Abstract
Acute inhibition is a powerful technique to test proteins for direct roles and order their activities in a pathway, but as a general gene-based strategy, it is mostly unavailable in mammalian systems. As a consequence, the precise roles of proteins in membrane trafficking have been difficult to assess in vivo. Here we used a strategy based on a genetically encoded fluorescent protein that generates highly localized and damaging reactive oxygen species to rapidly inactivate exit from the endoplasmic reticulum (ER) during live-cell imaging and address the long-standing question of whether the integrity of the Golgi complex depends on constant input from the ER. Light-induced blockade of ER exit immediately perturbed Golgi membranes, and surprisingly, revealed that cis-Golgi-resident proteins continuously cycle to peripheral ER-Golgi intermediate compartment (ERGIC) membranes and depend on ER exit for their return to the Golgi. These experiments demonstrate that ER exit and extensive cycling of cis-Golgi components to the cell periphery sustain the mammalian Golgi complex.
Key words: Golgi, Endoplasmic reticulum, Killer Red, Membrane trafficking
Introduction
The mammalian Golgi complex is composed of stacks of flattened cisternae linked through tubules to form a contiguous juxtanuclear ribbon. The stacked cisternae reflect the compartmentalization of Golgi enzymes for the processing of transiting cargo (Mellman and Simons, 1992; Puthenveedu and Linstedt, 2005). Cargo movement through the stack has been variously suggested to occur in vesicles, in maturing cisternae, or by tubular connections between cisternae (Malhotra et al., 1989; Mollenhauer and Morre, 1991; Glick et al., 1997; Allan and Balch, 1999; Pelham and Rothman, 2000; Trucco et al., 2004). Although not exclusive of other processes, maturation has gained recent support (Emr et al., 2009). This model starts with the creation of cargo-containing cis-Golgi cisternae. The cargo stays within these cisternae and is sequentially processed when the cis enzymes are replaced with medial enzymes, which are subsequently replaced with trans enzymes. As cisternae progress through the Golgi stack, this change in enzyme composition is mediated by recycling vesicles. For example, cis enzymes are removed by recycling vesicles that bud from cis cisternae, whereas medial enzymes are delivered by fusion of another set of recycling vesicles, which originate from maturing medial cisternae. Thus formation of new cis cisternae is predicted to rely on both input from the ER and the recycling of cis-Golgi residents to a pre-Golgi compartment. The extent of Golgi residents cycling to the ER remains controversial (Cole et al., 1998; Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001; Pecot and Malhotra, 2004; Rhee et al., 2005; Pecot and Malhotra, 2006). Further, if Golgi residents cycle, it is unclear whether they do so in the same vesicles as well-known recycling proteins such as the KDEL receptor and members of the p24 family (Martinez-Menarguez et al., 1999; Lanoix et al., 2001; Malsam et al., 2005). Although yeast Golgi cisternae have been visualized undergoing the predicted composition changes (Losev et al., 2006; Matsuura-Tokita et al., 2006; Glick and Nakano, 2009) and mammalian Golgi cisternae support movement of large cargo trapped inside (Bonfanti et al., 1998), the role of constant membrane input from the ER to form new cisternae and the existence of robust enzyme cycling between cisternae have not been convincingly demonstrated. This is probably due to temporal and spatial resolution constraints. In mammalian systems, few approaches allow acute inactivation and as a consequence the immediate effects of a blockade of membrane trafficking are largely unknown. Similarly, the reason that recycling vesicles have not been directly imaged might be because available live-cell imaging approaches cannot resolve vesicles within the short distances of their travel between cisternae.
Improvements in fluorescence microscopy offer an exciting avenue to address these shortcomings, particularly when combined with acute manipulations of the underlying molecular machinery. Therefore, we sought a technique for rapid protein inactivation while carrying out live-cell imaging experiments to better understand Golgi dynamics. Chromophores can inactivate proteins upon illumination by producing reactive oxygen species that can generate adducts, break peptide bonds and induce crosslinking (Jay, 1988; Liao et al., 1994; Surrey et al., 1998). Further, with a half maximal reactive distance of approximately 4 nm, the damage can be largely limited to a target protein and its interacting partners (Bulina et al., 2006). This approach has been of limited value because targeting proteins of interest mostly relied on microinjection of fluorophore-conjugated antibodies (Surrey et al., 1998; Jay and Sakurai, 1999). However, the development of Killer Red, a derivative of the Hydrazoa jellyfish chromoprotein anm2CP that generates at least 1000-fold increase in reactive oxygen species over green fluorescence protein (GFP), presents the possibility of simply expressing Killer Red fusion proteins (Bulina et al., 2006; Carpentier et al., 2009; Pletnev et al., 2009; Serebrovskaya et al., 2009; Destaing et al., 2010; Teh et al., 2010).
Here, we used Killer Red to impose an abrupt block in ER export and determine its effect on the Golgi complex. Our results indicate that the Golgi is sustained by both the constant input of membrane from the ER and a surprising level of peripheral cycling of cis-Golgi components.
