Abstract
The evolution of host resistance to parasites, shaped by associated fitness costs, is crucial for epidemiology and maintenance of genetic diversity. Selection imposed by multiple parasites could be a particularly strong constraint, as hosts either accumulate costs of multiple specific resistances or evolve a more costly general resistance mechanism. We used experimental evolution to test how parasite heterogeneity influences the evolution of host resistance. We show that bacterial host populations evolved specific resistance to local bacteriophage parasites, regardless of whether they were in single or multiple-phage environments, and that hosts evolving with multiple phages were no more resistant to novel phages than those evolving with single phages. However, hosts from multiple-phage environments paid a higher cost, in terms of population growth in the absence of phage, for their evolved specific resistances than those from single-phage environments. Given that in nature host populations face selection pressures from multiple parasite strains and species, our results suggest that costs may be even more critical in shaping the evolution of resistance than previously thought. Furthermore, our results highlight that a better understanding of resistance costs under combined control strategies could lead to a more ‘evolution-resistant’ treatment of disease.
Keywords: fitness trade-off, apparent competition, phage therapy, Pseudomonas syringae, coevolution
1. Introduction
Parasites are an ever-present force in nature and, through their negative effects on host fitness, can exert strong selection on host populations for increased resistance. Multiple studies have demonstrated parasite-mediated selection for host resistance both in the laboratory [1–8] and in natural populations [9–12]. Despite this propensity of hosts to evolve increased resistance, many natural populations are polymorphic with respect to resistance traits and disease remains ubiquitous. One reason for this might be that the evolution of resistance to local parasites carries a fitness cost for the host, such as reduced fecundity, that decreases the likelihood of fixation of resistance in a population. Indeed, costs associated with resistance to parasites have been demonstrated in a wide array of hosts, including bacteria [13], protozoa [6], nematodes [7], insects [2,3,5], crustaceans [11], molluscs [8,14], flowering plants [15,16], birds [17] and mammals [18]. A number of recent reviews, though, have emphasized that these costs are commonly not observed [19,20]. One explanation for this inconsistency in apparent fitness costs could be that resistance to a single parasite, as measured in most experimental studies, is not always costly, but that resistance to many different parasites, which is the common problem in nature, carries a more pronounced cost. Hence, a better understanding of fitness costs associated with evolving resistance against multiple parasites may provide key insight into how trade-offs maintain variation in resistance and shape the coevolutionary process.
There are at least two ways in which costs of resistance might increase with a host's breadth of resistance (i.e. the number of parasite genotypes and/or species that it successfully resists). First, resistance to an increasing number of parasite types might incur additive fitness costs, whereby each specific resistance carries its own costs that are compounded over time. This scenario might apply if resistance to one parasite does not correlate with resistance to another. Second, a diverse parasite assemblage could select for a more general resistance mechanism, which is itself more costly (but potentially less effective against a specific parasite) than single specific resistance mechanisms [21]. Notably, the existence of both specific and general resistance mechanisms could confound estimates of resistance costs against single parasites. This is because costly specific resistance and non-specific resistance are predicted to be negatively correlated: selection is unlikely to simultaneously favour both strategies. As such, high levels of specific resistance may appear to have fitness benefits in the absence of parasites against a background of hosts with more general resistance [21]. Thus, to fully appreciate the importance of fitness costs associated with resistance in shaping phenotypic diversity, we must examine resistance-fitness trade-offs in environments with more complex parasite-mediated selection.
One powerful way to explore the context-dependent nature of fitness costs is to use experimental evolution. This approach has provided clear evidence for costs of resistance in a number of systems [2,3,5–8,16,22] and allows for direct measurement of costs resulting either from pleiotropy or linkage and for distinction between genetic constraint and phenotypic correlation [23]. Interactions between bacteria and their viral parasites (phages) have proven particularly useful model systems for understanding host–parasite interactions over ecological and evolutionary timescales [24,25]. In these tightly paired interactions, phages bind to bacterial cells, inject their own genetic material and hijack the bacterial replication machinery in order to replicate. Lytic phages are obligate killers of their host cells, as they require cell lysis for the release of viral progeny and subsequent transmission. Therefore, they have the potential to impose strong selection on bacterial populations. Although phages are known to be abundant in many natural habitats [26], it is still unclear how important they may be as drivers of bacterial diversity. Laboratory studies have shown that phage populations can adapt rapidly and specifically to target bacterial hosts [27]; a process that is further shaped by heterogeneous environmental conditions [28–30]. However, bacterial resistance to co-occurring phages is widespread in natural populations [31], suggesting either that phage-mediated selection for host resistance is high or that phages are not adapting to bacterial hosts. This latter explanation is less likely given recent work showing phage adaptation to local bacterial populations in natural populations in the soil [32] and in the plant phyllosphere [33].
Bacterial resistance to phages can evolve via a number of different mechanisms, which vary in their specificity. For example, both clustered regularly interspaced short palindromic repeats (CRISPR)-mediated suppression of viral reproduction and modification of phage attachment sites are typically specific [34–36], whereas complete loss of receptors [22] or overproduction of exopolysaccharides, such as alginate [37], are likely to be more general. Many of these resistance mechanisms impose a significant fitness cost [22,38,39], but these costs can vary across environments and the degree of competition for resources [40,41]. Given the potential use of phages as biocontrol against pathogenic bacteria, a clear understanding of costs associated with evolved resistance could lead to more effective management strategies and the design of potent phage ‘cocktails’ against which resistance is too costly to maintain in the absence of treatment [42–44].
We use experimental evolution between the plant-pathogenic bacterium, Pseudomonas syringae, and naturally occurring lytic phages to test the prediction that hosts evolving with multiple parasites pay larger fitness costs (assayed as decreased population growth) for evolved resistance than hosts evolving with a single parasite. We find that while hosts in single-parasite environments are able to evolve resistance in an apparently cost-free fashion, hosts in multiple-parasite environments pay a significant cost of resistance that is not clearly associated with an increased cross-resistance to novel parasites.
