Abstract
We compare the receptor-based mechanisms that a small RNA virus and a larger DNA virus have evolved to drive the fusion of viral and cellular membranes. Both systems rely on tight control over triggering the concerted refolding of a trimeric fusion protein. While measles virus entry depends on a receptor-binding protein and a fusion protein only, the herpes simplex virus is more complex and requires four viral proteins. Nevertheless, in both viruses a receptor-binding protein is required for triggering the membrane fusion process. Moreover, specificity domains can be appended to these receptor-binding proteins to target virus entry to cells expressing a designated receptor. We discuss how principles established with measles and herpes simplex virus can be applied to targeting other enveloped viruses, and alternatively how retargeted envelopes can be fitted on foreign capsids.
INTRODUCTION
The Latin virus refers to poison and other noxious substances, but viruses have long been modified to serve as vectors for gene delivery. Viruses genetically modified to destroy cancer cells have reached advanced clinical trials, and one of them is an approved therapeutic[1]. Thus the transformation of viruses from agents of disease into therapeutics is already clinical reality. New developments that accelerate this transformation are based on the combination of three principles: targeting, arming and shielding[2]. We discuss here how the entry of enveloped viruses can be retargeted to specific cells for either oncolytic or gene delivery purposes. We review how one enveloped RNA virus and one enveloped DNA virus have been retargeted, and how principles established with these two systems can be applied to all enveloped viruses.
Of note, while a few viruses with post-entry tropism modifications are in advanced (phase III) clinical trials as oncolytics, viruses with retargeted entry have not yet reached the clinical trial stage. In particular, entry targeting of icosahedral viruses is complicated by the cumulative constraints of capsid symmetry. In view of these considerations, it is not surprising that to date, the number of viruses that have been successfully retargeted at the entry level remains small. However, these constraints do not apply to those enveloped RNA viruses that have helical nucleocapsids, and affect only indirectly the envelopes of the large DNA viruses. The virus family Paramyxoviridae, with a negative strand RNA genome, and the family Herpesviridae, with a double stranded DNA genome, have been retargeted to recognize and enter cells using designated receptors. In this review, we will cover the proposed entry mechanisms for measles virus (MV), a member of the paramyxovirus family, and of herpes simplex virus I (HSV), a member of the herpesviruses. We will discuss the similarities and differences between HSV and MV entry mechanisms, and how these systems have been modified to achieve retargeted entry.
Measles Virus (MV) Entry Mechanism
MV is a member of the paramyxovirus family, which includes prevalent human viruses such as mumps, parainfluenza and respiratory syncytial virus, and emerging viruses such as Hendra and Nipah. These viruses have similar two-glycoprotein entry complexes, but the receptor-binding protein is named HN (hemagglutinin-neuraminidase) when it interacts with oligosaccharide receptors, and either H or G protein when it interacts with protein receptors.
MV fuses its envelope with the host-cell membrane at neutral pH. The receptor binding and membrane fusion functions are separated on two proteins, the tetrameric hemagglutinin (H) and the trimeric fusion (F) proteins, respectively (Figure 1). MV uses two receptors sequentially to infect and spread through selected organs of the human body. In the airway's lumen, wild type MV-H binds to signaling lymphocyte activation molecule (SLAM, see Figure 1B) on alveolar macrophages and dendritic cells, which ferry the infection through the epithelial barrier [3]. Vigorous MV replication in primary and secondary lymphatic organs follows [4]. After several days progressively strong infection of nectin-4 expressing airway epithelial cells is documented [5] [6] [7]. The attenuated vaccine strains of MV have also gained the use of ubiquitous CD46 as a receptor.
Figure 1.
MV fusion complex.