Results
Targeted acute inactivation blocks ER exit
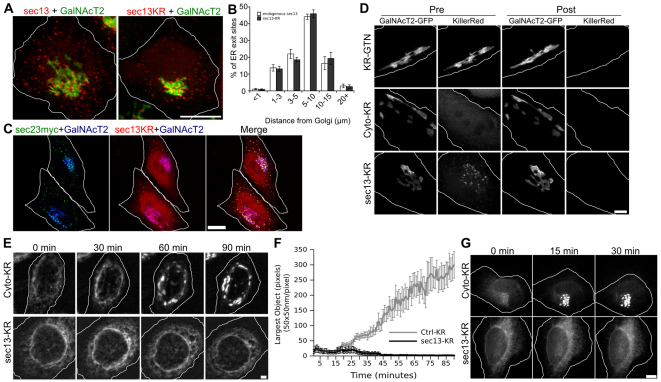
To impose an acute block of ER exit during live-cell imaging, we fused Killer Red to the coatomer protein (COP) II coat subunit Sec13 (Sec13–KR). As controls, we also generated a cytosolic version of Killer Red (Cyto–KR) and a version fused to the Golgi-localizing membrane anchor of giantin (KR–GTN). The constructs were expressed in cells expressing a GFP-tagged version of the Golgi enzyme N-acetylgalactosaminyl transferase-2 (GalNAcT2–GFP), which localizes to all Golgi cisternae (Storrie et al., 1998). Upon expression, the localization of Sec13–KR was similar to endogenous Sec13 in both its appearance (Fig. 1A) and in its quantified distribution relative to the Golgi (Fig. 1B). Further, Sec13–KR colocalized with the COPII adaptor Sec23, confirming its localization at ER-exit sites (Fig. 1C). Experiments were restricted to cells expressing Killer Red fusion proteins at comparable levels to GalNAcT2–GFP and with proper localization (Fig. 1D, pre-bleach). Nevertheless, increased levels of protein expression (up to 50% higher expression by fluorescence intensity) were non-toxic and yielded similar results. The antioxidant Trolox was added as a quenching agent to further reduce the likelihood of off-target effects (Bisby et al., 1999). Trolox concentration and irradiation time and power were optimized using the Golgi-localized control construct to achieve bleaching without affecting Golgi morphology. As shown, illumination of the entire cell with a mercury arc lamp through a 535–585 nm filter for 30 seconds at 2 W/cm2 in 375 μM Trolox bleached the Killer Red fluorescence of each target protein whereas the GalNAcT2–GFP fluorescence remained (Fig. 1D, post-bleach). Because Sec13 functions in a complex, there is a reasonable expectation that bleaching would damage not only the transfected construct, but also its endogenous partners, including Sec31 (Bulina et al., 2006). Previous work using knockdown of Sec13 failed to block ER exit suggesting the importance of complete inactivation of the Sec13–Sec31 complex (Townley et al., 2008).
Fig. 1.
Acute inactivation at the onset of Golgi assembly. (A) HeLa cells stably expressing GalNAcT2–GFP as a Golgi marker were fixed and stained with antibodies against Sec13 (left) or transiently transfected with Sec13–KR and fixed. Endogenous Sec13 and Sec13KR expression show similar patterns of ER exit sites. Scale bar: 10 μm. (B) Distribution of these exit sites was quantified by distance of each site from the center of the Golgi. The percentage of total ER exit sites within each radius is shown for both proteins. (C) Cells were transiently co-transfected with Sec13–KR and Sec23–Myc and fixed and stained with anti-Myc antibodies. Scale bar: 10 μm. (D) Pre- and post-bleach images for the indicated Killer Red constructs. HeLa cells stably expressing GalNacT2–GFP as a Golgi marker were transiently transfected with Killer Red constructs and, after 2 days, were placed in medium containing 375 μM Trolox. Cells were selected based on comparable Killer Red and GalNacT2–GFP fluorescence signals and proper localization of the constructs. Pre-bleach images of the selected cells were acquired in each channel and then the cells were irradiated with 535–580 nm light at an intensity of 2 W/cm2 for 30 seconds followed by the immediate acquisition of post-bleach images in each channel. Scale bar: 2 μm. (E) Time course of Golgi assembly after bleaching of the indicated Killer Red constructs. Cells were transfected with Killer Red constructs and, after 2 days, GalNAcT2–GFP was redistributed to the ER by exposure to 2 μg/ml brefeldin A for 15 minutes, followed by washing in PBS and immediate mounting in imaging medium for a 30 seconds bleaching of the Killer Red fluorescence. Imaging of GalNAcT2–GFP was then at 1 minute intervals for 90 minutes. Max value Z-projections are shown for the indicated time points and the corresponding full time course is shown in supplementary material Movie 1. Scale bar: 2 μm. (F) Golgi assembly was measured for the indicated constructs by determining the largest fluorescent object present at each time point with the threshold set above the level of GalNAcT2–GFP fluorescence in the ER (n=10, mean ± s.e.m.). (G) Time course of VSVG–GFP transit from the ER to Golgi initiated by temperature shift immediately before bleaching of the indicated Killer Red constructs. Imaging of VSVG–GFP was at 1 minute intervals for 90 minutes. Max value Z-projections are shown for the indicated time points. Scale bar: 2 μm.
To test the effectiveness of Sec13–KR in blocking ER exit, we used an assay in which the Golgi assembles out of the ER upon washout of brefeldin A (BFA), an inhibitor of Arf1 activation that reversibly redistributes Golgi enzymes into the ER (Lippincott-Schwartz et al., 1989; Peyroche et al., 1999). Immediately after drug washout, the Killer Red fusion proteins were bleached for 30 seconds and GalNAcT2–GFP was imaged for 90 minutes (Fig. 1E, supplementary material Movie 1). Cells expressing the control constructs, Cyto–KR or KR–GTN, showed full assembly of the Golgi with kinetics similar to untransfected cells. In striking contrast, bleaching of Sec13–KR blocked the exit of GalNAcT2–GFP from the ER, indicating the inactivation of this essential COPII subunit. Golgi assembly was quantified by determining the size of the largest fluorescent Golgi object at each time point and confirmed a robust and reproducible block by Sec13–KR inactivation (Fig. 1F). ER-exit blockade was also demonstrated using the temperature-sensitive vesicular stomatitis virus G protein (VSVG) transport assay (Presley et al., 1997). Cells expressing VSVG–GFP were shifted from 40°C to 32°C and simultaneously photo-inactivated. Control cells transported VSVG–GFP to the Golgi by 15 minutes, whereas VSVG–GFP was trapped in the ER in Sec13–KR-inactivated cells, where it remained for at least 90 minutes (Fig. 1G).
Acute dependence of Golgi integrity on ER exit
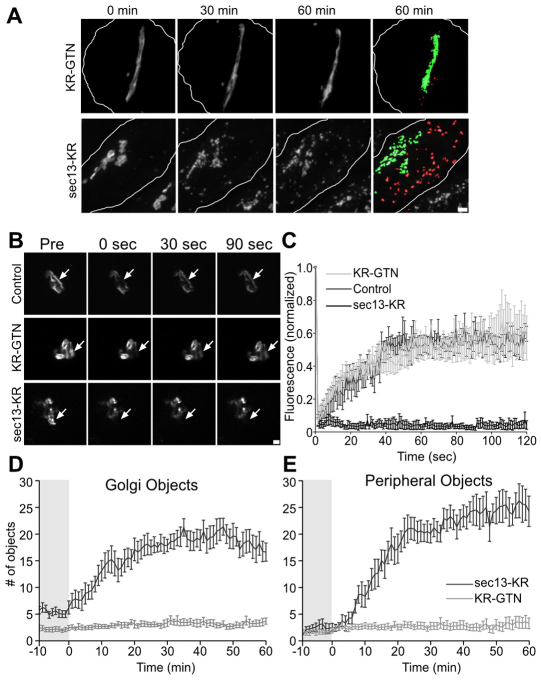
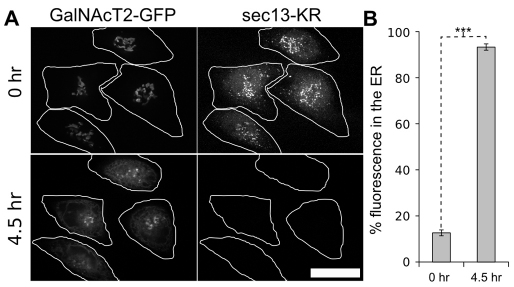
Having established its effectiveness, we used Sec13–KR inactivation to test the acute dependence of the Golgi complex on ER exit. Golgi labeled with GalNAcT2–GFP was imaged for 10 minutes to establish a base line. The control and Sec13–KR constructs were then bleached, and the Golgi was followed for another 60 minutes. Bleaching of KR–GTN yielded little effect on the stability or dynamics of the Golgi ribbon, whereas photobleaching of Sec13–KR had an immediate effect, causing both a fragmentation of the Golgi ribbon and a concomitant accumulation of GalNAcT2–GFP in small peripheral objects (Fig. 2A, supplementary material Movie 2). Integrity of the ribbon was assayed using fluorescence recovery after photobleaching. Whereas KR–GTN inactivated cells showed diffusion of GalNAcT2–GFP equivalent to untreated cells, Sec13–KR inactivated cells exhibited little or no GalNAcT2–GFP diffusion into the bleached zone, indicating loss of ribbon architecture (Fig. 2B,C). To analyze the appearance of GalNAcT2–GFP in the cell periphery after Sec13–KR inactivation, we first used an algorithm that highlights objects contiguous with the original Golgi in green and any peripheral objects that arise in red (Fig. 2A last panels, supplementary material Movie 3). Golgi objects corresponded to pixel sets that intersected with pixel sets in previous frames extending back in time to the original pixel set defining the Golgi and peripheral objects corresponded to all other above-threshold objects. The fluorescence in each group was quantified (Fig. 2D,E), confirming the reproducibility of both the fragmentation of the Golgi ribbon and the appearance of peripheral objects with each effect exhibiting a t1/2 of ≈10 minutes. Although there was extensive redistribution to the periphery, GalNAcT2–GFP was not detectable in the ER until the post-inactivation incubation was extended to 4.5 hours, at which time most of the fluorescence was present in the ER (Fig. 3). A slow rate of ER redistribution in response to an ER exit block is in accord with previously published longer-term experiments (Storrie et al., 1998; White et al., 1999), whereas the immediate effects in disrupting Golgi integrity and causing redistribution to peripheral punctae have not been previously observed. To provide further evidence that ER exit was blocked by Sec13–KR and did not contribute to the appearance of the peripheral punctae, we specifically bleached the Golgi GalNAcT2–GFP immediately after Sec13–KR inactivation. These cells showed no GalNActT2–GFP accumulating in the periphery (supplementary material Movie 4) confirming a Golgi origin for the peripheral fluorescence rather than exit of the residual ER pool.
Fig. 2.
Acute inactivation at steady state. (A) Time course of Golgi integrity after bleaching of the indicated Killer Red constructs. Cells transfected with Killer Red constructs were imaged to observe GalNAcT2–GFP at 1 minute intervals for 10 minutes before a 30 second bleaching and then for a further 60 minutes. Max value Z-projections are shown for the indicated time points and the 60 minute time point is also shown after pseudo-coloring to indicate objects that visibly detached from the Golgi (green) and objects that appeared in the periphery (red). The corresponding full time course is in supplementary material Movie 2 (gray scale) and Movie 3 (pseudo-colored). Scale bar: 2 μm. (B) Analysis of Golgi integrity by fluorescence recovery after photobleaching. Cells were subjected to no treatment, inactivation of KR–GTN or inactivation of Sec13–KR. After 15 minutes images were acquired before and after a small region of GalNAcT2–GFP fluorescence on the Golgi was bleached (arrows) and the indicated time points are shown. Scale bar: 2 μm. (C) Fluorescence was measured in bleached zone and is plotted versus time (n=5 cells, mean ± s.e.m.). (D,E) Golgi disassembly was measured by determining the number of total (D) and peripheral (E) fluorescent objects present at each time point using the ImageJ ‘Analyze particles’ function with the threshold set above the level of GalNAcT2–GFP fluorescence in the ER (n=10, mean ± s.e.m.). Pre- and post-bleach periods are indicated.
Fig. 3.
Golgi at 4.5 hours after Sec13–KR inactivation. (A) Representative Z-slices of GalNAcT2–GFP are shown before and 4.5 hours after inactivation. Scale bar: 10 μm. (B) The percentage fluorescence in the ER was calculated by measuring total cell fluorescence and subtracting any above-threshold fluorescent objects with the threshold manually set to exclude the nuclear envelope but retain remnant Golgi objects (n=8 cells, mean ± s.e.m.). Statistical significance was measured using the paired Student's t-test (***P=0.0000003).