2. Methods
(a). Experimental evolution
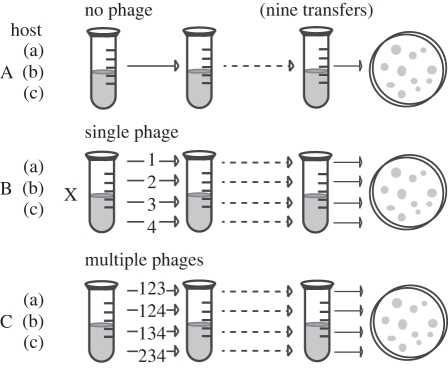
We examined the evolution of bacterial resistance to multiple parasitic bacteriophages using the plant-pathogenic bacterium, P. syringae pathovar (PV) tomato, and four lytic phages from environmental samples (isolated from disparate locations and sources and donated by OmniLytics Inc., Sandy, UT, USA). Pseudomonas syringae is a gram-negative, rod-shaped bacterium that has polar flagella and is known to induce disease symptoms on tomato plants. Replicate host lines of bacteria were generated from freezer stocks of three previously characterized P. syringae pv tomato strains: DC3000 [45], PT23 [46] and 08241 (from Christine Smart, Cornell University). For each of these, we isolated three individual colonies to generate replicate experimental populations that were initially genetically homogeneous. Each replicate population was then split into nine selection lines: one no-phage control, four single-phage treatments, and four multiple-phage treatments (figure 1).
Figure 1.
Experimental evolution methods. Three individual bacterium, representing each of three bacterial genotypes, were used to generate replicate populations experiencing one of three selection regimes (no-phage, single-phage or multiple-phage environment) over the course of approximately 130 bacterial generations. Numbers represent phage treatment (i.e. single phage or combinations of three out of the four phages).
We generated six independent phage inocula by first amplifying each phage clone through a separate strain of P. syringae pv tomato (A9 from Christine Smart, Cornell University) and then using this host to estimate phage densities of each inocula. This was done by spotting a dilution series of each phage inoculum in a grid formation onto a lawn of the host bacterium, grown in soft agar (King's medium B (KB); 10 g l–1 glycerol, 20 g l–1 proteose peptone no. 3 (Becton Dickinson UK Ltd.), 1.5 g l–1 K2HPO4 · 3H2O, 1.5 g l–1 MgSO4 · 7H2O and 0.6% w/v agar). Phage plaque formation within each spot was compared with bacterial growth across the lawn, and counted at the dilution for which individual plaques were visible. Using these estimates, we then diluted each inoculum to approximately 1000 plaque forming units per microlitre and measured the infectivity of each inoculum against a panel of 12 independent laboratory strains of P. syringae, representing six different pathovars (data not shown). Based on these infectivity profiles, we confirmed that the six phage inocula were phenotypically different from one another in terms of host range and that each was infective to the three focal bacterial genotypes used in experiment. Four of these phage inocula were then haphazardly chosen for use in experimental evolution (figure 1), while the other two were later used to estimate cross-resistance of each population to novel phages.
We initiated the experiment by adding 50 µl of each replicate bacterial host (grown overnight in KB broth at 28°C) into individual Eppendorf tubes containing either phage or phage-free buffer (M9; 1 mM thiamine hydrochloride, 0.4% glycerol, 0.2% casamino acids, 2 mM MgSO4, 0.1 mM CaCl2) and 1 ml of soft KB agar (0.6% w/v), cooled to 40°C. Tubes were vortexed and incubated for 48 h at 28°C. For the single-phage treatments, 3 µl of inocula was added and for the multiple-phage treatments, 1 µl of each of three inocula was added (according to treatment; figure 1). After 48 h, lines were transferred to new Eppendorf tubes, containing 50 µl of buffer and the appropriate phage treatment as described above, by taking five individual stabs of soft agar from each tube with a 100 µl pipette tip and adding it to buffer in the new tube. Cooled soft KB agar was then added and lines were vortexed and incubated for another 48 h. This process was repeated eight more times.
(b). Cross-inoculation experiment
We ended the experiment after the ninth experimental transfer (at approx. 130 bacterial generations). At that time, we sampled each line by taking five stabs, as described previously, and plated a dilution series of the bacteria on hard KB agar (1.2% w/v) to isolate 12 individual colonies (randomly chosen) from each replicate population. We then measured the resistance of six individual bacterial isolates (chosen at random from the 12 colonies) to each of the four phages used in the experiment, as well as the two additional phages to which none of the bacteria had been exposed (as described previously). Specifically, we sought to compare resistance to sympatric phages, with which a given bacterial population had been evolving, with resistance to allopatric phages to which the population had not previously been exposed. We measured resistance by examining growth of a small sample of each bacterium that had been streaked across a line of phage. Where growth was inhibited, we characterized the bacterial isolate as susceptible to the given phage. This allowed us to estimate the proportion of individuals within each population that had evolved resistance to each phage and to test for specific versus general resistance of bacterial lines.
(c). Bacterial fitness measures
At the beginning and again at the end of the experiment, we quantified growth of replicate bacterial populations in the presence and in the absence of each of the phages. We analysed fitness of each bacterial population in two ways: (i) using population growth rate (i.e. maximal rate of change in optical density (OD) during the log growth phase, Vmax (mOD min–1)) and (ii) estimating final bacterial densities after 24 h. This was done by taking optical density measurements at 600 nm using a microplate spectrophotometer (Molecular Devices SpectroMax M2e). Measurements were taken every 5 min at an incubation temperature of 24°C with 5 s shaking prior to each read over 24 h. For the ancestral growth experiment, we combined 10 µl of a diluted overnight (1 : 1000) of each ancestral bacterial strain and either buffer (no-phage control) or 3 µl of phage inocula to 150 µl of KB broth in a 96-well plate.
