Abstract
The hepatitis C virus (HCV) is a major medical problem with at least 130 million infected individuals worldwide. Over the last decade multiple host factors required for HCV cell entry have been identified, but a detailed understanding of their mechanistic interplay remains elusive. Nonetheless, recent advances in defining species-specific barriers of HCV transmission have allowed the identification of a minimal set of entry factors that are required for HCV infection of rodent cells and has culminated in an animal model that recapitulates HCV entry in vivo. A detailed understanding of the viral uptake pathway is imperative to define new drug targets allowing for more effective intervention against this devastating disease.
Introduction
HCV, the prototypical member of the Hepacivirus genus within the family Flaviviridae, is associated with the majority of newly diagnosed hepatocellular carcinomas in the United States [1,2] and is currently the leading cause of liver transplants worldwide [3]. An HCV vaccine is not available and current therapies are poorly effective [4,5] HCV-associated liver transplantation is only a palliative procedure due to universal infection of the graft after transplantation, often resulting in more rapid fibrosis progression and subsequent graft failure [6]. Even transient therapies inhibiting HCV cell entry could prevent graft infection and greatly improve the effectiveness of liver transplantation. Such therapeutic advances targeting this stage of the viral life cycle will require a much more solid understanding of HCV cell entry than is currently available. Here, we discuss recent advances in our understanding of molecules involved in viral uptake and their role in HCV tissue and species tropism, and we provide a putative mechanism of the HCV uptake pathway.
The HCV cell entry pathway
The HCV virion is comprised of a nucleocapsid core surrounded by a host-derived membrane bearing the E1 and E2 HCV glycoproteins, which mediate the majority of cell entry processes of the virion, including cell binding, endocytosis, and fusion in the low pH environment of early endosomes [7–15]. Both of these glycoproteins contain single transmembrane domains at their carboxy-termini that not only function to anchor them in membranes, but also contain ER-localization motifs[16,17]. The biogenesis of these proteins is closely linked and heavily influence by their coexpression (reviewed in [18]. The individual functions of these glycoproteins have not been completely delineated; while E2 has at least been shown to bind to some of the HCV cell surface entry factors, little is known about the roles of E1, mostly due to difficulties in purifying recombinant versions of this protein. Indeed, it has not been clearly defined which glycoprotein is responsible for mediating virion fusion.
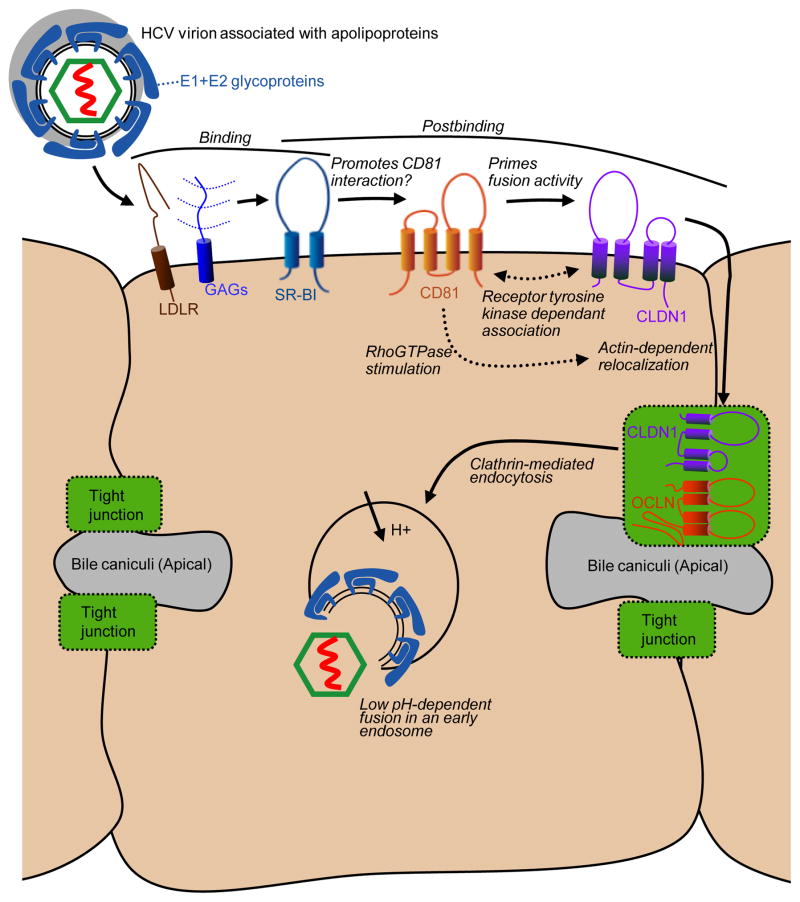
The HCV entry process into human hepatocytes requires numerous host proteins including glycosaminoglycans (GAGs) [14,19], the low density lipoprotein receptor (LDL-R) [20–23], the high density lipoprotein receptor scavenger receptor class B type I (SR-BI, also known as CLA-1 and officially designated SCARB1) [24], tetraspanin CD81 [25], and two tight junction (TJ) proteins, claudin-1 (CLDN1) [26] and occludin (OCLN) [27,28] (Fig. 1). With so many cellular factors involved, it is clear the HCV does not follow