GalNAcT2–GFP cycles to the cell periphery
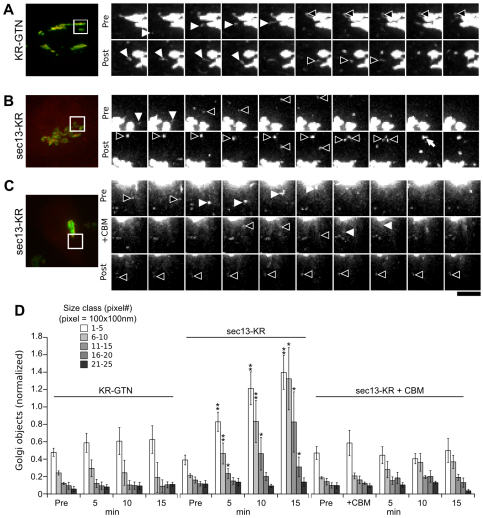
The redistribution of GalNAcT2–GFP to peripheral punctae upon ER exit blockade suggests that it constantly cycles to peripheral structures and depends on ER exit for its return to the Golgi. In an attempt to visualize GalNAcT2–GFP cycling directly, we carried out faster time-lapse imaging using binning and greater laser power to boost sensitivity. During a 15 minute pre-inactivation period, the enhanced imaging revealed a multitude of small, dim GalNAcT2–GFP objects leaving the Golgi and arriving at the Golgi. Fig. 4A–C presents images cropped around the Golgi region to highlight a few examples where objects left the Golgi and moved towards the periphery (black arrowheads) or moved from the periphery inward to the Golgi (white arrowheads). The full extent of these events is better observed in supplementary material Movie 5. Following this imaging, either KR–GTN or Sec13–KR was then inactivated and the cells were imaged for another 15 minutes. In the case of the KR–GTN control, inward and outward traffic persisted (Fig. 4A) and the distribution of objects remained constant (Fig. 4D). By contrast, after Sec13–KR inactivation there was a loss of inward-directed motility (Fig. 4B), and a significant and progressive increase in peripheral object size (Fig. 4D) because some objects approached each other and appeared to fuse (Fig. 4B, white arrow). To test the role of the COPI vesicle coat in generating the GalNAcT2–GFP objects leaving the Golgi, this experiment was repeated in the presence of 1,3-cyclohexanebimethylamine, which causes COPI membrane dissociation and blocks COPI function (Hu et al., 1999; Puthenveedu and Linstedt, 2001; Zhang et al., 2009). This treatment blocked the Sec13–KR-induced increase of peripheral GalNAcT2–GFP objects (Fig. 4C,D, supplementary material Movie 6). To further test the dependence on Arf-dependent COPI function, we added BFA to cells immediately after inactivation of Sec13–KR. In the 15 minute post-inactivation period there was extensive tubulation and then collapse of the Golgi into the ER. Peripheral punctae did not arise during tubulation and were not evident after Golgi collapse (supplementary material Movie 7). Thus, GalNAcT2–GFP continuously cycled to the cell periphery with the outward path requiring COPI and the inward return dependent on ER exit. Respectively, these dependencies probably reflect COPI-mediated budding of GalNAcT2-containing vesicles from the Golgi and a demand for new membrane and/or transport factors.
Fig. 4.
Evidence for GalNAcT2–GFP cycling. (A–C) Time course of structures detaching from the Golgi or moving back to the Golgi during pre- and post-bleaching periods in untreated cells expressing KR–GTN (A) or Sec13–KR (B), or in cells treated with 1,3-cyclohexanebimethylamine for 5 minutes before Sec13–KR inactivation (C). GalNAcT2–GFP was imaged every 15 seconds with 4× binning of the acquired signal for 10 minutes before (Pre) and 10 minutes after (Post) a 30 second bleaching. Max value Z-projections are shown for consecutive time points with inward objects indicated by white arrowheads and outward by black arrowheads. The corresponding full time courses are in supplementary material Movies 5, 6. Scale bar: 1 μm. (D) The distribution of small Golgi objects in size classes is shown for the indicated time points following bleaching of the KR–GTN control or the Sec13–KR construct. Values are the number of objects normalized to the value in the pre-inactivation segment (n=10, mean ± s.e.m., *P<0.005 and **P<0.0005).
Cycling is through the ER-Golgi intermediate compartment
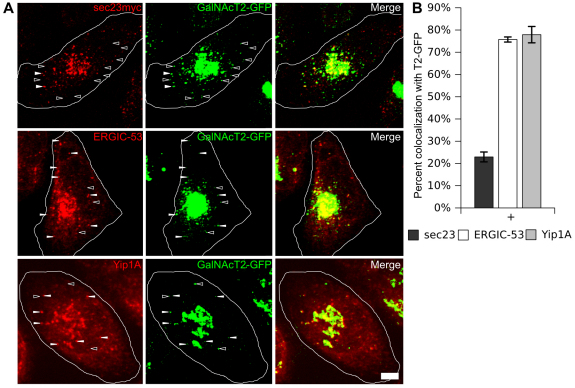
Previously it was hypothesized that cycling vesicles containing Golgi components fuse with ERGIC membranes (Love et al., 1998; Lin et al., 1999; Marra et al., 2001; Puthenveedu and Linstedt, 2001), therefore we wished to test whether the peripheral GalNAcT2–GFP objects were positive for ERGIC markers. However, the ERGIC collapses in the absence of ER exit (Aridor and Balch, 2000; Lee and Linstedt, 2000). This was confirmed in the case of Sec13–KR inactivation after 30 minutes (supplementary material Fig. S1); nevertheless, at 15 minutes after inactivation, the ERGIC markers ERGIC53 and Yip1A were still evident in peripheral punctae, so we asked whether these were positive for trapped GalNacT2–GFP. As a control, a parallel set of cells was stained at the same time point to visualize ER-exit sites using transfected, Myc-tagged Sec23. After thresholding the images, the Golgi region was ignored and all peripheral GalNacT2–GFP objects were scored for overlap with ERGIC53, Yip1A, or Sec23 punctate fluorescence. Whereas few GalNAcT2–GFP punctae colocalized with Sec23, there was extensive colocalization with ERGIC markers ERGIC53 and Yip1A (Fig. 5A,B). Thus, GalNAcT2–GFP probably cycles to ERGIC membranes, which in the absence of continued ER exit become depleted, leading to accumulation of GalNAcT2–GFP in the cell periphery.
Fig. 5.
GalNAcT2 redistribution to the ER-Golgi intermediate compartment. (A) Cells were fixed for 15 minutes after inactivation of Sec13–KR and stained for transfected Sec23–Myc or endogenous ERGIC-53. Max-value Z-projections are shown with representative GalNAcT2–GFP-positive objects which co-label peripheral punctae with Sec23–Myc or ERGIC-53 (filled arrowheads) and or lack Sec23 or ERGIC-53 (empty arrowheads). Scale bar: 2 μm. (B) GalNAcT2–GFP objects were scored for colocalization with Sec23 or ERGIC-53 based on overlap of punctate fluorescence above background in single Z-sections (n=15 cells objects >250).
Cycling involves cis- but not trans-Golgi proteins
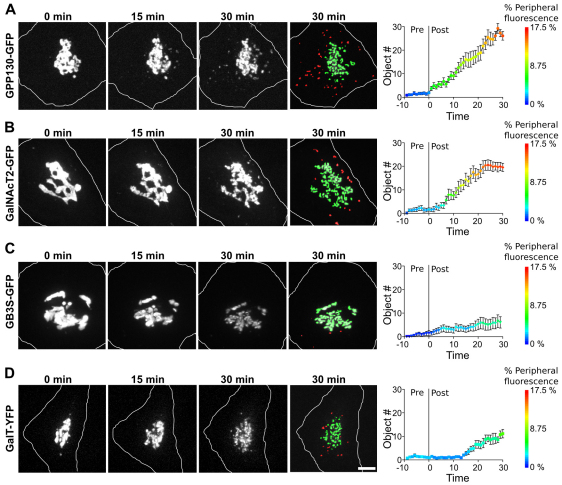
Next, we tested whether additional Golgi markers traffic in the pathway made evident by inactivation of Sec13–KR to see whether there might be preferential involvement of cis-Golgi proteins. First, we compared GalNAcT2–GFP to the cis-Golgi protein GPP130–GFP. Each responded to Sec13–KR inactivation by rapid accumulation in peripheral punctae with GPP130–GFP slightly preceding GalNAcT2–GFP (Fig. 6A,B, supplementary material Movies 8, 9). Redistribution was also confirmed for endogenous cis-localized proteins. Endogenous giantin, GPP130 and GRASP65 each accumulated in GalNAcT2–GFP-positive peripheral punctate structures after Sec13–KR inactivation (supplementary material Fig. S2). By contrast, two trans-Golgi markers, galactosyltransferase tagged with yellow fluorescent protein (GalT–YFP) and globotriaosylceramide synthase (GB3S–GFP), showed delayed and relatively minimal accumulation in peripheral punctae (Fig. 6C,D, supplementary material Movies 8, 9). Thus, if these trans components cycle, their cycling is probably restricted to the Golgi region and is therefore not discernible. These experiments strongly suggest that cycling of cis-Golgi-localized proteins to ERGIC membranes sustains Golgi biogenesis.
Fig. 6.
Preferential peripheral cycling of cis Golgi components. (A–D) Cells expressing GPP130–GFP (A), GalNAcT2–GFP (B), GB3S–GFP (C) or GalT–GFP (D) were imaged at 1 minute intervals for 10 minutes before a 30 second bleaching of Sec13–KR and then for a further 30 minutes. Max value Z-projections are shown for the indicated time points and the 30 minute time point is also shown after pseudo-coloring to indicate objects that visibly detached from the Golgi (green) and objects that appeared in the periphery (red). The corresponding full time course is in supplementary material Movie 7 (gray scale) and Movie 8 (pseudo-colored). Scale bar: 2 μm. Quantification of the number of peripheral objects (y-axis) and percent of total fluorescence in peripheral objects (color axis) is shown for each construct (n=10 cells, mean ± s.e.m.).
Discussion
These experiments demonstrate the technical feasibility of using a 30 second photo-inactivation of a crucial component in a dynamic system while carrying out live-cell fluorescent imaging. Although specificity of inactivation using Killer Red is not absolute, several considerations increase confidence in our conclusions. Chromophore-assisted light inactivation has a successful history and is based on validated physical theory (Surrey et al., 1998; Jay and Sakurai, 1999). The Killer Red variation of the technique increases specificity because it more precisely targets the chromophore. It also avoids microinjection, which is less reproducible and might impair cell viability. Although microinjection can, in principle, impose an immediate block, the need to allow cell recovery could explain why previous reports of blockade of ER exit using a dominant-negative version of Sar1 involved analysis 4–6 hours following microinjection (Storrie, 1998; Miles, 2001). The inactivation done in the steady state analysis used the same parameters and set-up as the Golgi biogenesis and VSVG–GFP transport assays confirming its effectiveness. Finally, our conclusions do not require highly specific inhibition of the single targeted component Sec13. Indeed, this would be unlikely given the proximity of Sec13 to its binding partners at an ER exit site. Rather, the conclusions rest on effective inhibition of ER export – a process that depends on Sec13 and its binding partners. Given that the approach we describe is based on a genetically encoded fluorescent protein whose localization and expression level are readily verified, we expect Killer-Red-mediated inactivation during live imaging to be widely adopted and of particular significance where temporally or spatially restricted inactivation of proteins and their binding partners is required.
Acute inactivation revealed the Golgi to be sustained by both ER exit and a surprisingly high rate of continuous cycling of cis-Golgi components to ERGIC membranes. These observations fulfill previously unmet expectations of the maturation model in which new cisternae are constantly built by the fusion of cargo-bearing membrane from the ER and cis-enzyme-bearing recycling membrane from the Golgi. Certain trafficking components were previously visualized in ERGIC-directed tubules but the tubules were devoid of Golgi enzymes (Marra et al., 2001), except when BFA was used to induce their appearance (Mardones et al., 2006). Significantly, COPI was required to generate the cycling membranes, whereas ER export was required for their inward return to the Golgi. Arguably, ER export is required to contribute membrane and transport factors that sustain the ERGIC and are essential for the progression ERGIC membranes towards biogenesis of cis-Golgi cisternae.
Our results help resolve controversies concerning cycling of Golgi proteins through the ER (Altan-Bonnet et al., 2006; Patterson et al., 2008; Budnik and Stephens, 2009; Emr et al., 2009; Wei and Seemann, 2009). Evidence in support of cycling through the ER (Cole et al., 1998; Storrie et al., 1998; Miles et al., 2001; Ward et al., 2001; Rhee et al., 2005) can be reinterpreted as showing the importance of ER exit in supporting Golgi enzyme cycling through the ERGIC with redistribution into the ER being a secondary, longer-term effect. Evidence against cycling through the ER, such as lack of Golgi enzyme trapping by ER-localized binding partners (Pecot and Malhotra, 2004; Pecot and Malhotra, 2006), although supported by our findings, missed the importance of ER exit in sustaining the robust cycling of enzymes through the ERGIC.
In conclusion, in contrast to longer-term inhibition experiments, photo-inactivation revealed that the mammalian Golgi complex is sustained by ER exit and extensive Golgi protein recycling.
Materials and Methods
Cell imaging
Cells were transferred to imaging medium (MEM, 7% FBS, 375 μM Trolox) and placed on a 37°C heated stage under a blood gas mixture (5% CO2, 95% air) on a Zeiss Axiovert 200 with a heated 100× Plan-Apo NA 1.4 oil objective (Zeiss, Thornwood, NY) attached to an UltraView spinning-disk confocal system (Perkin-Elmer, Shelton, CT). Cells expressing properly localized Killer Red constructs at brightness levels matching GalNacT2–GFP were identified. Cells expressing low levels or levels high enough to cause gross mistargeting were ignored because, upon photo-inactivation, the former failed to exhibit an effect and the latter sometimes resulted in cell death. Bleaching of Killer Red fluorescence was by epifluorescence light through a 535–585 nm filter at 2 W/cm2 for 30 seconds. Capture parameters before and after bleaching are indicated in the figure legends. Enhanced images were taken using 4× binning to increase camera sensitivity. FRAP experiments were carried out in OptiMEM containing 7% FBS and 375 μM Trolox at 37°C under ambient CO2. Acquisition was performed on an Andor Revolution XD system (Andor Technology, Belfast, UK). Killer Red was bleached at 100% laser power (561 nm) for 90 μseconds per pixel and GFP was bleached at 80% laser power (488 nm) for 20 μseconds per pixel. To stain inactivated cells, cells on glass-bottomed dishes were returned to a 37°C incubator for the indicated times after pre- and post-inactivation images had been acquired and then the cells were fixed in 3% paraformaldehyde at room temperature or a methanol/acetone (2:1 mixture) at −20°C. Antibodies were: rabbit anti-GRASP65 (Bachert and Linstedt, 2010), rabbit anti-Yip1A (Dykstra et al., 2010) mouse anti-GPP130 (Linstedt et al., 1997), mouse anti-α-tubulin (Sigma), mouse anti-giantin (Linstedt and Hauri, 1993), mouse anti-ERGIC53 (Schweizer et al., 1990), mouse anti-Myc (Evan et al., 1985) and Cy5-labeled secondary antibodies (Invitrogen).
Cloning
Sec13–KR was constructed by PCR amplifying Killer Red from pKillerRed-N (Evrogen) and inserting it into the XhoI and XbaI sites of pCS2-Sec13 (gift from Tina Lee, Carnegie Mellon University, Pittsburgh, PA). Killer Red was also cloned into the BamH1 and EcoR1 sites of pCS2 to generate Cyto–KR. For KR–GTN, the sequence encoding the last 100 amino acids was amplified and cloned into the EcoRI and XbaI sites of Cyto–KR. GPP130–GFP was as described (Mukhopadhyay et al., 2010). GalT–YFP was created by inserting the localization domain of GalT (residues 1–75) with YFP into the pIRES vector. GB3S–GFP was created by insertion into pEGFP.
Cell culture and transfection
HeLa cells stably expressing GalNAcT2–GFP were cultured in MEM (Thermo Scientific) with 10% FBS (Atlanta Biologicals). Before experiments, cells were grown to 30–50% confluency in a 60 mm dish, and transfected with JetPEI (PolyPlus) according to the manufacturer's protocol. After 24 hours, the cells were placed onto 35 mm coverslips, and at 36–72 hours post transfection the coverslips were mounted in an imaging chamber or 35 mm glass-bottomed dish (Matek). Brefeldin A (Sigma) was added at 2 μg/ml where indicated. Where indicated, 1,3-cyclohexanebimethylamine (Acros Organics) was added at 10 mM final concentration.
Analysis
ImageJ was used to carry out all analysis. Distribution of ER-exit sites was measured using the ‘Analyze Particles’ plug-in to find the centroid of each object and its distance from the Golgi was then measured. The measurements per cell were then binned and presented in a histogram. To determine the largest object size during Golgi assembly, images were thresholded to exclude ER-localized GalNAcT2–GFP fluorescence (value set using the 0 minutes time point) and the ‘Analyze Particles’ function was used to find the largest object in each cell for each time point. FRAP recovery was performed using the ‘FRAP Profiler’ plug-in. Statistical analysis used a one-tailed paired t-test. To determine the number and size of Golgi objects after steady-state inactivation, the images were enhanced using the spot-enhancing-filter plug-in (Sage et al., 2005), manually thresholded to maximize object recovery (single threshold/series), and analyzed using the ‘Analyze Particles’ function. To pseudo-color objects based on continuity with the starting Golgi, the spot-enhanced and thresholded images were converted to binary and object continuity over time, that is overlapping pixels, was determined using the ‘3D Object counter’ plug-in. All objects contiguous with the starting Golgi were added as a green layer on top of the original grayscale image and all other objects were added as a red layer. To quantify redistribution to the ER, the images were background subtracted followed by determination of total fluorescence less the fluorescence in objects above a threshold manually set to exclude the ER. The threshold was set above the level of the nuclear envelope fluorescence, which left only remnant Golgi fluorescence. To determine colocalization of peripheral GalNAcT2 punctae, the images were thresholded above background leaving only the peripheral punctae and the Golgi fluorescence. The latter was ignored and the number of peripheral punctae that were positive for ERGIC53, Yip1A, or Sec23 was quantified. To determine size distribution of cycling objects, the 4× binned images were thresholded and analyzed using the ‘Analyze Particles’ function with max object size set to 25 pixels.
Acknowledgements
We thank Manojkumar Puthenveedu and especially Collin Bachert for invaluable assistance and Tina Lee, Debrup Sengupta, Gerry Apodaca, Chuck Ettenshohn, Jon Minden and Jeff Brodsky for comments on an early version of the manuscript.
Footnotes
Funding
Funding was provided by the National Institutes of Health [grant numbers GM56779 and GM084111] to A.D.L. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.094441/-/DC1
References
- Allan B. B., Balch W. E. (1999). Protein sorting by directed maturation of Golgi compartments. Science 285, 63-66 [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet N., Sougrat R., Liu W., Snapp E. L., Ward T., Lippincott-Schwartz J. (2006). Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell 17, 990-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Balch W. E. (2000). Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 275, 35673-35676 [DOI] [PubMed] [Google Scholar]
- Bachert C., Linstedt A. D. (2010). Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285, 16294-16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisby R. H., Morgan C. G., Hamblett I., Gorman A. A. (1999). Quenching of singlet oxygen by Trolox C, ascorbate, and amino acids: effects of pH and temperature. J. Phys. Chem. A 103, 7454-7459 [Google Scholar]
- Bonfanti L., Mironov A. A., Jr, Martinez-Menarguez J. A., Martella O., Fusella A., Baldassarre M., Buccione R., Geuze H. J., Mironov A. A., Luini A. (1998). Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 95, 993-1003 [DOI] [PubMed] [Google Scholar]
- Budnik A., Stephens D. J. (2009). ER exit sites-localization and control of COPII vesicle formation. FEBS Lett. 583, 3796-3803 [DOI] [PubMed] [Google Scholar]
- Bulina M. E., Chudakov D. M., Britanova O. V., Yanushevich Y. G., Staroverov D. B., Chepurnykh T. V., Merzlyak E. M., Shkrob M. A., Lukyanov S., Lukyanov K. A. (2006). A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95-99 [DOI] [PubMed] [Google Scholar]
- Carpentier P., Violot S., Blanchoin L., Bourgeois D. (2009). Structural basis for the phototoxicity of the fluorescent protein KillerRed. FEBS Lett. 583, 2839-2842 [DOI] [PubMed] [Google Scholar]
- Cole N. B., Ellenberg J., Song J., DiEuliis D., Lippincott-Schwartz J. (1998). Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 140, 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Planus E., Bouvard D., Oddou C., Badowski C., Bossy V., Raducanu A., Fourcade B., Albiges-Rizo C., Block M. R. (2010). beta1A integrin is a master regulator of invadosome organization and function. Mol. Biol. Cell 21, 4108-4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra K. M., Pokusa J. E., Suhan J., Lee T. H. (2010). Yip1A structures the mammalian endoplasmic reticulum. Mol. Biol. Cell 21, 1556-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S., Glick B. S., Linstedt A. D., Lippincott-Schwartz J., Luini A., Malhotra V., Marsh B. J., Nakano A., Pfeffer S. R., Rabouille C., et al. (2009). Journeys through the Golgi–taking stock in a new era. J. Cell Biol. 187, 449-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. (1985). Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5, 3610-3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. S., Nakano A. (2009). Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 25, 113-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. S., Elston T., Oster G. (1997). A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 414, 177-181 [DOI] [PubMed] [Google Scholar]
- Hu T., Kao C. Y., Hudson R. T., Chen A., Draper R. K. (1999). Inhibition of secretion by 1,3-Cyclohexanebimethylamine), a dibasic compound that interferes with coatomer function. Mol. Biol. Cell 10, 921-933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay D. G. (1988). Selective destruction of protein function by chromophore-assisted laser inactivation. Proc. Natl. Acad. Sci. USA 85, 5454-5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay D. G., Sakurai T. (1999). Chromophore-assisted laser inactivation (CALI) to elucidate cellular mechanisms of cancer. Biochim. Biophys. Acta 1424, M39-M48 [DOI] [PubMed] [Google Scholar]
- Lanoix J., Ouwendijk J., Stark A., Szafer E., Cassel D., Dejgaard K., Weiss M., Nilsson T. (2001). Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J. Cell Biol. 155, 1199-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Linstedt A. D. (2000). Potential role for protein kinases in regulation of bidirectional endoplasmic reticulum-to-Golgi transport revealed by protein kinase inhibitor H89. Mol. Biol. Cell 11, 2577-2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J. C., Roider J., Jay D. G. (1994). Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc. Natl. Acad. Sci. USA 91, 2659-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. C., Love H. D., Gushue J. N., Bergeron J. J., Ostermann J. (1999). ER/Golgi intermediates acquire Golgi enzymes by brefeldin A-sensitive retrograde transport in vitro. J. Cell Biol. 147, 1457-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. (1993). Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell 4, 679-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Mehta A., Suhan J., Reggio H., Hauri H. P. (1997). Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell 8, 1073-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. (2006). Golgi maturation visualized in living yeast. Nature 441, 1002-1006 [DOI] [PubMed] [Google Scholar]