We measured growth rates of the evolved lines by inoculating into a single tube all 12 individually isolated bacterial colonies per line (to separate the bacteria from any phages left in the sample), growing them overnight in KB broth, and diluting each 1 : 1000 to initiate the growth experiments as described above. Growth plates were blocked such that one of each of the three replicate populations per bacterial strain and treatment was used in each run, with a total of three separate runs.
(d). Statistical analyses
All analyses were run using PASW Statistics v. 18, Release v. 18.0.3. We checked the suitability of our measure of quantitative resistance using Spearman rank order correlation to compare the proportion (out of six colonies tested) of resistant individuals from each experimental population with the growth rate of that population in the presence of sympatric phage (averaged across three sympatric phages for the multiple-phage treatment). We next compared evolved resistance across treatments using generalized linear models with binomial errors and controlling for overdispersion. We first examined the total resistance evolved to each of the six phages tested using the number of resistant bacterial colonies, out of the six tested, as the dependent variable and including as categorical factors in the model: bacterial genotype, experimental replicate nested within genotype, phage identity (i.e. the type of phage crossed with each bacterium), and selection regime. We then used planned contrasts, controlling for multiple comparisons using the sequential Sidak method, to compare the resistance evolved among the three selection regimes. We next examined the specificity of evolved resistance to sympatric versus allopatric phages using this model with the addition of phage sympatry as a categorical factor. The no-phage control populations were excluded from this latter analysis as they had no associated sympatric phages. For confirmation, we also ran both analyses as linear mixed models with proportion of resistant colonies as the dependent variable, experimental replicate included as a random factor, and bacterial strain treated as a fixed factor, as each represents an independently isolated laboratory strain and does necessarily represent natural diversity.
Finally, we compared the fitness of each population in the absence of phage by running general linear models across the three treatments for both continuous variables, growth rate and final density, using the mean of two runs. These analyses included bacterial genotype and phage treatment as fixed effects and replicate nested within genotype as a random effect. Planned comparisons among the selection regimes were run using least significant difference tests. In addition, we compared the breadth of resistance (i.e. the proportion of phages to which individuals from each line were resistant, averaged across individuals within a population and arcsine square root transformed) to population fitness using an analysis of covariance. Either population growth rate or final density was the dependent variable, bacterial genotype was a fixed effect, bacterial replicate was a random term, and breadth of resistance was the covariate.
3. Results
At the start of the experiment, the presence of phage significantly decreased the final density, but not the growth rate, of each ancestral bacterial population. There was no significant difference among the four phages in their effect on bacterial growth rate (see the electronic supplementary material for details; figure S2). The presence of phage in the environment also significantly changed the relative fitnesses of the three bacterial genotypes, whereby the ancestral strain that grew the best in the absence of phage did not have a competitive advantage in the presence of phage (electronic supplementary material, figure S1).
(a). Evolution of resistance to phages
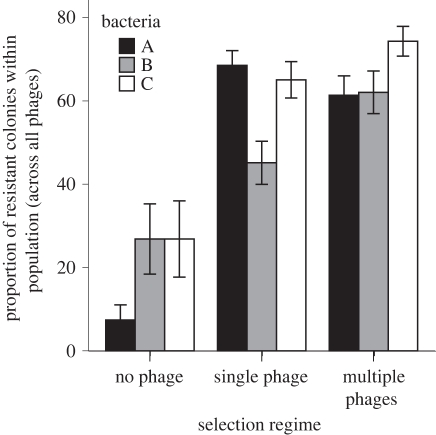
Over the course of the experiment all lines that had evolved in the presence of one or more phages showed increased resistance (figure 2). Our quantitative measure of resistance was a good predictor of how well each population grew in the presence of phage, as there was a strong positive correlation between the proportion of resistant individuals in a given population that were resistant to their sympatric phages and the final population density achieved in the presence of those phages (ρ = 0.477, n = 72, p < 0.001; excluding control populations because they did not have sympatric phages). The proportion of resistant bacteria was also positively correlated with the maximum growth rate of populations in the presence of sympatric phage (ρ = 0.545, n = 72, p < 0.001).
Figure 2.
Evolved resistance across replicate bacterial lines of each genotype according to selection regime. Six individual bacterial clones (i.e. colonies) from each bacterial population were streaked across a given phage inoculum and the mean proportion of resistant colonies per population is shown. Error bars represent ±1 s.e.m.
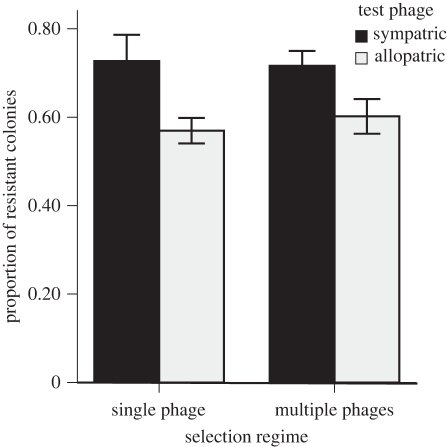
Bacterial populations under phage-mediated selection were significantly more resistant to phages than were the no-phage control lines, and the bacterial genotypes differed significantly in their response to selection (table 1 and figure 2). For those bacterial populations evolving with one or more phages, bacterial hosts evolved a specific resistance to their sympatric phages that was higher than the evolved resistance to allopatric phages (figure 3). However, there was no overall difference between bacteria from single- and multiple-phage treatments either for their overall resistance to phage or in terms of the specificity of their evolved resistance to sympatric versus allopatric phages (table 2). The conclusions drawn from both of these statistical analyses were not changed when conducted using general linear models with proportion of resistant colonies as the dependent variable.
Table 1.