A. Schematic of MV F and H proteins indicating cytoplasmic tail (CT), transmembrane (TM), stalk (S) and globular head regions. For the F-protein the fusion peptide (FP, blue shading) and the F1 and F2 subunits that are the result of an activating proteolytic cleavage are indicated. The first residue of the F2 protein is His29; residues 1–28 constitute the signal peptide. The inter-subunit disulfide bond between F1 and F2 is indicated (s–s). The inter-dimer disulfide bonds in the H-dimer are indicated by red lines (C139 and C154). Targeting ligands are appended to the C-terminus of the H-protein as indicated by an arrow. B. Left panel. The H-tetramer is indicated as a combination of a schematic (stalk) and crystal structure[45] (two H-head dimers). Each monomer of the H-tetramer is shaded in a unique color which is extended to the stalk schematic. The C-terminal residue of the crystal structure is indicated by a grey shaded space-filling amino acid (residue 607). The MV F-trimer has been modeled based on the crystal structure of the related human paramyxovirus 5 F-trimer[46]. Each monomer has been shaded in a different color for clarity. The fusion peptide has been shaded blue. Middle panel. The SLAM-binding footprint has been shaded orange and a schematic used to depict the approximate SLAM-binding location. Right panel. The nectin4-binding footprint is indicated by cyan shading. The proposed location of nectin4-binding is indicated by a schematic. Each oval represents an Ig-like domain. Only two receptor molecules are drawn for simplicity, but up to four receptor molecules may bind one H-tetramer to trigger fusion.
The current model for MV entry considers that any interaction with a membrane-anchored receptor that results in concerted movement of the H heads will trigger refolding of the trimeric F protein and subsequent membrane fusion[8–10]. In particular, H exists as a non-covalent dimer of dimers with the two subunits of each dimer linked by two disulfide bonds in the stalk (Figure 1A and Figure 1B, left panel). The F-protein is in contact with the H-stalk prior to receptor engagement by the globular H-head. Using engineered disulfide bonds to constrain movement of the H-head dimers, it was shown that the H-heads must move relative to each other after receptor binding to transmit the fusion-triggering signal to F [8]. Signal transmission may occur through an interaction between the H-stalk and the F-protein. Upon triggering, F undergoes dramatic refolding to insert its fusion peptide in the target membrane, resulting in membrane fusion.
Paramyxovirus Retargeting
Paramyxoviruses have been an ideal starting point to develop targeted cell entry because receptor attachment and fusion functions are separated on two proteins. Viruses that use protein-based receptors, like those of the morbillivirus genus including MV, have proven to be easier to retarget. Over a decade ago, it was shown that MV can enter cells through designated receptors when specificity domains are displayed on the flexible C-terminus of H [11]. The presence of these domains on the MV envelope has minimal effect on the use of natural receptors. However, recognition of the natural MV receptors can be selectively disrupted by mutations within the mapped receptor binding interfaces[12], generating completely retargeted vectors only able to enter cells through the designated receptor[13].
To characterize the mechanism of fusion triggering, hexahistidine tags were inserted in different locations in the top half of the H-protein [8]. The tag is recognized by a membrane-anchored single-chain antibody expressed on Vero-His cells. It was shown that these hexahistidine “handles,” when added to locations in close proximity to the binding footprints of the natural receptors, can be used by the membrane-anchored artificial receptor to trigger fusion, proving that “key-in-lock” specificity during receptor engagement is not required in this system. Strikingly, even addition of the hexahistidine handle to the flexible C-terminus sustains cell entry. All these observations are consistent with a torsional model of triggering. It is likely that binding to natural and artificial receptors ultimately triggers fusion through de-stabilization of the H-stalk interaction with F-trimers.
Based on this compliant, H-head movement based entry system, MV retargeting has been achieved through a broad range of cell surface molecules, including proteins that span the membrane only once, in type I or type II orientations, or several times. These receptors can be monomeric, homo- or hetero-oligomeric, and have a wide range of functions [14]. Equally important is the ability of the virus to accommodate the specificity ligands, which range in size from the compact 60-residue epithelial growth factor to single-chain antibodies with nearly 250 residues [14].
At least two factors contribute to the retargeting efficiency observed with MV: 1) the separation of receptor binding and membrane fusion functionality to different glycoproteins; and 2) the proposed entry mechanism that does not depend on key-in-lock receptor specificity. In this context, any specificity domain capable of transmitting torsional or extensional force on the H tetramer following receptor binding should function as a retargeting specificity domain. The versatility of the MV entry system therefore reflects the elegant simplicity of its mechanism, and future retargeting approaches that preserve the mobility of the H-heads in response to receptor binding should see continued success.
Herpes Simplex Virus (HSV) Entry Mechanism
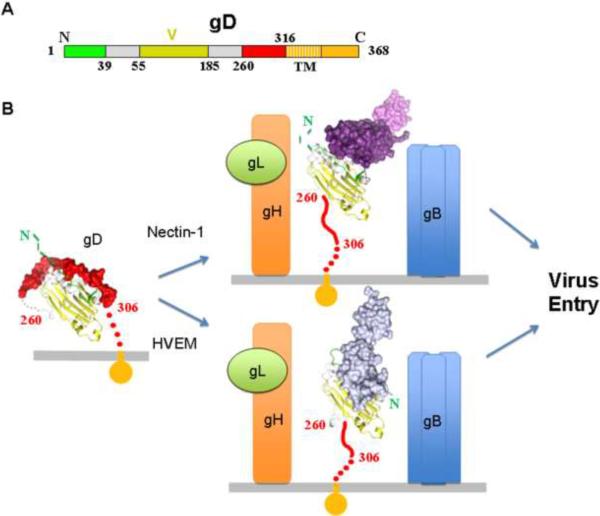
HSV entry is more complex than MV entry: it can occur via fusion of the viral envelope with either the plasma or endocytic membranes [15] (see also chapter by Campadelli-Fiume in this issue [16]). Five glycoproteins are implicated in HSV entry, gC, gB, gD and the heterodimer gH/gL [17]. The virus initially attaches to cells through the interaction of gC and gB with cell surface proteoglycans [18]. However, attachment is not sufficient to allow entry and requires the interaction between the receptor-binding protein gD and a specific cell surface receptor (Figure 2). The latter event triggers the membrane fusion process that is mediated by gB and gH/gL. Importantly, the gB trimer and gH/gL heterodimer are conserved across the Herpesviridae family (Figure 2B, right panel) whereas the receptor-binding protein varies amongst the family members.
Figure 2.
Involvement of the HSV receptor-binding protein gD in entry.
A. Schematic representation of HSV-1 gD. Numbering for gD starts at lysine 1 of the mature glycoprotein. The gD IgG core, labeled V for the Ig V-fold, is colored in yellow, residues forming the HVEM binding hairpin are in green and residues 260 to 316 that are displaced by receptor binding are in red. The gD Ig core has been shown to be replaceable when used for retargeting the virus. B. Structural schematic showing the triggering of the entry process mediated by gD. The same color code as in panel A was used. Regions that were not determined and presumably flexible in the crystal structures are shown as dotted lines. The left panel shows gD in the unliganded state where residues 260–306 are folded against the core structure. Nectin-1 or HVEM receptors bind to gD causing unwrapping of this C-terminal region (central panel). Exposure of this region mediates gH/gL (orange/green, respectively) and gB (blue trimer) interactions with concomitant membrane fusion and viral entry. The location of the C-terminal region of gD (residues 260–316) was not determined in the gD/Nectin-1 and gD/HVEM structures and is modeled as an extended coil pointing towards the viral membrane. Similarly the N-terminal 20 amino acids of gD were not localized in the unliganded gD and in the gD/Nectin-1 complex.
HSV gD (Figure 2B, left panel) can bind to three different receptors [15]. The herpesvirus entry mediator (HVEM), an immune modulator expressed on fibroblasts and ocular cell types was the first described HSV receptor [19] (Figure 2B, bottom right panel). The cell adhesion molecule nectin-1 is the primary receptor for HSV on neurons, keratinocytes and epithelial cells [20–22] (Figure 2B, top right panel). Finally, gD can also bind heparan sulfate specifically modified by 3-O-sulfotransfereases to trigger entry [23]. In addition, gB and the gH/gL heterodimer may also bind cellular tethering factors that serve to enhance entry [24].
In the last ten years detailed structural information has become available for the ectodomains of all four essential HSV viral entry glycoproteins (gD, gB and gH/gL) providing key insights into the HSV entry mechanism [25,26]. Co-crystal structures of gD in complex with each of the two proteinaceous receptors nectin-1 [27] and HVEM[28] are now available and a comparison between gD-receptor complexes versus unliganded gD structures have provided critical insight into the receptor-mediated activation of HSV fusion.
gD is composed of an Ig-like core that is wrapped by proline rich N- and C-terminal extensions. Binding to HVEM is associated with refolding of the gD 20 N-terminal amino acids to form a hairpin that contacts the receptor (Figure 2, bottom panel) [28]. Importantly, in this complex the gD N-terminal hairpin occupies the space filled by the C-terminal region of the gD-ectodomain in the unliganded molecule. However, the 20 N-terminal gD residues are dispensable for nectin-1 binding and HSV entry [29]. Instead, nectin-1 binds to an epitope that is buried by the same region of the C-terminal extension that is displaced upon formation of the gD/HVEM complex (Figure 2, upper panel) [27]. Thus, the interaction of gD with either receptor disrupts the association of the Ig-core and C-terminal residues causing unwrapping and exposure of the last 50-residues of the C-terminal extension (also referred as the pro-fusion domain) [30] [31].
After receptor binding the pro-fusion domain has been proposed to interact with the gH/gL heterodimer and/or the gB trimer [32]. Receptor-binding may also promote formation of specific contacts between gH/gL and gB. All these interactions converge towards the same event: they trigger gB to insert its fusion loops into the target cell membrane and to refold to a post-fusion structure concomitantly leading to membrane fusion. Thus, both HSV and MV fusion mechanisms rely on the tight control of the concerted triggering of a trimeric fusion protein. In both viral systems specific triggering can be achieved by at least three different receptors, setting a precedent for activation through targeted receptors.
Herpes Simplex Virus Retargeting
Herpesviruses require more proteins to accomplish cell entry compared to paramyxoviruses. As a result more aspects of the HSV entry mechanism remain to be elucidated. For example the role of gH/gL in the membrane fusion process is not understood and how the profusion domain triggers the fusion process remains to be determined. Notwithstanding the outstanding questions about the entry mechanism, the principles underlying retargeting of HSV are similar to those underlying MV retargeted entry. Similar to MV, the HSV receptor-binding protein gD is sufficiently plastic to allow insertion close to the N-terminus of targeting ligands such as interleukin 13 (IL-13) [33], urokinase plasminogen activator (uPA) [34] [35] and a single-chain antibody (scFv) to human epidermal growth factor receptor-2 [36] (HER2). This is especially significant as the HER2 scFv is comparable in size to gD, demonstrating that gD can tolerate large insertions and remain functional. Detargeting HSV was achieved analogous to the MV paradigm by mutation or deletion of gD residues known to be involved in receptor binding. These include the HVEM and heparan sulfate binding sites that map to amino acid residues 6–38. Nectin-1 binding was prevented by mutations in the binding site or by steric occlusion by inserting the targeting ligand in front of its interacting surface [36].
Strikingly, it was recently shown that gD function was maintained and complete detargeting achieved when the entire Ig-core of gD was replaced by a scFv antibody to HER2 [37]. This finding is consistent with and highlights the critical role of the gD pro-fusion domain in the triggering of the fusion process (Figure 2A). However, the current data cannot assess whether in these chimeric gDs the C-terminal region is displaced upon interaction with the retargeting receptor or is already exposed previous to binding resulting in a pre-activated gD. While the latter may be true for these engineered forms of gD, it may not be the mechanism that wild-type HSV uses. Exposure of the C-terminal region prior to receptor binding may diminish selectivity for target cells. For instance, if the C-terminus were always exposed, the virus could enter cells that express only a gB or a gH/gL receptor with the risk of inefficient entry or even fusion with the wrong cell compartment. Unmasking of the C-terminus only in the presence of a bona fide gD-receptor has the advantage of initiating the process only at the right time/place. It is likely that HSV has evolved to release the gD C-terminal region and initiate cell entry only upon receptor binding to ensure efficient tropism in the host.
In conclusion, the fact that separate HSV proteins are responsible for receptor-binding and fusion function allows for efficient retargeted entry. Moreover, the HSV receptor-binding protein is also sufficiently plastic to accommodate large insertions. The fact that the Ig-core of gD is dispensable for fusion-support function should allow the substitution of a large variety of targeting ligands and the development of versatile therapeutic gene delivery vectors. Mechanistic information, once available will facilitate the design of additional re-targeting strategies.
Perspectives
Remarkably, the structurally diverse receptor-binding proteins of MV and HSV both bind to the same location on their respective Ig-like receptors, SLAM and nectin-1. More generally, viruses of several families bind near the tip of one side of the most membrane-distal domain of Ig-superfamily proteins involved in cell adhesion [38]. Binding to cell adhesion molecules may favor virus spread by opening intracellular junctions, and reflect early adaptation of viruses to multicellular systems.
Both MV and HSV entry mechanisms rely principally on one oligomeric protein for binding, which then transmits a triggering signal to a trimeric fusion protein. Signal transmission is direct and involves the stalk of the binding protein for MV, whereas for HSV more than one triggering pathways may exist. In both systems viral attachment may involve multiple interactions: receptors may bind to different oligomers, and to individual subunits within each oligomer. Engagement of all the subunits within an oligomer may require their re-orientation and reconfiguration. Simultaneous re-configuration of multiple oligomers would result in de-stabilization of the envelope glycoprotein lattice, concerted re-folding of trimeric fusion proteins, and membrane fusion.
How easy is it to apply the principles of re-targeting delineated here to viruses other than MV or HSV? Among paramyxoviruses, the H-protein of the closely MV-related canine distemper virus, and that of the more distantly related Tupaia paramyxovirus, have been retargeted by display of single-chain antibodies [39] [40]. However, no success has been reported yet on re-targeting of sialic acid-binding HN-proteins, which are used by a majority of paramyxoviruses. Interestingly, recent mechanistic studies of fusion triggering [8], together with several other observations [41,42] and structural studies of the HN ectodomain [10] have suggested that the details of the activation mechanisms of H/G- versus HN-based fusion systems may differ. Once again, these observations highlight the importance of a deeper understanding of the molecular mechanisms of receptor binding and fusion triggering for the development of targeted cell-entry. Analogously, it should be relatively straightforward to retarget the closely related alphaherpesvirus HSV-2 based on the same principles established for HSV-1.
Finally, modification of the glycoproteins of certain enveloped virus families towards retargeting has been difficult. However, in an alternative experimental approach the retargeted MV glycoproteins have been incorporated in place of the original glycoprotein in the envelope of a lentiviral vector. These “pseudotyped” HIV nucleocapsids deliver genetic material very specifically to different target cell types [43]. To sustain efficient incorporation of the MV glycoproteins it was sufficient to precisely trim their respective cytoplasmic tails [44]. Analogously, after proper fitting of the retargeted glycoproteins of MV or related paramyxoviruses [39] [40] in the envelope of other viruses, chimeric vectors can be generated for systemic inoculation to achieve targeted gene transfer or oncolysis. Thus, re-targeting of enveloped virus families whose entry systems are complex or not well understood can be approached based on envelope exchange.
Highlights
Targeting: addition of specificity domains to the receptor-binding protein
Entry targeting of viral vectors improves efficacy of oncolytic and gene therapies
Enveloped viruses measles and herpes simplex virus I have been successfully retargeted
Envelope exchange for viruses with prohibitive entry constraints
Acknowledgements
The authors would like to thank Ethan C. Settembre for help with Figure 2. RC was funded by National Institutes of Health R01 CA90636.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Galanis E. Cancer: Tumour-fighting virus homes in. Nature. 2011;477:40–41. doi: 10.1038/477040a. [DOI] [PubMed] [Google Scholar]
- 2.Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 2008;6:529–540. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira CSA, Frenzke M, Leonard VHJ, Welstead GG, Richardson CD, Cattaneo R. Measles virus infection of alveolar macrophages and dendritic cells precedes spread to lymphatic organs in transgenic mice expressing human signaling lymphocytic activation molecule (SLAM, CD150) J. Virol. 2010;84:3033–3042. doi: 10.1128/JVI.01559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemon K, De Vries RD, Mesman AW, Mcquaid S, Van Amerongen G, Yüksel S, Ludlow M, Rennick LJ, Kuiken T, Rima BK, et al. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathogens. 2011;7:e1001263. doi: 10.1371/journal.ppat.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludlow M, Rennick LJ, Sarlang S, Skibinski G, McQuaid S, Moore T, de Swart RL, Duprex WP. Wild-type measles virus infection of primary epithelial cells occurs via the basolateral surface without syncytium formation or release of infectious virus. J. Gen. Virol. 2010;91:971–979. doi: 10.1099/vir.0.016428-0. [DOI] [PubMed] [Google Scholar]
- 6.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathogens. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011 Nov. 2 doi: 10.1038/nature10639. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VHJ, Braun W, Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 10.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. USA. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider U, Bullough F, Vongpunsawad S, Russell SJ, Cattaneo R. Recombinant measles viruses efficiently entering cells through targeted receptors. J. Virol. 2000;74:9928–9936. doi: 10.1128/jvi.74.21.9928-9936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: Identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 2004;78:302–313. doi: 10.1128/JVI.78.1.302-313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura T, Peng K-W, Harvey M, Greiner S, Lorimer IAJ, James CD, Russell SJ. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat. Biotech. 2005;23:209–214. doi: 10.1038/nbt1060. [DOI] [PubMed] [Google Scholar]
- 14.Navaratnarajah CK, Leonard VH, Cattaneo R. Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr. Top. Microbiol. Immunol. 2009;329:59–76. doi: 10.1007/978-3-540-70523-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campadelli-Fiume G, Menotti L, E A, Gianni T. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr. Opin. Virol. 2012;2 doi: 10.1016/j.coviro.2011.12.001. accepted. [DOI] [PubMed] [Google Scholar]
- 17.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 20.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 21.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krummenacher C, Baribaud F, Ponce de Leon M, Baribaud I, Whitbeck JC, Xu R, Cohen GH, Eisenberg RJ. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology. 2004;322:286–299. doi: 10.1016/j.virol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue BY, Liu J, Shukla D. Role for 3-O-sulfated heparan sulfate as the receptor for herpes simplex virus type 1 entry into primary human corneal fibroblasts. J. Virol. 2006;80:8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, et al. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 26.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. Structure of Herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathogens. 2011;7:e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 29.Krummenacher C, Baribaud I, Ponce de Leon M, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 2000;74:10863–10872. doi: 10.1128/jvi.74.23.10863-10872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni T, Amasio M, Campadelli-Fiume G. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 2009;284:17370–17382. doi: 10.1074/jbc.M109.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou G, Ye GJ, Debinski W, Roizman B. Engineered herpes simplex virus 1 is dependent on IL13Ralpha 2 receptor for cell entry and independent of glycoprotein D receptor interaction. Proc. Natl. Acad. Sci. USA. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamiyama H, Zhou G, Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13:621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 35.Zhou G, Roizman B. Separation of receptor-binding and profusogenic domains of glycoprotein D of herpes simplex virus 1 into distinct interacting proteins. Proc. Natl. Acad. Sci. USA. 2007;104:4142–4146. doi: 10.1073/pnas.0611565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menotti L, Cerretani A, Hengel H, Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J. Virol. 2008;82:10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menotti L, Nicoletti G, Gatta V, Croci S, Landuzzi L, De Giovanni C, Nanni P, Lollini PL, Campadelli-Fiume G. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl. Acad. Sci. USA. 2009;106:9039–9044. doi: 10.1073/pnas.0812268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dermody TS, Kirchner E, Guglielmi KM, Stehle T. Immunoglobulin superfamily virus receptors and the evolution of adaptive immunity. PLoS Pathogens. 2009;5:e1000481. doi: 10.1371/journal.ppat.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springfeld C, von Messling V, Tidona CA, Darai G, Cattaneo R. Envelope targeting: hemagglutinin attachment specificity rather than fusion protein cleavage-activation restricts Tupaia paramyxovirus tropism. J. Virol. 2005;79:10155–10163. doi: 10.1128/JVI.79.16.10155-10163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miest TS, Yaiw K-C, Frenzke M, Lampe J, Hudacek AW, Springfeld C, von Messling V, Ungerechts G, Cattaneo R. Envelope-chimeric entry-targeted measles virus escapes neutralization and achieves oncolysis. Mol. Ther. 2011;19:1813–1820. doi: 10.1038/mt.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future virol. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee B, Ataman ZA. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol. 2011;19:389–399. doi: 10.1016/j.tim.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anliker B, Abel T, Kneissl S, Hlavaty J, Caputi A, Brynza J, Schneider IC, Münch RC, Petznek H, Kontermann RE, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat. Methods. 2010;7:929–935. doi: 10.1038/nmeth.1514. [DOI] [PubMed] [Google Scholar]
- 44.Funke S, Maisner A, Mühlebach MD, Koehl U, Grez M, Cattaneo R, Cichutek K, Buchholz CJ. Targeted cell entry of lentiviral vectors. Mol. Ther. 2008;16:1427–1436. doi: 10.1038/mt.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. USA. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin H-S, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]