98:7841. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis J.E. Carbillet J.P. Caplan A.I. Charbord P. The STRO-1+ marrow cell population is multipotential. Cells Tissues Organs. 2002;170:73. doi: 10.1159/000046182. [DOI] [PubMed] [Google Scholar]
- 22.Ignatius A. Blessing H. Liedert A., et al. Tissue engineering of bone: effects of mechanical strain on osteoblastic cells in type I collagen matrices. Biomaterials. 2005;26:311. doi: 10.1016/j.biomaterials.2004.02.045. [DOI] [PubMed] [Google Scholar]
- 23.Ma P.X. Zhang R.Y. Synthetic nano-scale fibrous extracellular matrix. J Biomed Materl Res. 1999;46:60. doi: 10.1002/(sici)1097-4636(199907)46:1<60::aid-jbm7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Cummings C.L. Gawlitta D.N. Robert M., et al. Properties of engineered vascular constructs made from collagen, fibrin and collagen–fibrin mixtures. Biomaterials. 2004;25:3699. doi: 10.1016/j.biomaterials.2003.10.073. [DOI] [PubMed] [Google Scholar]
- 25.Young R.G. Butler D.L. Weber W., et al. Use of mesenchymal stem cells in a collagen matrix for Achilles tendon repair. J of Orthop Res. 1998;16:406. doi: 10.1002/jor.1100160403. [DOI] [PubMed] [Google Scholar]
- 26.Wei S. Yang D. Wang D. Phimphilai M. Krebsbach P.H. Franceschi R. T. In vitro and in vivo synergistic interactions between the Runx2/Cbfa1 transcription factor and bone morphogenetic protein-2 in stimulating osteoblast differentiation. J Bone Miner Res. 2003;18:705. doi: 10.1359/jbmr.2003.18.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barak Y. Nelson M.C. Ong E.S. Jones Y.Z. Ruiz-Lozano P. Chien K.R. Koder A. Evans R.M. PPAR gamma is required for placental, cardiac and adipose tissue development. Mol Cell. 1999;4:585. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre V. Behringer R.R. de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9:S69. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi I. Nuckolls G.H. Takahashi K. Tanaka O. Semba I. Dashner R. Shum L. Slavkin H. C. Compressive force promotes Sox9, type II collagen and aggrecan and inhibits IL-1 beta expression resulting in chondrogenesis in mouse embryonic limb bud mesenchymal cells. J Cell Sci. 1998;111:2067. doi: 10.1242/jcs.111.14.2067. [DOI] [PubMed] [Google Scholar]
- 30.Huang C.Y. Hagar K.L. Frost L.E. Sun Y. Cheung H.S. Effects of cyclic compressive loading on chondrogenesis of rabbit bone-marrow derived mesenchymal stem cells. Stem Cells. 2004;22:313. doi: 10.1634/stemcells.22-3-313. [DOI] [PubMed] [Google Scholar]
- 31.Campbell J.J. Lee D.A. Bader D.L. Dynamic compressive strain influences chondrogenic gene expression in human mesenchymal stem cells. Biorheology. 2006;43:455. [PubMed] [Google Scholar]
- 32.Sumanasinghe R.D. Bernacki S.H. Loboa E.G. Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP-2) mRNA expression. Tissue Eng. 2006;12:3459. doi: 10.1089/ten.2006.12.3459. [DOI] [PubMed] [Google Scholar]
- 33.Silver F.H. Siperko L.M. Seehra G.P. Mechanobiology of force transduction in dermal tissue. Skin Res Technol. 2003;9:3. doi: 10.1034/j.1600-0846.2003.00358.x. [DOI] [PubMed] [Google Scholar]