Generalized linear model examining evolved resistance across experimental populations. (Resistance to each of six test phages was measured for six bacterial colonies per population, with contrasts comparing the three selection regimes (no phage, single phage and multiple phages). Significant effects are highlighted in bold.)
| χ2 | d.f. | p | |
|---|---|---|---|
| bacterial genotype | 8.282 | 2 | 0.016 |
| replicate (bacterial genotype) | 45.751 | 6 | <0.001 |
| phage identity | 44.885 | 5 | <0.001 |
| selection regime | 54.343 | 2 | <0.001 |
| no phage versus single phagea | 1 | <0.001 | |
| no phage versus multiple phagesa | 1 | <0.001 | |
| single phage versus multiple phagesa | 1 | 0.094 | |
| bacterial genotype × phage identity | 9.522 | 10 | 0.483 |
| bacterial genotype × selection regime | 15.712 | 4 | 0.003 |
| selection regime × phage identity | 8.227 | 10 | 0.607 |
aPlanned contrasts with sequential Sidak correction applied.
Figure 3.
Bacterial resistance to phages with which the bacterial lines were evolved (black, sympatric) and phages not present during evolution (grey, allopatric). Control populations are not included, as they had no associated sympatric phages. The mean proportion of resistant colonies per population is shown. Error bars represent ±1 s.e.m.
Table 2.
Generalized linear model examining specificity of evolved resistance for populations evolved with phages (i.e. excluding the control populations). (Resistance to each of six test phages was measured for six bacterial colonies per population. There was no interaction effect between selection regime and phage sympatry, indicating that both treatments evolved specific resistance to their sympatric phage(s). Significant effects are highlighted in bold.)
| χ2 | d.f. | p | |
|---|---|---|---|
| bacterial genotype | 12.048 | 2 | 0.002 |
| replicate (bacterial genotype) | 37.825 | 6 | <0.001 |
| phage identity | 99.045 | 5 | <0.001 |
| selection regime | 0.283 | 1 | 0.594 |
| phage sympatry | 4.567 | 1 | 0.033 |
| phage identity × phage sympatry | 0.423 | 3 | 0.935 |
| bacterial genotype × phage sympatry | 2.843 | 2 | 0.241 |
| selection regime × phage sympatry | 0.721 | 1 | 0.396 |
(b). Costs of resistance examined in the absence of phage
We tested for costs associated with evolved resistance by comparing population growth of all bacterial lines in a phage-free environment and found that bacterial fitness differed among the three selection regimes (figure 4). There was a significant effect of both bacterial genotype (F2,10 = 9.318, p = 0.005) and selection regime (F2,66 = 7.202, p = 0.001) on the final population density achieved, but no interaction between the two (F4,66 = 1.798, p = 0.140). The bacterial populations evolved in the presence of multiple phages grew less well than either the control populations (p = 0.004) or the populations evolved in the presence of single phages (p = 0.002). There was no significant difference between the no-phage and single-phage treatments (p = 0.313). Similarly, there was a significant effect of bacterial genotype (F2,17 = 18.050, p < 0.001) and selection regime (F2,66 = 6.041, p = 0.004) on bacterial growth rate, but no interaction between the two (F4,66 = 1.078, p = 0.374). Again, bacterial populations evolved in the presence of multiple phages grew less well than either the control populations (p = 0.021) or the populations evolved in the presence of single phages (p = 0.002). There was no significant difference between the no-phage and single-phage treatments (p = 0.713). Thus, both estimates of population fitness suggest that the no-phage and single-phage treatments grew just as well as one another but better than populations evolving with multiple phages.
Figure 4.
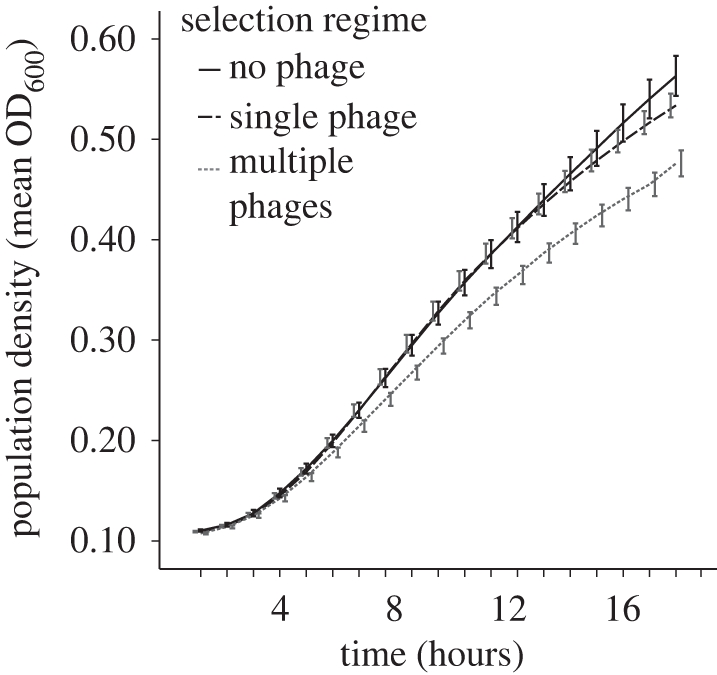
Population growth, in the absence of phage, of experimental strains at the end of the experiment. Bacterial density was measured every 5 min at an optical density (OD) of 600 nm, and mean growth over each hour (±1 s.e.m.) for 20 h is shown.
Given the degree of cross-resistance attained during experimental coevolution in both single- and multiple-phage treatments, we did not expect a simple relationship between the breadth of resistance and the cost associated with resistance. Nevertheless, we directly tested for this relationship by comparing the breadth of resistance, measured as the proportion of phages (out of the six tested) to which each bacterial line was resistant, with bacterial fitness. Bacteria with greater breadth of resistance were significantly less fit with regard to final population density than those with narrower resistance (F1,69 = 5.226, p = 0.025) and this effect did not differ among bacterial genotypes (interaction between genotype and breadth of resistance, F2,69 = 2.317, p = 0.106). The relationship between breadth of resistance and population growth rate was also significant (F1,69 = 4.674, p = 0.034; interaction between genotype and breadth of resistance, F2,69 = 1.437, p = 0.245). Hence, in line with the previous results, there was evidence for a negative association between growth rate and breadth of resistance.
4. Discussion
We set out to test whether evolving resistance to multiple parasites simultaneously was less likely and/or more costly than evolving resistance to a single parasite. Using experimental evolution of bacteria and phages, we found that hosts in heterogeneous parasite environments pay a higher cost for their evolved resistance than hosts in single-parasite environments (figure 4), without gaining an increased general resistance against novel parasites (figure 3). Bacterial populations from multiple-phage environments were able to evolve specific resistance to their sympatric phages just as readily as populations from single-phage environments, and individuals from both treatments showed a strong degree of cross-resistance to novel, allopatric phages (figure 3). Thus, although the bacteria from multiple-phage environments evolved a somewhat greater breadth of resistance, and may have paid accumulating costs of specific resistances, they were no more resistant to novel phages than those from single-phage environments. This suggests that phage-mediated selection led simultaneously to specific resistance against local parasites and to an increased general resistance, which may be less effective overall but that inadvertently protects against novel phages. This specific resistance supports previous evidence that parasites are important for shaping diversity as host populations diverge from one another in response to local parasite-mediated selection [4,6,7,12,22,47].
There is a great deal of empirical work documenting that bacterial resistance to local phages can evolve over very short timescales [27,30,35]. Although the exact mechanism of resistance in our experimental populations is unknown, one likely explanation given the high degree of cross-resistance observed is that the phages are binding to the same molecule on the bacterial surface. Thus, selection for resistance to one phage-type would lead to alteration or loss of specific receptors and, as a consequence, also confer resistance to the other phages [13,24,35]. However, given that the cross-resistance observed was not as strong as the specific resistance evolved, the phages must be binding to the receptors in slightly different ways. Alternatively, phage-mediated selection might have led to both a general resistance mechanism (such as alginate production [37]) that confers resistance to allopatric phages and a more specific resistance (such as restriction modification or CRISPR-related interference [34]) that confers resistance to the sympatric phage.
The fact that we only observed fitness costs associated with resistance in multiple-phage environments is consistent with the accumulation of multiple costly resistance mutations, each conferring specific resistance to sympatric phages. This interpretation is also supported by the observed negative correlation between breadth of resistance and population fitness. However, an alternative explanation is that decreased population size of bacterial hosts in multiple-phage environments led to a decreased efficacy of selection (i.e. lack of compensatory mutation owing to fewer mutational events and increased drift). A similar argument has been used to explain the decreased cost of antibiotic resistance in bacterial populations with higher migration rates [48]. Either way, the costs are associated with evolutionary change in response to parasites and, given that each population was started from a single individual bacterium, represent the effect of mutational change rather than selection on standing variation. Previous work examining epistatic interactions among bacterial resistance mutations in Escherichia coli evolved against each of two phage types found that resistance to both phages simultaneously was more costly than resistance to each phage individually, but less costly than expected under an additive model (although this result was highly dependent on the assay environment) [38]. Similarly, previous work on the evolution of multiple antibiotic resistance has documented strong synergistic epistasis among alleles that confer resistance in E. coli [49] and antagonistic epistasis between resistance mutations in Pseudomonas aeruginosa [50].
There has been a large focus on the fitness costs associated with resistance for two primary reasons. First, costly resistance would help explain the tremendous diversity of defence strategies seen in natural populations and the often polymorphic resistance phenotypes observed against local parasites [51,52], and second, understanding the costs of resistance to parasites (and to phages in particular) can help inform predictions about the evolution of a pathogen's resistance to drug treatment. The use of phage cocktails to treat disease has received a good deal of attention [43,53] and there is some evidence that using multiple phages reduces bacterial population sizes more effectively than single-phage treatment [42,44]. Our results add to this by suggesting that, although resistance is just as likely to evolve against multiple phages, the use of phage cocktails could select for resistant mutants that are less fit and thus more likely to be lost over time (especially once treatment is removed) than the use of single phages. In addition, we found no evidence that using multiple phages would select for a more general resistance mechanism against future, novel phages. Together with previous work showing how variation in host resistance is shaped for populations coevolving as a geographical mosaic [30,54], it is clear that evolution of host resistance will depend on environmental heterogeneity, the diversity and strength of antagonistic pressures faced by local populations, migration rates and the local diversity of parasite populations.
Not all studies examining costs of resistance have found evidence for them; a review of resistance trade-offs across 88 plant studies found that only about half of the data suggest costs of resistance [19] and a review across 31 arthropod studies found that less than two-third of studies document significant costs of resistance [20]. The incongruent results among studies could be explained by the presence of relatively cost-free mechanisms of resistance in some systems, the difficulty in measuring fitness under laboratory conditions or the context-dependent nature of trade-offs. For example, some studies find evidence for costs of resistance when examining relative fitness (e.g. comparing population growth of resistant and susceptible genotypes when put in direct competition), but no evidence for costs associated when comparing absolute fitness of the two [5,13]. In addition, mutations conferring resistance might only be costly in the short-term, becoming relatively cost-free as compensatory mutations accumulate [39,40]. Our work suggests a third possibility: that costs associated with resistance to a single parasite might not be generalizable to more complex parasite pressures, and thus that trade-offs might be more common than previously thought once natural parasite diversity is incorporated.
In addition to the evolutionary implications of these results, they support a direct effect of phages on relative fitness of bacterial hosts; the three bacterial genotypes chosen for this experiment initially differed in terms of their fitness relative to one another, with bacterial genotype ‘C’ having a clear advantage in the absence, but not the presence, of phages (electronic supplementary material, figure S1). This adds to a long-standing body of work on how parasites can alter competitive interactions among host genotypes and species [55,56] and be critical determinants of population and community composition [39,57,58]. In addition to this direct effect of phages on relative fitness, our results also indicate that the outcome of phage-mediated selection differed among the bacterial genotypes examined, and therefore that phages can play an important role in shaping competitive interactions among hosts over coevolutionary timescales.