- Love H. D., Lin C. C., Short C. S., Ostermann J. (1998). Isolation of functional Golgi-derived vesicles with a possible role in retrograde transport. J. Cell Biol. 140, 541-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V., Serafini T., Orci L., Shepherd J. C., Rothman J. E. (1989). Purification of a novel class of coated vesicles mediating biosynthetic protein transport through the Golgi stack. Cell 58, 329-336 [DOI] [PubMed] [Google Scholar]
- Malsam J., Satoh A., Pelletier L., Warren G. (2005). Golgin tethers define subpopulations of COPI vesicles. Science 307, 1095-1098 [DOI] [PubMed] [Google Scholar]
- Mardones G. A., Snyder C. M., Howell K. E. (2006). Cis-Golgi matrix proteins move directly to endoplasmic reticulum exit sites by association with tubules. Mol. Biol. Cell 17, 525-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P., Maffucci T., Daniele T., Tullio G. D., Ikehara Y., Chan E. K., Luini A., Beznoussenko G., Mironov A., De Matteis M. A. (2001). The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3, 1101-1113 [DOI] [PubMed] [Google Scholar]
- Martinez-Menarguez J. A., Geuze H. J., Slot J. W., Klumperman J. (1999). Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell 98, 81-90 [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007-1010 [DOI] [PubMed] [Google Scholar]
- Mellman I., Simons K. (1992). The Golgi complex: in vitro veritas? Cell 68, 829-840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles S., McManus H., Forsten K. E., Storrie B. (2001). Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155, 543-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer H. H., Morre D. J. (1991). Perspectives on Golgi apparatus form and function. J. Electron Microsc. Tech. 17, 2-14 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Bachert C., Smith D. R., Linstedt A. D. (2010). Manganese-induced trafficking and turnover of the cis-Golgi glycoprotein GPP130. Mol. Biol. Cell 21, 1282-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. H., Hirschberg K., Polishchuk R. S., Gerlich D., Phair R. D., Lippincott-Schwartz J. (2008). Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell 133, 1055-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot M. Y., Malhotra V. (2004). Golgi membranes remain segregated from the endoplasmic reticulum during mitosis in mammalian cells. Cell 116, 99-107 [DOI] [PubMed] [Google Scholar]
- Pecot M. Y., Malhotra V. (2006). The Golgi apparatus maintains its organization independent of the endoplasmic reticulum. Mol. Biol. Cell 17, 5372-5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Rothman J. E. (2000). The debate about transport in the Golgi – two sides of the same coin? Cell 102, 713-719 [DOI] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C. L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285 [DOI] [PubMed] [Google Scholar]
- Pletnev S., Gurskaya N. G., Pletneva N. V., Lukyanov K. A., Chudakov D. M., Martynov V. I., Popov V. O., Kovalchuk M. V., Wlodawer A., Dauter Z., et al. (2009). Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. J. Biol. Chem. 284, 32028-32039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. (1997). ER-to-Golgi transport visualized in living cells. Nature 389, 81-85 [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. (2001). Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 155, 227-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. (2005). Subcompartmentalizing the Golgi apparatus. Curr. Opin. Cell Biol. 17, 369-375 [DOI] [PubMed] [Google Scholar]
- Rhee S. W., Starr T., Forsten-Williams K., Storrie B. (2005). The steady-state distribution of glycosyltransferases between the Golgi apparatus and the endoplasmic reticulum is approximately 90:10. Traffic 6, 978-990 [DOI] [PubMed] [Google Scholar]
- Sage D., Neumann F. R., Hediger F., Gasser S. M., Unser M. (2005). Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 14, 1372-1383 [DOI] [PubMed] [Google Scholar]
- Schweizer A., Fransen J. A., Matter K., Kreis T. E., Ginsel L., Hauri H. P. (1990). Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur. J. Cell Biol. 53, 185-196 [PubMed] [Google Scholar]
- Serebrovskaya E. O., Edelweiss E. F., Stremovskiy O. A., Lukyanov K. A., Chudakov D. M., Deyev S. M. (2009). Targeting cancer cells by using an antireceptor antibody-photosensitizer fusion protein. Proc. Natl. Acad. Sci. USA 106, 9221-9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., White J., Rottger S., Stelzer E. H., Suganuma T., Nilsson T. (1998). Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505-1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T., Elowitz M. B., Wolf P. E., Yang F., Nedelec F., Shokat K., Leibler S. (1998). Chromophore-assisted light inactivation and self-organization of microtubules and motors. Proc. Natl. Acad. Sci. USA 95, 4293-4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh C., Chudakov D. M., Poon K. L., Mamedov I. Z., Sek J. Y., Shidlovsky K., Lukyanov S., Korzh V. (2010). Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol. 10, 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley A. K., Feng Y., Schmidt K., Carter D. A., Porter R., Verkade P., Stephens D. J. (2008). Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 121, 3025-3034 [DOI] [PubMed] [Google Scholar]
- Trucco A., Polishchuk R. S., Martella O., Di Pentima A., Fusella A., Di Giandomenico D., San Pietro E., Beznoussenko G. V., Polishchuk E. V., Baldassarre M., et al. (2004). Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat. Cell Biol. 6, 1071-1081 [DOI] [PubMed] [Google Scholar]
- Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K., Lippincott-Schwartz J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. H., Seemann J. (2009). Mitotic division of the mammalian Golgi apparatus. Semin. Cell Dev. Biol. 20, 810-816 [DOI] [PubMed] [Google Scholar]
- White J., Johannes L., Mallard F., Girod A., Grill S., Reinsch S., Keller P., Tzschaschel B., Echard A., Goud B., et al. (1999). Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 147, 743-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lee S. Y., Beznoussenko G. V., Peters P. J., Yang J. S., Gilbert H. Y., Brass A. L., Elledge S. J., Isaacs S. N., Moss B., et al. (2009). A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc. Natl. Acad. Sci. USA 106, 163-168 [DOI] [PMC free article] [PubMed] [Google Scholar]