the classic view of a virus that interacts with a single cellular receptor. It is likely that the incoming HCV virion uses the factors in a sequential manner to perform a range of functions along the cell entry process. GAGs may mediate initial, perhaps low affinity, interactions between the HCV virion and the cell surface [14,19]. LDLR may also be involved in cell binding, perhaps mediated by virion-associated apolipoproteins [23]. SR-BI antibody blocking studies indicate this protein plays a role in the binding of HCV to host cells, perhaps subsequent to GAG interactions, and is also involved in a post cell binding event [29–32]. The lipid uptake activity of SR-BI is also required for HCV cell entry, although this requirement may be indirect and reflect the involvement of cellular cholesterol levels to promote signalling events, HCV entry factor associations, and/or suitable target membranes for fusion (reviewed in [33]).
Figure 1.
Potential route of HCV entry into polarized hepatocytes.
E1 and E2 = HCV envelope glycoproteins 1 and 2, LDLR = low density lipoprotein receptor, GAGs = glycosaminoglycans, SR-BI = scavenger receptor type B class 1, CLDN1 = claudin 1, and OCLN = occludin.
Antibody blocking studies reveal that CD81 plays a post-cell binding role in HCV cell entry [14]. Furthermore, although CD81 can interact with a soluble form of E2, it does not appear to be able to directly interact with mature HCV virions, suggesting that prior host cell interactions may alter the virion conformation to reveal CD81 binding sites. This hypothesis is supported by the isolation mutations within E1 and E2 that enhance the ability of HCV to utilize the normally poorly functional mouse CD81 ortholog [34]. These mutations appears to influence the glycoprotein conformation on the virion to promote a more open conformation than the wild type virion, as the mutant virus is more susceptible to E2 antibody neutralization, and displays higher affinity interactions with CD81 and reduced dependence on SR-BI and OCLN.
While HCV requires low pH for membrane fusion, the free virion is not inactivated by low pH treatment, which suggests that interactions between the virion and cellular factors primes the glycoproteins to respond to the low pH environment of the endosome. This hypothesis was supported by early experiments that showed that HCV can fuse directly at the cell surface when endosome acidification is blocked and cells are briefly washed with a low pH buffer. However this fusion event requires that cells with bound virions are incubated at 37°C prior to the low pH wash, suggesting that virus requires postbinding interactions with cellular factors to become fusogenic [10]. Recent studies suggest that CD81 may act to prime the fusogenic activity of the HCV glycoproteins [35]. The mouse CD81 adapted HCV mutant described above does not require incubation with cells at 37°C to fuse directly with the cell surface, suggesting it may already exhibit a more open or “pre-primed” conformation [34]. In addition, cells that lack CD81 do not support cell surface fusion, however this block can rescued by adding soluble recombinant CD81 protein [35]. Furthermore, simultaneous treatment of cell free virions with both a low pH and soluble CD81 impairs infectivity, likely by triggering irreversible release of the viral fusion peptide in the absence of a host membrane [35].
CLDN1 likely plays a post-binding role in the HCV cell entry process, but exactly where and when this factor participates is not clear. Blocking experiments with antibodies directed against CLDN1 indicate this protein is used late in the entry process, around the time of virion internalization [26], or with kinetics of usage to overlap with those of CD81 [36,37]. A lack of reagents to directly block OCLN has prevented the examination of when this factor participates in the entry process.
Whether the HCV virion directs binds to either CLDN1 or OCLN has not been conclusively demonstrated, however if such an interaction is required for HCV cell entry it is not clear how such tight junction proteins become accessible to incoming virions, as the tight junction region is not accessible to HCV circulating in the bloodstream. The entry process of group B coxsackieviruses (CVB), which follows an elaborate route of polarized cell entry and has become a paradigm for this type of viral entry [38], may shed light on HCV cell entry. The primary CVB receptor, decay accelerating factor (DAF), is located on the luminal, or apical, surface of epithelial cells and directly available to incoming virus. A secondary entry factor, coxsackie-adenovirus-receptor (CAR), is TJ-associated and normally inaccessible to virions. However, CVB binding to DAF on the exposed cell surface stimulates intracellular c-Abl signaling that promotes the actin-dependent relocation of the virion/DAF complex to the tight junction region, where an interaction with CAR leads to virion endocytosis. Based on the expected localization of HCV entry factors, a similar stepwise entry cell entry pathway is conceivable for HCV, where virion interactions with basolaterally exposed factors mediate early binding events as well as post binding events that mediate the translocation of virions to CLDN1 and OCLN within normally inaccessible tight junctions. In support of this model, CD81 engagement, either by CD81 antibodies or soluble E2 protein, has been shown to activate Rho GTPases to mediate actin-dependant relocalization of such CD81 complexes to cell junction regions [39]. On the other hand, it remains possible that HCV does not even enter tight junction regions during entry. Indeed, evidence suggests that, in nonpolarized cells and partially polarized HepG2 cells, CLDN1 may interact with CD81 outside of cell junctions prior to infection and influence the cell entry process [40,41]. However, cell polarity can negatively regulate this interaction, as interactions between CLDN1 and both CD81 and the closely related tetraspanin CD9 was greatly reduced in highly polarized Caco-2 and MDCK cells [42]. Lupberger et al. recently reported that two receptor tyrosine kinases, epidermal growth factor receptor (EGFR) and ephrin receptor A2, are required for HCV cell entry, and possibly act by modulating the interaction between CD81 and CLDN1[43]. This scenario would be consistent with previous reports that EGFR activation influences CLDN1 localization and function [44].
The above evidence suggests the following hypothetical HCV cell entry model (Fig. 1). The HCV virion exists in a closed conformation prior to interaction with the host cell. Glycans and apoliproteins shield the majority of the HCV glycoproteins from immune surveillance, leaving only minimal GAG and SR-BI interacting domains exposed. These interactions result in exposure of the CD81 binding site, and interaction with this protein in turn primes the low-pH fusion activity of the virion. However, according to this model, fusogenic virus must still translocate to the tight junctions in order to be endocytosed. This could occur through an association between CD81 and CLDN1, either pre-existing or stimulated by virion-mediated activation of receptor tyrosine kinase signalling, and subsequent actin-dependant transport of the virion into the tight junction region, where endocytosis may be mediated by interactions with OCLN. While this may be an attractive model, fully defining this process and how cell polarity influences the HCV cell entry process will likely require additional cell systems that are more polarized than those currently available and that support robust infection with cell culture grown HCV.
HCV entry contributes to tissue and species tropism
Several clinical observations have fostered speculations on the existence of putative extrahepatic sites of HCV infection. For example, HCV infected patients frequently show encephalopathy and neuropsychiatric disorders suggesting an involvement of the central nervous system [45]. Indeed, it has been shown that HCV can enter human peripheral neuro-blastoma and -epithelioma cells in vitro [46,47] and HCV RNA has been detected brain tissue from HCV infected individuals [48]. However, analysis of post mortem collected specimens is challenging because of the high propensity of sample contamination and direct evidence of active HCV RNA replication in these tissues has not been provided. In addition, chronic HCV carriers frequently exhibit signs of various lymphoproliferative disorders. HCV RNA has indeed been observed in association with hematopoietically derived cells including B and T lymphocytes, monocytes, and dendritic cells. This could be interpreted as the reflection of a productive entry event. However, it has been demonstrated that at least cell-culture produced HCV cannot enter any subset of peripheral blood mononuclear cells [49], suggesting that HCV particles and/or RNA may adhere to these cells but not efficiently enter. HCV protein has also been found in epithelial cells of intestinal specimens collected from HCV infected individuals [50]. In cell culture, the colorectal adenocarcinoma Caco-2 cell line supports viral uptake [51]. While these data are intriguing it remains to be unambigously demonstrated that HCV can enter non-parenchymal cells in vivo and, more importantly, that these putative extrahepatic sites have any clinical relevance.
Determinants governing HCV tissue tropism are poorly understood. While all stages of the HCV life cycle appear to be specifically attuned to the hepatocyte environment, host cell entry appears to play an important, and at least the earliest, role in governing the HCV hepatotropism. Although the minimal set of HCV cell entry factors are present in various nonliver mammalian tissues, suboptimal expression levels and/or an inadequate subcellular localization may limit their ability to promote HCV cell entry. CD81 is ubiquitously expressed in almost all nucleated cells. OCLN is present in all tight junction complexes present in polarized cell layers. However, the OCLN transcript is subject to post-transcriptional splicing creating considerable mRNA diversity. These alternatively spliced forms are differentially abundant across tissues and have different capacities to support viral entry [52], which might contribute to HCV tissue tropism and possibly modify the outcome of HCV infection in humans. CLDN1 and SR-BI are present in multiple tissues but the combination of both at high levels is only present in the liver, which has led to the hypothesis that these two entry factors primarily control HCV tissue specificity at the entry level.
While the abundance of entry factors in different tissues likely affects viral tropism, negative regulators of entry expressed in non-hepatic cells may also contribute to restricting HCV tissue tropism. Immunoglobulin superfamily member 8 (Igsf8), also known as EWI-2int or CD316, binds to CD81 and has recently been demonstrated to function as a dominant negative inhibitor of HCV entry in vitro [53]. Igsf8 is not expressed in hepatocytes, but is present in several non-hepatic cell lines and is abundant in B and T cells as well as the brain. Ectopic expression of Igsf8 in a hepatoma cell line (Huh-7) blocks HCV entry by inhibiting the interaction between CD81 and the viral glycoproteins. This finding suggests that, in addition to the presence of specific uptake factors in hepatocytes, the lack of a specific inhibitor may contribute to the hepatotropism of HCV. It is also conceivable that HCV can potentially enter other non-hepatic cell types, but is unable to efficiently establish RNA replication due to the lack of liver specific host factors that are required for HCV replication, such as microRNA-122 [54].
HCV has a narrow host range infecting only humans and chimpanzees. Although the basis of this highly restricted species tropism is incompletely understood, analyzing the viral life cycle in mouse cells and in mice has helped to shed some light on species-specific barriers of HCV transmission. Although all four proteins are required for efficient HCV cellentry into mouse cells, the murine orthologs of SR-BI and CLDN1 function as efficiently as the human verions [27]. Conversely, human CD81 and OCLN define the species barrier during the viral uptake process. Several residues within the second extracellular loop of CD81 have been demonstrated to be involved in binding of HCV E2, and they are not or only partially conserved across species [55,56]. The regions in OCLN conferring species tropism have been identified in the second extracellular loop of this protein [27,57]. It was recently demonstrated that mice can indeed be engineered to take up HCV in a viral glycoprotein-dependent fashion by simply expressing human CD81 and OCLN [58]. While this new genetically model holds great promise to dissect HCV entry in vivo it remains to be determined whether mice can be coaxed to support the entire viral life cycle. Downstream of viral entry, HCV RNA replication is extremely inefficient in mouse cells, thereby creating an additional barrier in species tropism. However, using a transcomplementation-based system, mouse hepatoma cell lines have been shown to be capable of supporting assembly and release of infectious HCV particles [59]. This result provides hope that, if the block to HCV RNA replication in mouse cells can be overcome, the ambitious goal of generating a tractable HCV mouse model can indeed be achieved. Such a mouse model would open up unprecedented opportunities to dissect viral pathogenesis and to evaluate drug and vaccine candidates [60].
Conclusions
HCV entry is a highly complex process that has yet has to be fully characterized. Multiple cellular and viral factors have been implicated in the viral uptake process, but their exact role and the kinetics of their engagement remain incompletely understood. HCV entry appears to contribute to both tissue and species tropism. Dissection of viral uptake has not only been instrumental to engineering new animal models for HCV infection, but will continue to shed light on new and possibly more suitable antiviral drug targets.
Highlights.
HCV utilizes a complex set of host factors to facilitate uptake into its host cell
Detailed mechanism of HCV entry remains incompletely understood
Human CD81 and OCLN are required for HCV infection of mouse cells and mice
Blocking HCV entry constitutes an attractive therapeutic approach in particular to prevent universal graft re-infection after liver transplantation
Acknowledgments
We wish to thank Carina Storrs for careful editing of this manuscript. This work was supported in part by award number RC1DK087193 (A.P.) from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases, Infectious Disease Society of America Astella Young Investigator Award (A.P.), award number R00AI077800 from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (M.J.E.), and the Pew Charitable Funds (M.J.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 2.Montalto G, Cervello M, Giannitrapani L, Dantona F, Terranova A, Castagnetta LA. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann N Y Acad Sci. 2002;963:13–20. doi: 10.1111/j.1749-6632.2002.tb04090.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown RS., Jr Hepatitis C and liver transplantation. Nature. 2005;436:973–978. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- *4.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *5.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. References 4 and 5 summarize the data of clinical trials demonstrating that the addition of directly acting antiviral significantly improved sustained virological response rates. [DOI] [PubMed] [Google Scholar]
- 6.Forman LM, Lewis JD, Berlin JA, Feldman HI, Lucey MR. The association between hepatitis C infection and survival after orthotopic liver transplantation. Gastroenterology. 2002;122:889–896. doi: 10.1053/gast.2002.32418. [DOI] [PubMed] [Google Scholar]
- 7.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn YS, Rice CM, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubuisson J, Hsu HH, Cheung RC, Greenberg H, Russell DR, Rice CM. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Op De Beeck A, Voisset C, Bartosch B, Ciczora Y, Cocquerel L, Keck Z, Foung S, Cosset FL, Dubuisson J. Characterization of functional hepatitis C virus envelope glycoproteins. J Virol. 2004;78:2994–3002. doi: 10.1128/JVI.78.6.2994-3002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol. 2006;80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. doi: 10.1371/journal.ppat.1000702. The authors developed a single particle tracking assay of infectious fluorescent HCV particles to examine the co-trafficking of HCV virions with cellular cofactors of endocytosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocquerel L, Duvet S, Meunier J-C, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocquerel L, Meunier J-C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavie M, Goffard A, Dubuisson J. Assembly of a functional HCV glycoprotein heterodimer. Curr Issues Mol Biol. 2007;9:71–86. [PubMed] [Google Scholar]
- 19.Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 20.Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 21.Agnello V, Abel G, Elfahal M, Knight GB, Zhang Q-X. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. Journal of Medical Virology. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Owen DM, Huang H, Ye J, Gale M., Jr Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21 :5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pileri P, Uematsu Y, Compagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- **26.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. This study describes the identification of CLDN1 as a HCV entry factor and implicates for the first time tight junction proteins in the HCV uptake process. [DOI] [PubMed] [Google Scholar]
- **27.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. This study demonstrates that expression of human CD81 and OCLN renders mouse cell lines permissive to HCV entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83 :2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An Interplay between Hypervariable Region 1 of the Hepatitis C Virus E2 Glycoprotein, the Scavenger Receptor BI, and High-Density Lipoprotein Promotes both Enhancement of Infection and Protection against Neutralizing Antibodies. J Virol. 2005;79:8217–8229. doi: 10.1128/JVI.79.13.8217-8229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A, et al. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84 :34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, Santini C, Luzzago A, Rice CM, Cortese R, et al. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J Virol. 2007;81:8063–8071. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 33.Dao Thi VL, Dreux M, Cosset FL. Scavenger receptor class B type I and the hypervariable region-1 of hepatitis C virus in cell entry and neutralisation. Expert Rev Mol Med. 2011;13:e13. doi: 10.1017/S1462399411001785. [DOI] [PubMed] [Google Scholar]
- *34.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. This study describes the adaptation of the HCV envelop to usage of mouse CD81. The resulting murine tropic HCV strain is an important proof of concept highlighting that a mouse model for HCV could be potentially be developed by viral adaptation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. Hepatitis C Virus Is Primed by CD81 Protein for Low pH-dependent Fusion. J Biol Chem. 2011;286:30361–30376. doi: 10.1074/jbc.M111.263350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, Soulier E, Royer C, Lambotin M, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 37.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–964. 964 e951–954. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 38.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, Hu K, Yuan F, Deng H, Hubscher SG, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol. 2008;82:5007–5020. doi: 10.1128/JVI.02286-07. The authors demonstrate for the first time a physical interactions between CD81 and CLDN1 in hepatic cells suggesting this being a critical step in the viral uptake process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, et al. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics. 2007;6:1855–1867. doi: 10.1074/mcp.M700183-MCP200. [DOI] [PubMed] [Google Scholar]
- **43.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589–595. doi: 10.1038/nm.2341. The authors identify two receptor tyrosine kinases that had not previously been implicated in the HCV entry process. These results also shed light on some of the signalling pathways that coordinate the interactions of at least some of the HCV entry factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh AB, Harris RC. Epidermal growth factor receptor activation differentially regulates claudin expression and enhances transepithelial resistance in Madin-Darby canine kidney cells. J Biol Chem. 2004;279:3543–3552. doi: 10.1074/jbc.M308682200. [DOI] [PubMed] [Google Scholar]
- 45.Forton DM, Taylor-Robinson SD, Thomas HC. Cerebral dysfunction in chronic hepatitis C infection. J Viral Hepat. 2003;10:81–86. doi: 10.1046/j.1365-2893.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 46.Burgel B, Friesland M, Koch A, Manns MP, Wedemeyer H, Weissenborn K, Schulz-Schaeffer WJ, Pietschmann T, Steinmann E, Ciesek S. Hepatitis C virus enters human peripheral neuroblastoma cells - evidence for extra-hepatic cells sustaining hepatitis C virus penetration. J Viral Hepat. 2010 doi: 10.1111/j.1365-2893.2010.01339.x. [DOI] [PubMed] [Google Scholar]
- 47.Fletcher NF, Yang JP, Farquhar MJ, Hu K, Davis C, He Q, Dowd K, Ray SC, Krieger SE, Neyts J, et al. Hepatitis C virus infection of neuroepithelioma cell lines. Gastroenterology. 2010;139:1365–1374. doi: 10.1053/j.gastro.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fishman SL, Murray JM, Eng FJ, Walewski JL, Morgello S, Branch AD. Molecular and bioinformatic evidence of hepatitis C virus evolution in brain. J Infect Dis. 2008;197:597–607. doi: 10.1086/526519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. Cell culture–produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deforges S, Evlashev A, Perret M, Sodoyer M, Pouzol S, Scoazec JY, Bonnaud B, Diaz O, Paranhos-Baccala G, Lotteau V, et al. Expression of hepatitis C virus proteins in epithelial intestinal cells in vivo. J Gen Virol. 2004;85:2515–2523. doi: 10.1099/vir.0.80071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mee CJ, Grove J, Harris HJ, Hu K, Balfe P, McKeating JA. Effect of cell polarization on hepatitis C virus entry. J Virol. 2008;82:461–470. doi: 10.1128/JVI.01894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohaar I, Ploss A, Korol E, Mu K, Schoggins JW, O'Brien TR, Rice CM, Prokunina-Olsson L. Splicing diversity of the human OCLN gene and its biological significance for hepatitis C virus entry. J Virol. 2010;84:6987–6994. doi: 10.1128/JVI.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rocha-Perugini V, Montpellier C, Delgrange D, Wychowski C, Helle F, Pillez A, Drobecq H, Le Naour F, Charrin S, Levy S, et al. The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS ONE. 2008;3:e1866. doi: 10.1371/journal.pone.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 55.Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flint M, von Hahn T, Zhang J, Farquhar M, Jones CT, Balfe P, Rice CM, McKeating JA. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80 :11331–11342. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michta ML, Hopcraft SE, