Abstract
Factors explaining why human immunodeficiency virus (HIV) enhances the risk of reactivated tuberculosis (TB) are poorly understood. Unfortunately, experimental models of HIV-induced reactivated TB are lacking. We examined whether cynomolgus macaques, which accurately model latent TB in humans, could be used to model pathogenesis of HIV infection in the lungs and associated lymph nodes. These experiments precede studies modeling the effects of HIV infection on latent TB. We infected two groups of macaques with chimeric simian–human immunodeficiency viruses (SHIV-89.6P and SHIV-KU2) and followed viral titers and immunologic parameters including lymphocytes numbers and phenotype in the blood, bronchoalveolar lavage cells, and lymph nodes over the course of infection. Tissues from the lungs, liver, kidney, spleen, and lymph nodes were similarly examined at necropsy. Both strains produced dramatic CD4+ T cell depletion. Plasma titers were not different between viruses, but we found more SHIV-89.6P in the lungs. Both viruses induced similar patterns of cell activation markers. SHIV-89.6P induced more IFN-γ expression than SHIV-KU2. These results indicate SHIV-89.6P and SHIV-KU2 infect cynomolgus macaques and may be used to accurately model effects of HIV infection on latent TB.
INTRODUCTION
Coinfection with human immunodeficiency virus (HIV) and Mycobacterium tuberculosis (Mtb) represents one of the most pressing health problems in modern times. Approximately one-third of the world’s population is infected with Mtb, the causative agent of tuberculosis (TB). Immunocompetent individuals are able to contain the infection in a subclinical state (hereafter referred to as latency) with a 10% lifetime risk of experiencing reactivated TB. In contrast, an individual coinfected with HIV and Mtb experiences a 10% annual risk of experiencing active or reactivated TB,1 making TB one of the most frequent killers of HIV-infected individuals.2 The basis for the high mortality of Mtb infection in HIV-positive individuals is unclear, but may include an HIV-induced decline in granuloma integrity, shifts in cytokine expression away from Mtb-protective Th1 responses toward Th2 responses, and loss of Mtb-specific CD4+ T lymphocytes.3
Simian immunodeficiency virus (SIV) is an HIV-like lentivirus that can infect macaques and has been used to model human HIV infection. SIV is a naturally occurring primate virus that can cause massive depletion of activated and replicating CD4+ T cells in susceptible monkeys.4 There are significant differences in the basic biology of SIV and HIV, however. While both SIV and HIV use the chemokine coreceptor CCR5 to infect macrophages (R5 tropism), SIV does not normally use the chemokine receptor CXCR4 as a cofactor to enter CD4+ T cells.5 Moreover, SIV infection can take months to years to deplete CD4+ T cell populations to numbers that are equivalent to humans AIDS.6 SIV/HIV chimeras (SHIV) have been developed that use CXCR4 to infect CD4+ T cells (X4 tropism) and cause rapid immunosuppression.5 Pathogenic SHIVs contain the HIV env, rev, tat, and vpu genes cloned onto the SIV backbone and can produce AIDS-like disease in macaques within weeks to months by eliminating naive and memory CD4+ T cell populations.5,7 These viruses infect lymphocytes and macrophages, including alveolar macrophages, and have been used to model the effects of HIV.8,9 These factors make SHIV experimentally attractive and biologically relevant tools for modeling the effects of HIV infection on pulmonary immune responses to pathogenic microorganisms.
A variety of animal models and experimental approaches have been applied to investigate TB, yet few models accurately represent the pathology and latency that occur in humans. Cynomolgus macaques (Macaca fasicularis) have been established as a model that mirrors aspects of human TB including granuloma morphology and the development of asymptomatic latent infections.10,11 Considering the applicability of cynomolgus macaques in TB research, we performed infections with SHIV strains alone to determine their usefulness for modeling immunologic outcomes of Mtb/HIV coinfection and reactivated TB following HIV infection. In this report, we describe the effects of a pathogenic X4-tropic SHIV isolate, SHIV-KU2,12 and a pathogenic dual (X4/R5) tropic SHIV isolate, SHIV-89.6P,13 on cynomolgus macaques by following immune markers and virus load over the course of infection. We found both virus strains caused rapid, substantial long-term depletion of CD4+ T lymphocytes, and virus RNA was detectable in a variety of tissues that are frequently infected by Mtb. Importantly, we found more SHIV-89.6P virus associated with the lungs than SHIV-KU2. Our findings suggest that SHIV-89.6P-infected cynomolgus macaques can provide a relevant model for examining the effect of HIV infection on latent TB.
MATERIALS AND METHODS
Experimental animals
Eight adult cynomolgus macaques of Vietnamese or Chinese origin weighing 4.1–6.5 kg and ranging from 4 to 8 years of age were used for these studies. Prior to commencement of the study, the animals were tested to ensure they were not infected with Mtb, SIV, simian type D retrovirus, or SHIV. Additional diagnostic procedures including physical examinations, differential blood count, serum chemistry profile, and thoracic radiography were performed to ensure the animals were free of underlying disease processes. Monkey 7004 was diagnosed early in the study with a nematode-like infection in the nose and was treated with Panacur (fenbendazole) for 7 days. All animals were prophylactically treated for Pneumocystis infection with Bactrim (sulfamethoxazole and trimethoprim) for 14 days. These studies followed all animal experimentation guidelines and all experimental manipulations and protocols were approved by the University of Pittsburgh School of Medicine Institutional Animal Care and Use Committee.
Experimental viral infection
Four animals were infected with SHIV-89.6P and four were infected with SHIV-KU2. SHIV-89.6P and SHIV-KU2 virus stocks were kind gifts from Dr. O. Narayan (University of Kansas Medical Center, Kansas City, KS). Virus stocks contained approximately 104 TCID50 units of virus, and animals were infected with approximately 5 × 103 TCID50 units of SHIV-KU2 or 1 × 104 TCID50 units of SHIV-89.6P via intravenous injection.
Bronchoalveolar lavage (BAL)
Animals were anesthetized with an intramuscular dose of ketamine (10 mg/kg, Phoenix Pharmaceuticals, St. Joseph, MO) or tiletamine-zolazepam (5–8 mg/kg, Telazol; Fort Dodge, Fort Dodge, IA). Animals also received atropine (0.04 mg/kg, Phoenix Pharmaceuticals) intramuscularly to reduce salivary secretions and inhibit bradycardia. Once sufficiently anesthetized, animals were placed in dorsal recumbency on an examination table and preoxygenated with 100% oxygen via face mask. To facilitate insertion of the bronchoscope into a segmental bronchus of the right middle or right caudal lung lobe, cetacaine (Bergen Brunswig, Carrollton, TX) was applied topically to the epiglottis and the vocal folds via a spray bottle, and 1–5 ml of 1% lidocaine (Bergen Brunswig) was applied to the mucosa of the trachea at the level of the carina via the biopsy channel of the bronchoscope. For BAL, the bronchoscope was wedged into the desired segmental bronchus, and four individual 10-ml aliquots of sterile 0.9% saline were instilled into and aspirated from the lung via the biopsy channel of the bronchoscope. Post-BAL animals were oxygenated via facemask and monitored closely until they recovered completely from anesthesia.
Percoll gradient isolation of peripheral blood mononuclear cells (PBMCs) from blood
Blood samples were collected via femoral venipuncture with Vacutainer needles (22 gauge), needle holders, and blood collection tubes while the animals were under ketamine or tiletamine-zolazepam anesthesia. Whole blood was centrifuged at 1500 × g for 15 min and the plasma was removed. Percoll gradients were prepared by mixing 7.95 ml of room temperature Percoll (Pharmacia), 1.5 ml sterile Ca2+- and Mg2+-free 10× phosphate-buffered saline (PBS), and 5.55 ml sterile water (53% Percoll, 10% PBS, 37% water) per 10 ml blood. Blood was diluted to a ratio of 1:3.5 with 35 ml of sterile Ca2+- and Mg2+-free 1× PBS and carefully layered over 15 ml of diluted Percoll. Erythrocytes were sedimented by centrifugation at 1000 × g for 30 min at room temperature with the centrifuge brake off. The buffy coat was subsequently removed and washed with 1× PBS. Viability of cells was determined by a standard trypan blue dye exclusion assay and viable cells were counted with a hemocytometer.
Necropsy procedures
Prior to necropsy, animals were anesthetized with ketamine or Telazol, bled maximally from the femoral vein with the Vacutainer system described previously, and humanely euthanized with an intravenous overdose of sodium pentobarbital. The trachea and attached heart–lung block were then removed from the thoracic cavity and placed in a sterile tray for dissection and evaluation. All pathological findings in different organs were recorded. Mediastinal and tracheobronchial lymph nodes were collected. Lung lobes, as well as other tissues, were sectioned and samples were homogenized to obtain cells for FACS and ELISPOT analysis. The brain was removed and sectioned and serial sections were examined for evidence of central nervous system involvement and encephalitis. Selected pieces of pulmonary, lymphatic, and other organ tissues were preserved in 10% formalin for determination of tissue virus burden.
Virus load determination
RNA was extracted from plasma, BAL, lymph nodes, and tissues using the Qiagen RNeasy kit (Qiagen, Valencia, CA). Plasma was isolated from EDTA-treated whole blood. Virus loads from BAL cells, lymph nodes, and tissues were quantified as virus RNA copies per 106s cells, while virus load from plasma was quantified as virus RNA copies per ml of plasma. Quantification of SHIV-89.6P and SHIV-KU2 RNA was performed using an adaptation of a quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) protocol detecting the SIV gag sequence following a modification of a protocol described by Leutenegger et al.,14 and transformed into a multiplex assay by addition of an internal control (eGFP), as described by Cook et al.15 The eGFP probe used in these experiments was labeled at the 5 terminus with the fluorescent reporter VIC and the quencher TAMRA at the 3 end while the gag probe was labeled at the 5 end with FAM and the quencher TAMRA at the 3 end. During RNA isolation, each plasma sample was spiked with 108 copies of eGFP RNA transcript to allow back calculation for efficiency of RNA isolation. Plasma virus levels were calculated based on Ct values determined from standard curves consisting of SIV RNA transcripts diluted 10-fold from 109 RNA copies to 102 RNA copies per sample. Standards were run in triplicate and assays where the correlation coefficient of the standard curve (R2) was less than 0.990 were not accepted. The detection threshold was determined to be 50 virus copies RNA/ml. Samples were prepared in triplicate and each 50 μl RT-PCR mixture contained 1× AmpliTaq Gold reaction buffer (Applied Biosystems Inc., Foster City, CA), 1 mM dNTP mixture, 800 nM SIV.510f (5′-GCCAGGATTTCAGGCACTGT-3′), 800 nM SIV.592r (5′-GCTTGATGGTCTCCCACACAA-3′), 125 nM SIV.535pr (5′-AAGGTTGCACCCCCTATGACATTAATCAGATGTTA-3′), 40 nM eGFPf (5′-GCAGTGCTTCAGCCGCTAC-3′), 40 nM eGFPr (5′-AAGAAGATGGTGCGCTCCTG-3′), 125 nM eGFPpr (5′-CCGACCACATGAAGCAGCACGACTT-3′), 1 μl ROX dye (Invitrogen), 5 mM MgCl2, 6 U MMLV reverse transcriptase (Applied Biosystems Inc.), 1.25 U AmpliTaq gold (Applied Biosystems Inc.), 4 U RNase Out (Invitrogen), and 10 μl RNA sample. Samples were assayed on an ABI Prism 7700 Sequence Detector (Applied Biosystems Inc.). Cycling conditions were 48°C for 35 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min.
Generation of dendritic cells (DC) from PBMCs
Percoll gradient isolated PBMCs were plated at approximately 9 × 106 cells in 1–2 ml of DC medium [Iscove’s modified Dulbecco’s media (Invitrogen) containing 1% L-glutamine, 1% HEPES, 1000 U/ml human GM-CSF (Sigma, St. Louis, MO), 1000 U/ml human interleukin (IL)-4 (Sigma), and 10% human AB serum (Gemini Bioproducts, Calabasas, CA)] in six-well plates and incubated at 37°C/5% CO2 for 7 days. Fresh DC medium was added after 3–4 days. DCs were harvested at days 5–7 by removing the supernatant, adding 3–4 ml of 1× PBS per well, and incubating the plate on ice for 15 min. Cells were resuspended with gentle pipetting, transferred to a 15-ml conical tube, and centrifuged at 900 × g for 8 min. Cells were then resuspended in 1 ml of RPMI–10% human AB serum and counted.
Measurement of virus-induced interferon (IFN)-γ response by ELISPOT
ELISPOT assays were prepared by stimulating isolated PBMCs, BAL, or cells from tissues obtained at necropsy with SIV/SHIV-specific proteins. Then 96-well multiscreen MAIS-IP plates (Millipore, Bedford, MA) were coated by incubating them at 4°C overnight with 6 μg/ml antihuman/monkey IFN-γ monoclonal antibody (mAb GZ4, Mabtech, Cincinnati, OH) in endotoxin-free PBS (D-PBS, Life Technologies, Gaithersburg, MD). Coated plates were washed three times with D-PBS and blocked for 1 h at 37°C with 100 μl/well RPMI-1640 containing 1% HEPES, 1% L-glutamine, and 10% human AB serum (hereafter referred to as ELISPOT medium). SIVmac239 (SIV) and SHIV-89.6P antigens used as stimulators were obtained as overlapping 15-mer peptides from the AIDS Research and Reference Reagent Program (National Institutes of Health, Germantown, MD). Number of peptides included and NIH catalog numbers are indicated in parentheses. SIV/SHIV peptides assayed by ELISPOT included Gag (Cat. #6204, 125 peptides), Nef (Cat. #6206, 21 peptides), Tat (Cat. #6207, 30 peptides), Rev (Cat. #6448, 23 peptides), Pol (Cat. #6443, 263 peptides), and SHIV-89.6P (SHIV) Env (Cat. #4827, 218 peptides). Gag, SHIV Env, and Pol peptide pools were too large to include all peptide fragments in a single pool; consequently we subdivided Gag into two pools (Gag A, Gag B) corresponding to amino acids 5211–5273 and 5274–5335, and SHIV Env into four peptide pools (SHIV Env A, B, C, and D) corresponding to amino acids 6528–6582, 6582–6636, 6637–6690, and 6691–6745. Pol was subdivided into four pools (Pol A, B, C, and D) corresponding to amino acids 5710–5775, 5776–5841, 5842–5907, and 5908–5972. Concentrated stocks of stimulators were prepared by dissolving the lyophilized peptides in water or dimethyl sulfoxide (DMSO; Sigma) to a concentration of 2 mg/ml.
Stimulators for ELISPOT were prepared by diluting concentrated stock in ELISPOT medium to concentrations of 2 μg/ml for individual proteins and 10 μg/ml for pooled peptides. Phorbol 12,13-dibutyrate (PDBu) + ionomycin (50 nM and 10 μM final concentration respectively; Sigma) and staphylococcal enterotoxin B (SEB) (4 μg/ml; Sigma) were used as positive controls. Medium was removed from the plate and 50 μl of diluted stimulator or negative control (media only or DMSO-containing medium) was added to each well in duplicate. Percoll gradient-isolated PBMCs were resuspended in ELISPOT medium at a density of 2 × 105 cells/well in 150 μl of ELISPOT medium and added to stimulator-containing wells bringing the final volume to 200 μl/well. Dendritic cells (described above) were added to BAL cells at a DC:BAL ratio of 1:20 in 200 μl SIV/SHIV stimulator-containing ELISPOT medium. The same DC:cell ratio was used in ELISPOT assays using lymph nodes and biopsied tissue cells. BAL and tissue cell ELISPOT assays also included a pair of dendritic cell-only wells and BAL/tissue cell-only wells (media only, no dendritic cells) as negative controls. Plates were incubated with antigen for 40 h at 37°C/5% CO2, and then washed for 10 min with distilled water followed by four washes with ELISPOT wash buffer [D-PBS containing 0.05% Tween-20 (Sigma)]. The plates were then incubated with 2.5 μg/ml biotinylated antihuman IFN-γ antibody (mAb7-B61-Biotin, Mabtech) for 2 h at 37°C/5% CO2 and washed four times with ELISPOT wash buffer. Plates were then incubated with streptavidin–HRP (1:100 dilution; Mabtech) at 37°C/5% CO2, washed four times with ELISPOT wash buffer, and developed with AEC peroxidase substrate kit solution (Vector Laboratories Inc., Burlingame, CA). Color development was stopped by washing the plates three times with distilled water, once with PBS, air dried, and read using an ELISPOT plate reader (Cellular Technology LTD, Cleveland, OH). Data were normalized by dividing the mean number of spots from duplicate antigen-treated wells with the mean number of spots from duplicate media-only control wells (media-only wells without dendritic cells for BAL samples) and expressed as percent of media-only control with standard error of the mean.
Flow cytometry
Each sample to be stained contained approximately 7.5 × 105 cells isolated as described above. Cells were stained with fluorescently conjugated anti-CD3ε (clone SP34), anti-CD16 (clone 3G8), anti-CD8 (clone SK1), anti-CD4 (clone SK3), anti-CD69 (clone FN50), anti-CD29 (clone HUTS-21), anti-CD20 (clone 2H7), anti-CD11c (clone3.9, Biosource International, USA), anti-CD14 (clone MφP9), anti-CD11b/Mac-1 (clone ICRF44), anti-CCR5 (clone CTC5; R&D Systems, Minneapolis, MN), anti-CXCR4 (clone 12G5), and anti-CXCR3 (clone 1C6/CXCR3) antibodies. All antibodies were directed against human proteins and purchased from BD Biosciences (San Jose, CA) unless otherwise indicated. Replicate samples stained with appropriate isotype controls were used as negative controls. Flow cytometry was performed using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and analyzed using CellQuest software (Becton Dickinson).
Enumeration of CD4+ T cells
CD4+ T cell percentages were derived by flow cytometric analysis of blood lymphocytes. Fluorophore-conjugated anti-CD3ε and anti-CD4 (clone SK3) antibodies were used to stain Percoll gradient isolated PBMCs. Absolute lymphocyte counts (expressed as lymphocytes per μl blood) were estimated using an automated hematology instrument (Department of Hematology, Children’s Hospital of Pittsburgh, Pittsburgh, PA) and the results of this procedure multiplied by the percent of CD4+CD3+ cells determined by flow cytometry to obtain absolute numbers of CD4+ T cells/μl blood.
Determination of anti-Env and anti-Gag antibody titers
Longitudinal serum samples were assayed for antibody response to HIV-IIIB envelope (Env) or core Gag SIV proteins. For determining Env-specific antibody responses, Immulon II plates (Dynex Technologies, Chantilly, VA) were coated with 100 μl/well of Con A (50 μg/ml diluted in PBS) and incubated for 1 h at room temperature. Plates were washed twice with 1 × PBS. Detergent-disrupted HIV-IIIB antigen (1 μg/well diluted in 1 × PBS containing 1% Triton X-100) was added and incubated for 1 h at room temperature to specifically capture viral Env protein. Plates were washed four times with 1 × PBS. Blotto (5% instant milk in PBS; 100 μl/well) was added and the plates were incubated either for 1 h at 25°C or 4°C overnight. To measure anti-Gag antibody titers, Immulon II plates were coated with detergent-disrupted SIV-B7 (1 μg/well diluted in 0.05 M sodium bicarbonate buffer, pH = 7.2, containing 0.05% Triton X-100) and incubated overnight at 25°C. Plates were washed twice with 1× PBS and then blocked using 5% blotto (100 μl/well) for 1 h at 25°C. To quantify the antibody titers bound to the HIV/SIV antigens, blotto was removed from the plates and 50 μl of 2-fold serial dilutions of monkey serum in blotto was added to each antigen-coated well. Plates were incubated for 1 h at room temperature. Plates were then washed five times with 1 × PBS and antimonkey IgG-HRP (50 μl/well, diluted 1:30,000 in blotto) was added to each well followed by a 1-h incubation at room temperature. Plates were washed five times with 1× PBS. Then 200 μl/well of room temperature TM blue substrate (Celliance, Milford, MA) was added and shaken for 20 min at room temperature. Color development was stopped by adding 50 μl/well of 1 N sulfuric acid. Absorbance was read at 450 nm.
RESULTS
SHIV infection and gross outcome of infection
We infected two groups of monkeys with SHIV-89.6P (monkeys 6704, 6904, 7004, and 7504) or SHIV-KU2 (monkeys 14804, 15404, 15504, and 15604) and followed a range of immunologic parameters over the course of infection to determine how these viruses affect the health of cynomolgus monkeys. Monkeys 6704 and 15604 were necropsied at predetermined time points 6 months and 1 year postinfection (pi), respectively. Monkeys 6904 and 15404 were euthanized and necropsied after developing respiratory distress during BAL procedures at 20.4 and 16.9 weeks pi, respectively. The remaining monkeys (7004, 7504, 14804, and 15504) were followed between 40 and 51 weeks pi and entered into subsequent studies, which will be detailed elsewhere. None of the monkeys displayed symptoms of simian AIDS (SAIDS)-like disease including anorexia, chronic diarrhea, SHIV-related encephalitis, or SAIDS-associated neurologic symptoms, but monkey 14804 presented with fatal lymphoma 1 week after exiting this study.
SHIV infection causes rapid depletion of CD4+ T cells
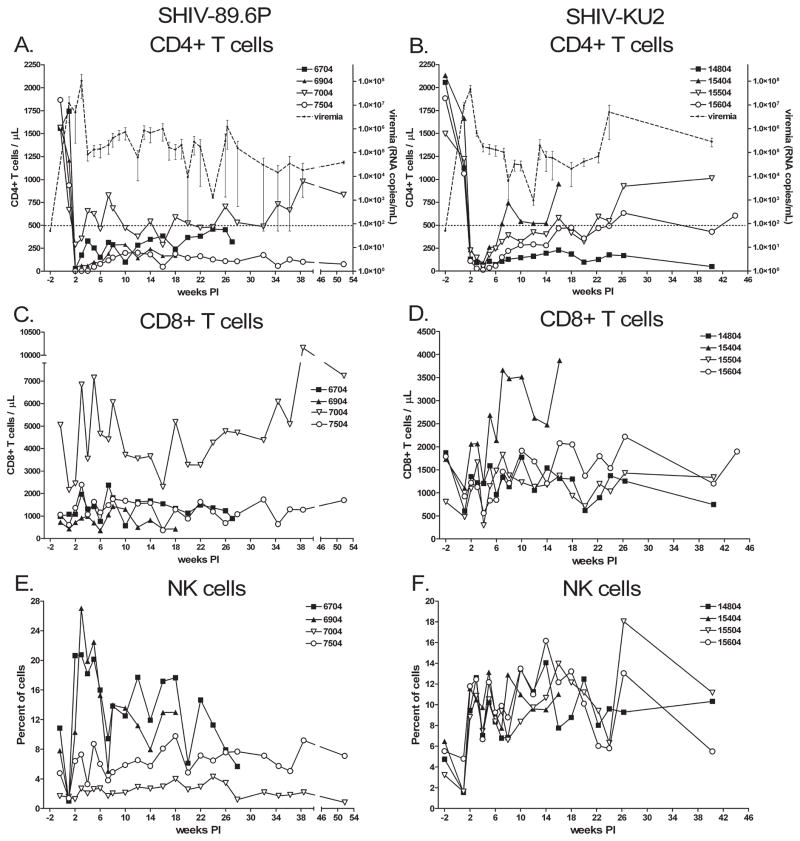
The proportions of CD4+ and CD8+ T cells in each monkey group were determined to examine how SHIV infection affected these cell populations. We found the kinetics of CD4+ T cell depletion followed similar patterns with either virus (Fig. 1A and B). Depletion occurred most rapidly in SHIV-89.6P-infected monkeys with maximal depletion observed 2 weeks pi (average 92 CD4+ T cells/μl, range 3–289) compared with 4 weeks pi for SHIV-KU2-infected monkeys (average 54 CD4+ T cells/μl, range 14–88). Shortly after reaching the nadir, cell numbers recovered to a new baseline of CD4+ T cells/μl that was 325 ± 19 cells/μl (standard error of 21 time points) and 374 ± 36 cells/μl (standard error of 15 time points) for SHIV-89.6P- and SHIV-KU2-infected animals, respectively.
FIG. 1.
Changes in CD4+ and CD8+ T lymphocyte populations in SHIV-infected cynomolgus macaques. The numbers of CD4+, CD8+ T cells and NK cells from SHIV-89.6P (A, C, and E, respectively) and SHIV-KU2 (B, D, and F, respectively) were determined by selectively gating on CD4+CD3+, CD8+CD3+, and CD8+CD16+ (NK cells) cells. The threshold for simian AIDS (CD4+ T cell numbers ≤500 cells/μl) is indicated by a horizontal dashed line in (A) and (B). Plasma virus load was determined at the same time as T lymphocyte numbers, and is indicated as the mean of replicate infections (dashed line with small closed circles). Error bars indicate standard error.
We determined the proportions of CD4+ T cells in the BAL fluid of all monkeys at 9 and 17 weeks pi, and the inguinal lymph nodes and tissues of two necropsied monkeys from each group. At 9 weeks pi, CD4+ T cells comprised 12.3 ± 3.2% and 14.8 ± 4.0% of cells within the lymphocyte gate in SHIV-89.6P- and SHIV-KU2-infected animals. There were significant declines in the proportion of CD4+ T cells in lymph nodes of SHIV89.6P-infected animals at 17 weeks pi, with 3.9 ± 1.1% of cells within the lymphocyte gate being CD3+CD4+ (Student’s t test, p = 0.046). Differences were observed between proportions of CD4+ cells in BAL fluid of SHIV-89.6P-and SHIV-KU2-infected animals at 17 weeks pi, with 3.9 ± 1.1% and 15.1 ± 3.5% of cells being CD3+CD4+, respectively (Mann-Whitney rank sum test, p = 0.057). The percent of CD4+ T cells in inguinal lymph nodes was determined at 4 weeks and 8 weeks with little change between the two time points in either infection (data not shown). The numbers of CD4+ T cells in spleen, lung, and lymph node tissues were determined from monkeys 6704 and 6904 at necropsy. We found 10–20% of T cells within the lymphocyte gate were CD4+, while numbers of CD4+ cells in the lung lobes were significantly lower (<1% of gated lymphocytes; data not shown). Monkey 6704 had even fewer CD4+ T cells in these tissues, with lymph nodes containing 4–10% of CD4+ cells and the lungs containing less than 0.5% CD4+ T cells (data not shown).
Plasma virus titers were determined every week pi for 16 weeks and every 2 weeks thereafter. Virus titer in plasma (measured as virus RNA copies/ml plasma) increased sharply following infection and reached peak viremia at a time that corresponded with the lowest number of CD4+ T cells (Fig. 1A and B). SHIV-89.6P-infected monkeys experienced peak viremia 3 weeks pi with 1.0 × 108 8.7 × 107 (mean ± SE) virus RNA copies/ml (Fig. 1A), while viremia in SHIV-KU2-infected monkeys occurred at 2 weeks pi and was lower with a peak of 4.6 × 107 ± 2.0 × 107 virus RNA copies/ml (Fig. 1B). After reaching peak viremia, virus burden subsided to 2.6 × 105 ± 5.7 × 104 virus RNA copies/ml for SHIV-89.6P and 4.4 × 105 ± 2.6 × 105 virus RNA copies/ml for SHIV-KU2-infected monkeys.
CD8+ T cell, B cell, natural killer cell, and monocyte numbers following SHIV infection
We assessed whether SHIV infection caused significant changes in the numbers of CD8+ T cells, B cells, natural killer cells (NK cells), and monocytes in the blood. CD8+ T cell numbers did not undergo changes that could be directly attributed to SHIV infection (Fig. 1C and D). Monkey 7004, which had the highest number of CD8+ T cells/μl, maintained a higher baseline number of CD4+ T cells (mean CD4+ T cell counts = 585 ± 37 cells/μl) postinfection and the best controlled plasma viremia. The population of B cells (defined here as CD14−CD20+ cells) and monocytes (defined here as CD11c+CD14+ cells) did not undergo changes that correlated with virus burden (data not shown). Numbers of NK cells (defined here as CD8+CD16+ cells) in SHIV-89.6P-infected animals 6704 and 6904 and all SHIV-KU2-infected monkeys underwent a slight contraction after infection followed by an increase to slightly higher than initial numbers for the remainder of the study (Fig. 1E and F). This response was observed more consistently, and occurred most uniformly in SHIV-KU2-infected monkeys.
Chemokine receptor expression on CD4+ and CD8+ T cells
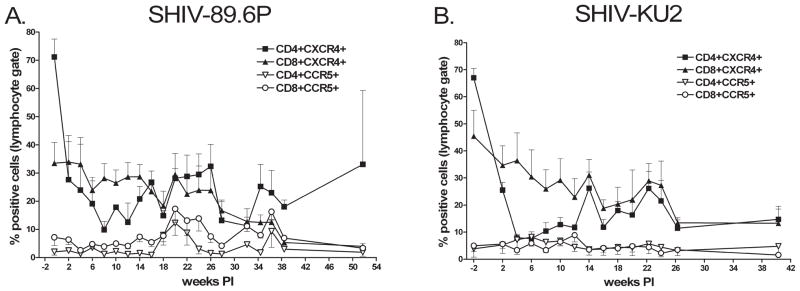
We used flow cytometry to measure the proportion of CD4+ and CD8+ cells expressing the chemokine receptors CXCR4 and CCR5 to determine how SHIV infection affected these subpopulations (Fig. 2). Both viruses caused precipitous declines in CXCR4+CD4+ T cells with minimum numbers of CXCR4+CD4+ for SHIV-89.6P-infected animals occurring 8 weeks pi (Fig. 2A) and 4 weeks pi for SHIV-KU2-infected animals (Fig. 2B). The proportion of CXCR4+CD4+ T cells recovered slightly, but in both cases remained depressed below the preinfection numbers. The numbers of CXCR4+CD8+ T cells steadily declined from the time of infection to the end of the study. We observed that CCR5+CD4+ and CCR5+CD8+ T cells were present in small numbers and populations did not undergo changes that correlated with viremia.
FIG. 2.
Changes in CXCR4 and CCR5 expressing lymphocytes following SHIV infection. Lymphocytes were selectively gated CD4+ and CD8+ cells and the proportion of CXCR4+ and CCR5+ cells was determined. (A) SHIV-89.6P-infected macaques. (B) SHIV-KU2-infected macaques. Data represent the mean of replicate infections. Lower error bars for CXCR4+ cells and upper error bars for CCR5+ cells indicating standard error have been omitted for clarity.
Changes in activation and memory marker expressing T cell populations
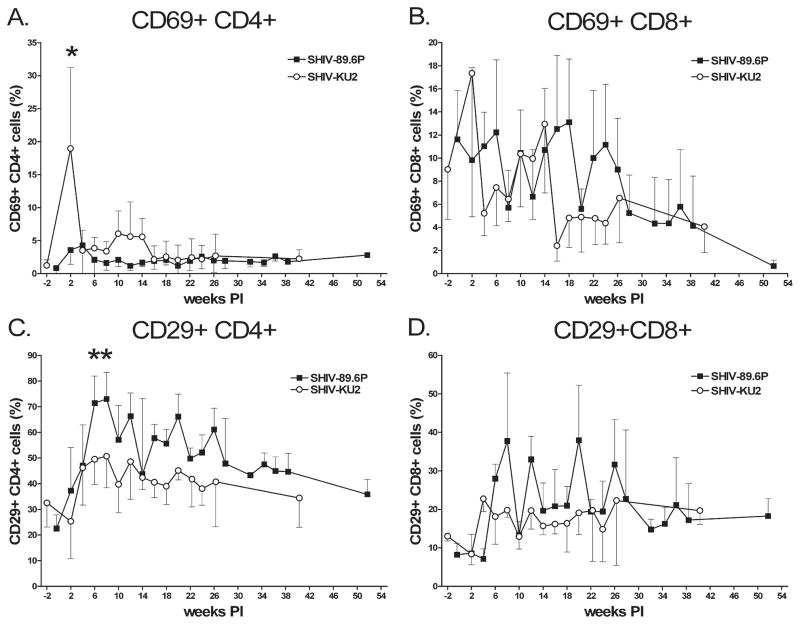
To determine whether there were differences in overall T cell activation patterns, expression of an early activation marker (CD69), and expression of a T cell memory marker (CD29) were followed. We noted an early increase in the number of CD69+CD4+ T cells in SHIV-KU2-infected monkeys that was significantly greater than the small increase noted in SHIV-89.6P-infected monkeys (Student’s two-tailed t test, p < 0.05) (Fig. 3A). Initial increases in CD69+CD4+ T cells were short lived, however, and we observed few of these cells beyond the early time points. In contrast, we observed negative trends in numbers of CD69+CD8+ T cells that was apparent with both virus strains (Fig. 3B). Virus infection initially caused increases in CD29+CD4+ cells with peak numbers at 8 weeks pi for both viruses, with significantly more CD29+CD4+ cells noted in SHIV-89.6P-infected monkeys than SHIV-KU2-infected monkeys 6 and 8 weeks pi (p < 0.05) (Fig. 3C). There were declines in CD29+CD4+ T cell numbers after this peak and by the end of the study period the numbers of CD29+CD4+ T cells were similar to initially observed numbers. Infection also caused a slight elevation in CD29+CD8+ cell numbers that was sustained for the duration of the study (Fig. 3D).
FIG. 3.
Activation status of CD4+ and CD8+ T lymphocytes following SHIV infection. CD69+ and CD29+ T lymphocytes from SHIV-89.6P (A and C, respectively) and SHIV-KU2 (B and D, respectively) were determined by selectively gating on activation marker-positive CD4+ and CD8+ cells. Data represent the mean value of replicate infections. Asterisks indicate statistically significant differences (p < 0.05). Upper or lower error bars indicating standard error have been omitted for clarity.
Virus burden in organs, lymph nodes, BAL cells, and lungs
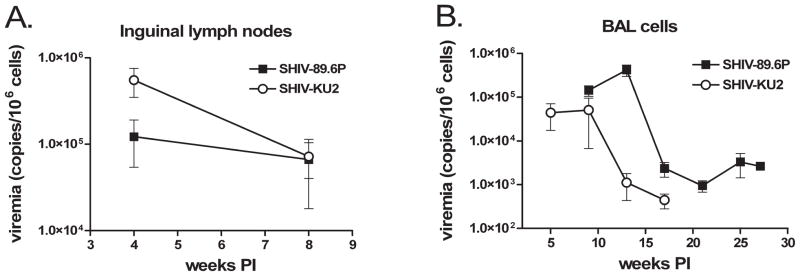
We measured virus burden in tissues from the spleen, kidneys, liver, lymph nodes, lungs, and BAL cells to determine whether there were virus-specific differences in tissue tropism and virus burden. We found tissue from the spleen, kidneys, and liver contained measurable quantities of virus RNA (Table 1). There was considerable variation between tissues from different animals that did not correlate with virus strain or length of infection. The lungs are the initial site of Mtb infection, and it is important for our future studies to know whether these viruses are in the lungs and whether there are virus-specific differences in lung burden. BAL provided a minimally invasive method to quantify virus in the lung environment. We found virus in BAL cells from monkeys of both groups, with higher levels of viral RNA in SHIV-89.6P-infected monkeys (Fig. 4A). Virus burden in BAL cells was highest early in infection and lower at later measurements (~103 virus RNA copies/106 cells), but we found more virus and longer persistence in SHIV-89.9P-infected animals. Similarly, lung tissue from SHIV-89.6P-infected monkeys contained greater quantities of viral RNA than lung tissue from SHIV-KU2-infected monkeys (Table 2). Thoracic lymph nodes are frequent sites of secondary Mtb infection, and viral replication in the lymph nodes may affect outcome of Mtb infection. Inguinal lymph nodes were biopsied at 4 and 8 weeks pi. There was more SHIV-KU2 RNA in lymph nodes than SHIV89.6P RNA at 4 weeks pi, but by 8 weeks pi this quantity had declined and both viruses were present in equivalent amounts (Fig. 4B). Hilar lymph nodes from animals 6704, 6904, and 15604 at the time of necropsy also contained significant quantities of viral RNA (Table 3).
Table 1.
Virus Load in Organs from SHIV-89.6P and SHIV-KU2-Infected Monkeys
| Monkey numbera | Weeks PI | Organb |
||
|---|---|---|---|---|
| Spleen | Kidney | Liver | ||
| 6704 | 27.6 | 84 | 2.0 × 103 | ≤50 |
| 6904 | 20.4 | 1.1 × 103 | 986 | 3.8 × 103 |
| 15604 | 43.0 | 5.9 × 104 | 2.1 × 106 | ≤50 |
Monkeys 6704 and 6904 belong to the SHIV-89.6P-infected group while monkey 15604 was infected with SHIV-KU2.
Virus load is expressed as viral copies per 106 cells from tissues obtained at necropsy.
FIG. 4.
Virus load in biopsied lymph nodes and BAL cells. (A) Inguinal lymph nodes were biopsied at 4 and 8 weeks, homogenized into single-cell suspensions, and the virus burden quantified per 106 cells by real-time PCR. (B) Cells from BAL procedures were centrifuged and the virus load determined per 106 cells by real-time PCR. Data represent the mean of replicate infections. Error bars indicate the standard error. Asterisks indicate statistically significant differences (p < 0.05).
Table 2.
Virus Load in Lymph Nodes from SHIV-89.6P and SHIV-KU2-Infected Monkeys
| Monkey numbera | Mediastinalb | Axillaryb |
Inguinalb |
Hilarb |
|||
|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | Left | Right | ||
| 6704 | 809 | ND | 4.6 × 103 | ND | 1.3 × 104 | 3.4 × 103 | 546 |
| 6904 | ND | ND | 9.9 × 106 | ND | ND | ND | 1.9 × 106 |
| 15604 | 1.1 × 106 | 4.0 × 104 | 3.1 × 105 | 4.1 × 106 | ND | ND | 3.1 × 105 |
Monkeys 6704 and 6904 belong to the SHIV-89.6P-infected group while monkey 15604 was infected with SHIV-KU2. The time of virus quantification is indicated in Table 1.
Virus load is expressed as virus copies per 106 cells from tissues obtained at necropsy. ND indicates where virus load was not determined.
Table 3.
Virus Load in Lungs from SHIV-89.6P and SHIV-KU2-Infected Monkeys
| Monkey numbera | BALb | Left lobeb |
Right lobeb |
||||
|---|---|---|---|---|---|---|---|
| Upper | Middle | Lower | Upper | Middle | Lower | ||
| 6704 | 2.6 × 103 | ≤50 | ND | ≤50 | ND | ND | ≤50 |
| 6904 | ND | 2.0 × 103 | ND | 3.2 × 103 | 805 | 2.0 × 103 | 244 |
| 15604 | ≤50 | ≤50 | ≤50 | 1.7 × 104 | ≤50 | ≤50 | ≤50 |
Monkeys 6704 and 6904 belong to the SHIV-89.6P-infected group while monkey 15604 was infected with SHIV-KU2. The time of virus quantification is indicated in Table 1.
Virus load is expressed as virus copies per 106 cells from tissues obtained at necropsy. ND indicates where virus load was not determined.
Measurement of anti-SHIV immune responses
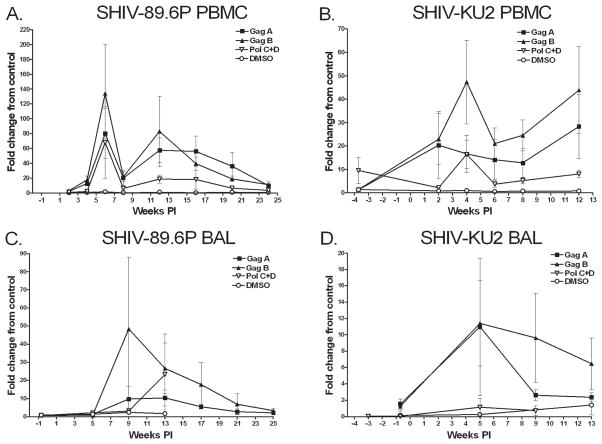
ELISPOT assays, stimulating with SHIV antigen peptide pools, were used to assess the frequency of IFN-γ-secreting cells in PBMCs, BAL fluid, and tissues. No response was detected in negative controls or dendritic cell-only wells while positive controls always elicited strong responses. The frequency of IFN-γ-producing cells in PBMCs or BAL fluid in response to the SIV/SHIV peptides Nef and Rev or pools of Tat, Pol, and SHIV Env peptides was very low (data not shown). IFN-γ-producing cells were observed primarily in response to Gag A, Gag B, Pol C + D peptide pools (Fig. 5). BAL cells from SHIV-89.6P-infected monkeys produced significantly more IFN-γ than BAL cells from SHIV-KU2-infected monkeys (Man–Whitney rank sum test; p = 0.015). Significant differences in the frequency of IFN-γ-secreting PBMCs were not noted between groups. Peptide-specific responses in the axillary lymph nodes at 4 and 8 weeks pi were generally low in both infections, with modest responses to Gag, Pol, and Tat peptide pools (data not shown). IFN-γ secretion from lung, spleen, and hilar lymph node tissue obtained at necropsy similarly displayed low responsiveness (data not shown).
FIG. 5.
IFN-γ expression by BAL cells and PBMCs following interaction with SHIV antigen-pulsed dendritic cells. Cells were seeded into 96-well plates and IFN-γ expression measured by ELISPOT assay. Results are expressed as the fold change from control where the number of spot-forming cells (SFCs) in wells containing SHIV antigen pulsed dendritic cells or DMSO only was compared to the number of SFCs produced by cells in wells receiving ELISPOT medium only. Data represent the mean of replicate SHIV-89.6P-infected monkeys (A and C) and SHIV-KU2-infected monkeys (B and D). Error bars indicate standard error.
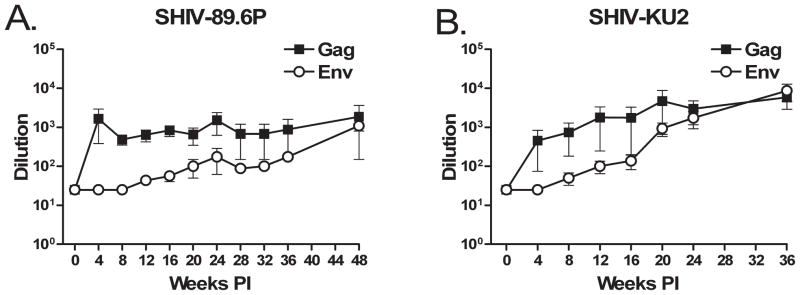
We determined antibody titers against HIV strain HIV-IIIB and SIV-B7 Gag proteins by ELISA every 4 weeks pi (Fig. 6). While antibody titers progressively increased over the course of infection, neither virus elicited strong production of antibodies directed against Env or Gag proteins. Anti-Gag antibody titers were higher in both groups; they peaked early in SHIV-89.6-infected animals, but did not increase significantly afterward (Fig. 6A). Titers against Env remained lower than anti-Gag titers, although by 36 weeks pi the antibody titers against Env and Gag were equivalent in SHIV-KU2-infected monkeys.
FIG. 6.
Production of anti-Gag and anti-Env antibodies. Plasma antibody titers were determined by Gag and Env ELISAs. Dilution indicates the reciprocal of the endpoint dilution beyond which bound antibody was no longer detectable. (A) SHIV-89.6P-infected animals. (B) SHIV-KU2-infected animals. Error bars indicate the standard error and in some cases are obscured by the data marker.
DISCUSSION
A variety of primate models have been established to reproduce the effects of human infection with HIV. While no model is ideal, SIV-infected rhesus macaques (Macaca mulatta) have been informative and display much of the pathology associated with HIV-related immunosuppression. Unfortunately, SIV depletes CD4+ T cells very slowly over a time frame, making it difficult to distinguish the disease caused by SIV and that related to coinfecting microorganisms. Pathogenic SHIV rapidly reduces CD4+ T cell numbers and may be a better system for investigating host–pathogen immune responses in a severely immunocompromised host. In addition, the rapid loss of CD4 T cells provides a more cost-effective and reproducible model of coinfections in a CD4-depleted host.
SHIV-infected rhesus macaques are frequently used to model HIV infection and their responses to SHIV have been intensively studied.4,7,12,13 In addition to modeling HIV, macaques have also been used as models of human TB. Reactivation of latent TB is a significant source of mortality in HIV-positive individuals, yet a model accurately representing Mtb–HIV coinfection has remained elusive. Although the directly comparative studies are small, and have not been performed with low level infectious doses, it appears that rhesus macaques may be more susceptible to Mtb-induced disease than cynomolgus macaques.11 However, following low-dose infection, cynomolgus macaques accurately mimic active, latent, and reactivated TB in humans.10,16 In fact, this is the only true animal model of latent TB. Therefore, we sought to determine whether SHIV-infected cynomolgus macaques experience immunosuppression and could be used to model the physiology of reactivated TB in HIV-infected humans.
We infected cynomolgus macaques with two pathogenic SHIV strains, SHIV-89.6P and SHIV-KU2, and followed the animals to see how the viruses affected their immune status. These SHIV strains cause substantial CD4+ T cell depletion in rhesus macaques within a few weeks of infection.13,17,18 In contrast, cynomolgus macaques of Mauritanian origin that have been reported to be relatively resistant to SHIV-89.6P caused CD4+ T cell depletion.19 Our results were consistent with the rhesus model of SHIV infection in that we observed significant CD4+ depletion and prolonged depression of CD4+ cell numbers following infection. Rhesus macaques from geographically distinct origins show significantly different responses to SIV infection,20 suggesting geography-related genetic differences may play roles in determining how susceptible animals are to SHIV infection. Given this, we suspect genetic differences associated with the Vietnamese or Chinese-origin macaques we used may explain the differences between our observations and the previously reported study. SHIV-89.6P infection depressed CD4+ T cells somewhat earlier and produced slightly higher plasma viremia than SHIV-KU2 infection. While the initial CD4+ T cell depression was slightly greater in SHIV-89.6P-infected monkeys, both viruses exhibited similar capacities to inhibit CD4+ T cell recovery. Neither virus strongly affected numbers of circulating CD8+ T cells, B cells, or monocytes and we observed slight increases in NK cell numbers similar to human HIV infection.21 SHIV-infected macaques frequently experience SAIDS-like symptoms within weeks to months of infection.6,13 Despite steep declines in CD4+ numbers, our monkeys did not experience obvious SAIDS-like symptoms. One SHIV-KU2-infected monkey (14804) succumbed to lymphoma, a pathology occasionally observed in SIV-immunosupressed cynomolgus macaques,22 suggesting infection had additional effects beyond depletion of CD4+ T cells.
We noted virus strain-specific differences in the generation of CD29+ memory T cells and IFN-γ expression. Infection caused an increase in numbers of CD29+CD4+ cells, particularly in SHIV-89.6P-infected monkeys. The increased memory response correlates with our ELISPOT data indicating SHIV-89.6P elicited greater IFN-γ production than SHIV-KU2. Expanded memory T cell populations were not sustained, however, and much of the early accumulation was lost by the end of the study. X4-tropic SHIV infections significantly impact naive CD4+ T cells populations in the peripheral blood and lymph nodes7 while dual tropic viruses also focus on memory CD4+ T cells in the GALT, spleen, and lymph nodes.7,23 Thus, once naive T cell populations are lost, declines in memory T cell populations herald further immunosuppression.24 The declines in memory CD4+ T cells seen here indicate both virus strains exacted significant long-term tolls on a variety of CD4+ T cell populations that may include cell populations important in containing Mtb infection.
Primary Mtb infection usually occurs in the lungs with dissemination to lymph nodes and other organs.25,26 Consequently, we wanted to determine whether these viruses localize in tissues that frequently become infected with Mtb, so that when coinfection studies are performed, the impact of Mtb on viral infection in the lungs, and vice versa, can be studied. Not surprisingly, both viruses were detectable in the lungs, BAL cells, lymph nodes, spleen, kidneys, and liver. Biopsied lung tissues and BAL cells consistently contained more SHIV-89.6P RNA than SHIV-KU2 RNA. Moreover, SHIV-89.6P virus titer persisted in the lungs longer than SHIV-KU2. SHIV-89.6P has dual (X4/R5) tropism and can infect both lymphocytes and macrophages,13 while SHIV-KU2 is X4 tropic 27 and restricted to a smaller subset of cells that may not include alveolar macrophages, the most abundant cell type in BAL fluid and the first cell type infected by Mtb. These differences in tropism may have direct consequences in Mtb-infected macaques as Mtb infection has been reported to upregulate CXCR4 expression on alveolar macrophages and provide an additional reservoir for virus in the lung after CD4+ T cell depletion.28
Cynomolgus macaques are outbred and genetically diverse, and we noted considerable monkey-to-monkey variation in the immunologic and virologic parameters we followed. The diversity of responses indicates these animals present many physiologic aspects and pathologic outcomes observed in human HIV infection. We predict there will be a similar range of responses to Mtb coinfection that resemble human responses, reinforcing the utility of this species as a model of Mtb/HIV coinfection. That said, we found both viruses exhibited similar effects in cynomolgus macaques. A primary and important difference was the persistence and quantity of virus in the lungs. Consequently, we anticipate that cynomolgus macaques infected with SHIV-89.6P will be the best model for examining the effects of virus-induced immunosuppression on latent TB.
Acknowledgments
We dedicate this work to Dr. Opendra “Bill’ Narayan (1936–2007) in recognition of his accomplishments and contributions to HIV/AIDS research. The authors thank Dr. Anita Trichel, Dr. Edwin Klein, and Dr. Saverio Capuano, as well as Andre Samuel and the veterinary technical staff at the University of Pittsburgh School of Medicine for procedures and care of the animals. We also thank Russell D. Salter and Sarita Singh for providing supporting data (data not shown, funding through NIH N01 AI-50018). SHIV peptides were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
References
- 1.Selwyn PA, Hartel D, Lewis VA, et al. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N Engl J Med. 1989;320:545–550. doi: 10.1056/NEJM198903023200901. [DOI] [PubMed] [Google Scholar]
- 2.Corbett EL, Watt CJ, Walker N, et al. The growing burden of tuberculosis: Global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 3.Lawn SD, Butera ST, Shinnick TM. Tuberculosis unleashed: The impact of human immunodeficiency virus infection on the host granulomatous response to Mycobacterium tuberculosis. Microbes Infect. 2002;4:635–646. doi: 10.1016/s1286-4579(02)01582-4. [DOI] [PubMed] [Google Scholar]
- 4.Hu SL. Non-human primate models for AIDS vaccine research. Curr Drug Targets Infect Disord. 2005;5:193–201. doi: 10.2174/1568005054201508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nath BM, Schumann KE, Boyer JD. The chimpanzee and other non-human-primate models in HIV-1 vaccine research. Trends Microbiol. 2000;8:426–431. doi: 10.1016/s0966-842x(00)01816-3. [DOI] [PubMed] [Google Scholar]
- 6.Joag SV. Primate models of AIDS. Microbes Infect. 2000;2:223–229. doi: 10.1016/s1286-4579(00)00266-5. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura Y, Brown CR, Mattapallil JJ, et al. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc Natl Acad Sci USA. 2005;102:8000–8005. doi: 10.1073/pnas.0503233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber SA, Gama L, Li M, et al. Longitudinal analysis of simian immunodeficiency virus (SIV) replication in the lungs: Compartmentalized regulation of SIV. J Infect Dis. 2006;194:931–938. doi: 10.1086/507429. [DOI] [PubMed] [Google Scholar]
- 9.Sui Y, Li S, Pinson D, et al. Simian human immunodeficiency virus-associated pneumonia correlates with increased expression of MCP-1, CXCL10, and viral RNA in the lungs of rhesus macaques. Am J Pathol. 2005;166:355–365. doi: 10.1016/S0002-9440(10)62259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capuano SV, Croix DA, Pawar S, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh GP, Tan EV, de la Cruz EC, et al. The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease. Nat Med. 1996;2:430–436. doi: 10.1038/nm0496-430. [DOI] [PubMed] [Google Scholar]
- 12.Joag SV, Li Z, Foresman L, et al. Characterization of the pathogenic KU-SHIV model of acquired immunodeficiency syndrome in macaques. AIDS Res Hum Retroviruses. 1997;13:635–645. doi: 10.1089/aid.1997.13.635. [DOI] [PubMed] [Google Scholar]
- 13.Reimann KA, Li JT, Veazey R, et al. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leutenegger CM, Higgins J, Matthews TB, et al. Real-time Taq-Man PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res Hum Retroviruses. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- 15.Cook RF, Cook SJ, Li FL, Montelaro RC, Issel CJ. Development of a multiplex real-time reverse transcriptase-polymerase chain reaction for equine infectious anemia virus (EIAV) J Virol Methods. 2002;105:171–179. doi: 10.1016/s0166-0934(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 16.Lin PL, Pawar S, Myers A, et al. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74:3790–3803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reimann KA, Watson A, Dailey PJ, et al. Viral burden and disease progression in rhesus monkeys infected with chimeric simian-human immunodeficiency viruses. Virology. 1999;256:15–21. doi: 10.1006/viro.1999.9632. [DOI] [PubMed] [Google Scholar]
- 18.Pal R, Venzon D, Letvin NL, et al. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimann KA, Parker RA, Seaman MS, et al. Pathogenicity of simian-human immunodeficiency virus SHIV-89. 6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J Virol. 2005;79:8878–8885. doi: 10.1128/JVI.79.14.8878-8885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcondes MC, Penedo MC, Lanigan C, et al. Simian immunodeficiency virus-induced CD4+ T cell deficits in cytokine secretion profile are dependent on monkey origin. Viral Immunol. 2006;19:679–689. doi: 10.1089/vim.2006.19.679. [DOI] [PubMed] [Google Scholar]
- 21.Beck JM. The immunocompromised host: HIV infection. Proc Am Thorac Soc. 2005;2:423–427. doi: 10.1513/pats.200507-077JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habis A, Baskin GB, Murphey-Corb M, Levy LS. Simian AIDS-associated lymphoma in rhesus and cynomolgus monkeys recapitulates the primary pathobiological features of AIDS-associated non-Hodgkin’s lymphoma. AIDS Res Hum Retroviruses. 1999;15:1389–1398. doi: 10.1089/088922299310098. [DOI] [PubMed] [Google Scholar]
- 23.Grivel JC, Elliott J, Lisco A, et al. HIV-1 pathogenesis differs in rectosigmoid and tonsillar tissues infected ex vivo with CCR5- and CXCR4-tropic HIV-1. AIDS. 2007;21:1263–1272. doi: 10.1097/QAD.0b013e3281864667. [DOI] [PubMed] [Google Scholar]
- 24.Ayash-Rashkovsky M, Chenine AL, Steele LN, et al. Coinfection with Schistosoma mansoni reactivates viremia in rhesus macaques with chronic simian-human immunodeficiency virus clade C infection. Infect Immun. 2007;75:1751–1756. doi: 10.1128/IAI.01703-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Molecular epidemiology of tuberculosis: Current insights. Clin Microbiol Rev. 2006;19:658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma SK, Mohan A, Sharma A, Mitra DK. Miliary tuberculosis: New insights into an old disease. Lancet Infect Dis. 2005;5:415–430. doi: 10.1016/S1473-3099(05)70163-8. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman TL, Stephens EB, Narayan O, Doms RW. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoshino Y, Tse DB, Rochford G, et al. Mycobacterium tuberculosis-induced CXCR4 and chemokine expression leads to preferential X4 HIV-1 replication in human macrophages. J Immunol. 2004;172:6251–6258. doi: 10.4049/jimmunol.172.10.6251. [DOI] [PubMed] [Google Scholar]