In conclusion, our work adds to the growing evidence that costs of resistance are context-dependent by demonstrating a key role of parasite diversity in shaping the fitness-resistance trade-off. Evolutionary costs have previously been shown to vary across environments, such as differing levels of resource availability [13], increasing biotic complexity [59], extreme weather conditions [18] or across temperatures [41,60]. Our results illustrate that the (co)evolutionary history of a population can also have significant impact on the observed trade-off between resistance to parasites and fitness. The increased cost of resistance was clearly linked to the evolutionary history of the bacteria (i.e. whether the bacteria was evolving with single or multiple phages) but was not as clearly related to the absolute degree of resistance attained given the large amount of cross-resistance observed. Thus, studies in which coevolutionary history is not taken into account might miss an important trade-off, as it is masked by the contemporary ecological pattern. Taken together, our results show that resistance against multiple phages is just as likely to occur as resistance against single phages, is not clearly associated with an increased general resistance, and may in fact carry higher fitness costs to the host.
Acknowledgements
We thank B. Carter for technical assistance with spectrophotometry, G. Preston for providing the bacterial strains, and OmniLytics, Inc. for providing the environmental phage isolates, F. Inglis, C. Metcalf, L. Ross and C. Schwind for valuable discussion, and the associate editor and two reviewers for helpful comments. Funding for the project was provided by the US National Science Foundation (B.K., NSF-DEB 0754399). A.B. also acknowledges support from the European Research Council.
References
- 1.Bérénos C., Schmid-Hempel P., Wegner M. K. 2009. Evolution of host resistance and trade-offs between virulence and transmission potential in an obligately killing parasite. J. Evol. Biol. 22, 2049–2056 10.1111/j.1420-9101.2009.01821.x (doi:10.1111/j.1420-9101.2009.01821.x) [DOI] [PubMed] [Google Scholar]
- 2.Boots M., Begon M. 1993. Trade-offs with resistance to a granulosis virus in the Indian meal moth, examined by a laboratory evolution experiment. Funct. Ecol. 7, 528–534 10.2307/2390128 (doi:10.2307/2390128) [DOI] [Google Scholar]
- 3.Fuxa J. R., Richter A. R. 1989. Reversion of resistance by Spodoptera frugiperda to nuclear polyhedrosis virus. J. Invertebr. Pathol. 53, 52–56 10.1016/0022-2011(89)90073-6 (doi:10.1016/0022-2011(89)90073-6) [DOI] [Google Scholar]
- 4.Koskella B., Lively C. M. 2007. Advice of the rose: experimental coevolution of a trematode parasite and its snail host. Evolution 61, 152–159 10.1111/j.1558-5646.2007.00012.x (doi:10.1111/j.1558-5646.2007.00012.x) [DOI] [PubMed] [Google Scholar]
- 5.Kraaijeveld A. R., Godfray H. C. J. 1997. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature 389, 278–280 10.1038/38483 (doi:10.1038/38483) [DOI] [PubMed] [Google Scholar]
- 6.Lohse K., Gutierrez A., Kaltz O. 2006. Experimental evolution of resistance in Paramecium caudatum against the bacterial parasite Holospora undulate. Evolution 60, 1177–1186 10.1111/j.0014-3820.2006.tb01196.x (doi:10.1111/j.0014-3820.2006.tb01196.x) [DOI] [PubMed] [Google Scholar]
- 7.Schulte R. D., Makus C., Hasert B., Michiels N. K., Schulenburg H. 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl Acad. Sci. USA 107, 7359–7364 10.1073/Pnas.1003113107 (doi:10.1073/Pnas.1003113107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster J. P., Woolhouse M. E. J. 1999. Cost of resistance: relationship between reduced fertility and increased resistance in a Schistosoma host–parasite system. Proc. R. Soc. Lond. B 266, 391–396 10.1098/rspb.1999.0650 (doi:10.1098/rspb.1999.0650) [DOI] [Google Scholar]
- 9.Bonneaud C., Balenger S. L., Russell A. F., Zhang J., Hill G. E., Edwards S. V. 2011. Rapid evolution of disease resistance is accompanied by functional changes in gene expression in a wild bird. Proc. Natl Acad. Sci. USA 108, 7866–7871 10.1073/pnas.1018580108 (doi:10.1073/pnas.1018580108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan A. B., Little T. J. 2007. Parasite-driven genetic change in a natural population of Daphnia. Evolution 61, 796–803 10.1111/j.1558-5646.2007.00072.x (doi:10.1111/j.1558-5646.2007.00072.x) [DOI] [PubMed] [Google Scholar]
- 11.Hasu T., Benesh D. P., Valtonen E. T. 2009. Differences in parasite susceptibility and costs of resistance between naturally exposed and unexposed host populations. J. Evol. Biol. 22, 699–707 10.1111/j.1420-9101.2009.01704.x (doi:10.1111/j.1420-9101.2009.01704.x) [DOI] [PubMed] [Google Scholar]
- 12.Shapiro O. H., Kushmaro A., Brenner A. 2010. Bacteriophage predation regulates microbial abundance and diversity in a full-scale bioreactor treating industrial wastewater. ISME J. 4, 327–336 10.1038/ismej.2009.118 (doi:10.1038/ismej.2009.118) [DOI] [PubMed] [Google Scholar]
- 13.Lenski R. E., Levin B. R. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125, 585–602 10.1086/284364 (doi:10.1086/284364) [DOI] [Google Scholar]
- 14.Langand J., Jourdane J., Coustau C., Delay B., Morand S. 1998. Cost of resistance, expressed as a delayed maturity, detected in the host–parasite system Biomphalaria glabrata/Echinostoma caproni. Heredity 80, 320–325 10.1046/j.1365-2540.1998.00291.x (doi:10.1046/j.1365-2540.1998.00291.x) [DOI] [Google Scholar]
- 15.Parker M. A. 1991. Local genetic differentiation for disease resistance in a selfing annual. Biol. J. Linn. Soc. 42, 337–349 10.1111/j.1095-8312.1991.tb00567.x (doi:10.1111/j.1095-8312.1991.tb00567.x) [DOI] [Google Scholar]
- 16.Alexander H. M., Antonovics J. 1995. Spread of anther-smut disease (Ustilago violacea) and character correlations in a genetically variable experimental population of Silene Alba. J. Ecol. 83, 783–794 10.2307/2261415 (doi:10.2307/2261415) [DOI] [Google Scholar]
- 17.Verhulst S., Dieleman S. J., Parmentier H. K. 1999. A tradeoff between immunocompetence and sexual ornamentation in domestic fowl. Proc. Natl Acad. Sci. USA 96, 4478–4481 10.1073/pnas.96.8.4478 (doi:10.1073/pnas.96.8.4478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham A. L., Hayward A. D., Watt K. A., Pilkington J. G., Pemberton J. M., Nussey D. H. 2010. Fitness correlates of heritable variation in antibody responsiveness in a wild mammal. Science 330, 662–665 10.1126/science.1194878 (doi:10.1126/science.1194878) [DOI] [PubMed] [Google Scholar]
- 19.Bergelson J., Purrington C. B. 1996. Surveying patterns in the cost of resistance in plants. Am. Nat. 148, 536–558 10.1086/285938 (doi:10.1086/285938) [DOI] [Google Scholar]
- 20.Labbe P., Vale P., Little T. 2010. Successfully resisting a pathogen is rarely costly in Daphnia magna. BMC Evol. Biol. 10, 355. 10.1186/1471-2148-10-355 (doi:10.1186/1471-2148-10-355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank S. A. 2000. Specific and non-specific defense against parasitic attack. J. Theor. Biol. 202, 283–304 10.1006/jtbi.1999.1054 (doi:10.1006/jtbi.1999.1054) [DOI] [PubMed] [Google Scholar]
- 22.Brockhurst M. A., Buckling A., Rainey P. B. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc. R. Soc. B 272, 1385–1391 10.1098/rspb.2005.3086 (doi:10.1098/rspb.2005.3086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong D., Pai A., Yan G. 2005. Costly resistance to parasitism: evidence from simultaneous quantitative trait loci mapping for resistance and fitness in Tribolium castaneum. Genetics 169, 2127–2135 10.1534/genetics.104.038794 (doi:10.1534/genetics.104.038794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohannan B. J. M., Lenski R. E. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377 10.1046/j.1461-0248.2000.00161.x (doi:10.1046/j.1461-0248.2000.00161.x) [DOI] [Google Scholar]
- 25.Buckling A., Maclean R. C., Brockhurst M. A., Colegrave N. 2009. The Beagle in a bottle. Nature 457, 824–829 10.1038/Nature07892 (doi:10.1038/Nature07892) [DOI] [PubMed] [Google Scholar]
- 26.Chibani-Chennoufi S., Bruttin A., Dillmann M.-L., Brussow H. 2004. Phage–host interaction: an ecological perspective. J. Bacteriol. 186, 3677–3686 10.1128/jb.186.12.3677-3686.2004 (doi:10.1128/jb.186.12.3677-3686.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckling A., Rainey P. B. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B 269, 931–936 10.1098/rspb.2001.1945 (doi:10.1098/rspb.2001.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brockhurst M., Buckling A., Rainey P. B. 2006. Spatial heterogeneity and the stability of host–parasite coexistence. J. Evol. Biol. 19, 374–379 10.1111/j.1420-9101.2005.01026.x (doi:10.1111/j.1420-9101.2005.01026.x) [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Pascua L. D. C., Buckling A. 2008. Increasing productivity accelerates host–parasite coevolution. J. Evol. Biol. 21, 853–860 10.1111/j.1420-9101.2008.01501.x (doi:10.1111/j.1420-9101.2008.01501.x) [DOI] [PubMed] [Google Scholar]
- 30.Forde S. E., Thompson J. N., Bohannan B. J. M. 2004. Adaptation varies through space and time in a coevolving host–parasitoid interaction. Nature 431, 841–844 10.1038/nature02906 (doi:10.1038/nature02906) [DOI] [PubMed] [Google Scholar]
- 31.Waterbury J. B., Valois F. W. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59, 3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos M., Birkett P. J., Birch E., Griffiths R. I., Buckling A. 2009. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 325, 833. 10.1126/science.1174173 (doi:10.1126/science.1174173) [DOI] [PubMed] [Google Scholar]
- 33.Koskella B., Thompson J. N., Preston G. M., Buckling A. 2011. Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria. Am. Nat. 177, 440–451 10.1086/658991 (doi:10.1086/658991) [DOI] [PubMed] [Google Scholar]
- 34.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 10.1126/science.1138140 (doi:10.1126/science.1138140) [DOI] [PubMed] [Google Scholar]
- 35.Lenski R. E. 1988. Dynamics of interactions between bacteria and virulent bacteriophage. Adv. Microb. Ecol. 10, 1–44 [Google Scholar]
- 36.Qimron U., Marintcheva B., Tabor S., Richardson C. C. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl Acad. Sci. USA 103, 19 039–19 044 10.1073/pnas.0609428103 (doi:10.1073/pnas.0609428103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammad A. M. M. 1998. Evaluation of alginate-encapsulated Azotobacter chroococcum as a phage-resistant and an effective inoculum. J. Basic Microbiol. 38, 9–16 (doi:10.1002/(SICI)1521-4028(199803)38:1<9::AID-JOBM9>3.0.CO;2-4) [DOI] [Google Scholar]
- 38.Bohannan B. J. M., Travisano M., Lenski R. E. 1999. Epistatic interactions can lower the cost of resistance to multiple consumers. Evolution 53, 292–295 10.2307/2640942 (doi:10.2307/2640942) [DOI] [PubMed] [Google Scholar]
- 39.Lenski R. E. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. II. Compensation for maladaptive effects associated with resistance to virus T4. Evolution 42, 433–440 10.2307/2409029 (doi:10.2307/2409029) [DOI] [PubMed] [Google Scholar]
- 40.Lennon J.T., Khatana S., Marston M., Martiny J. 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 1, 300–312 [DOI] [PubMed] [Google Scholar]
- 41.Quance M. A., Travisano M. 2009. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Eschericha coli. Evolution 63, 1406–1416 10.1111/j.1558-5646.2009.00654.x (doi:10.1111/j.1558-5646.2009.00654.x) [DOI] [PubMed] [Google Scholar]
- 42.Heringa S. D., Kim J. K., Jiang X., Doyle M. P., Erickson M. C. 2010. Use of a mixture of bacteriophages for biological control of Salmonella enterica strains in compost. Appl. Environ. Microbiol. 76, 5327–5332 10.1128/AEM.00075-10 (doi:10.1128/AEM.00075-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin B. R., Bull J. J. 2004. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2, 166–173 10.1038/nrmicro822 (doi:10.1038/nrmicro822) [DOI] [PubMed] [Google Scholar]
- 44.O'Flynn G., Ross R. P., Fitzgerald G. F., Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157: H7. Appl. Environ. Microbiol. 70, 3417–3424 10.1128/AEM.70.6.3417-3424.2004 (doi:10.1128/AEM.70.6.3417-3424.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buell C. R., et al. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA 100, 10 181–10 186 10.1073/pnas.1731982100 (doi:10.1073/pnas.1731982100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bender C. L., Cooksey D. A. 1986. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J. Bacteriol. 165, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buckling A., Rainey P. B. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420, 496–499 10.1038/Nature01164 (doi:10.1038/Nature01164) [DOI] [PubMed] [Google Scholar]
- 48.Perron G. G., Gonzalez A., Buckling A. 2007. Source–sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. R. Soc. B 274, 2351–2356 10.1098/rspb.2007.0640 (doi:10.1098/rspb.2007.0640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trindade S., Sousa A., Xavier K. B., Dionisio F., Ferreira M. G., Gordo I. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5, e1000578. 10.1371/journal.pgen.1000578 (doi:10.1371/journal.pgen.1000578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward H., Perron G. G., Maclean R. C. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22, 997–1003 10.1111/j.1420-9101.2009.01712.x (doi:10.1111/j.1420-9101.2009.01712.x) [DOI] [PubMed] [Google Scholar]
- 51.Antonovics J., Thrall P. H. 1994. The cost of resistance and the maintenance of genetic polymorphism in host–pathogen systems. Proc. R. Soc. Lond. B 257, 105–110 10.1098/rspb.1994.0101 (doi:10.1098/rspb.1994.0101) [DOI] [Google Scholar]
- 52.Bowers R. G., Boots M., Begon M. 1994. Life-history trade-offs and the evolution of pathogen resistance: competition between host strains. Proc. R. Soc. Lond. B 257, 247–253 10.1098/rspb.1994.0122 (doi:10.1098/rspb.1994.0122) [DOI] [PubMed] [Google Scholar]
- 53.Jones J. B., Jackson L. E., Balogh B., Obradovic A., Iriarte F. B., Momol M. T. 2007. Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 45, 245–262 10.1146/annurev.phyto.45.062806.094411 (doi:10.1146/annurev.phyto.45.062806.094411) [DOI] [PubMed] [Google Scholar]
- 54.Vogwill T., Fenton A., Buckling A., Hochberg M. E., Brockhurst M. A. 2009. Source populations act as coevolutionary pacemakers in experimental selection mosaics containing hotspots and coldspots. Am. Nat. 173, E171–E176 10.1086/597374 (doi:10.1086/597374) [DOI] [PubMed] [Google Scholar]
- 55.Hatcher M. J., Dick J. T. A., Dunn A. M. 2006. How parasites affect interactions between competitors and predators. Ecol. Lett. 9, 1253–1271 10.1111/j.1461-0248.2006.00964.x (doi:10.1111/j.1461-0248.2006.00964.x) [DOI] [PubMed] [Google Scholar]
- 56.Price P. W., Westoby M., Rice B., Atsatt P. R., Fritz R. S., Thompson J. N., Mobley K. 1986. Parasite mediation in ecological interactions. Annu. Rev. Ecol. Syst. 17, 487–505 10.1146/annurev.es.17.110186.002415 (doi:10.1146/annurev.es.17.110186.002415) [DOI] [Google Scholar]
- 57.Joo J., Gunny M., Cases M., Hudson P., Albert R., Harvill E. 2006. Bacteriophage-mediated competition in Bordetella bacteria. Proc. R. Soc. B 273, 1843–1848 10.1098/rspb.2006.3512 (doi:10.1098/rspb.2006.3512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohannan B. J. M., Lenski R. E. 2000. The relative importance of competition and predation varies with productivity in a model community. Am. Nat. 156, 329–340 10.1086/303393 (doi:10.1086/303393) [DOI] [PubMed] [Google Scholar]
- 59.Gómez P., Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332, 106–109 10.1126/science.1198767 (doi:10.1126/science.1198767) [DOI] [PubMed] [Google Scholar]
- 60.Burdon J. J., Müller W. J. 1987. Measuring the cost of resistance to Puccinia coronata Cda in Avena fatua L. J. Appl. Ecol. 24, 191–200 10.2307/2403797 (doi:10.2307/2403797) [DOI] [Google Scholar]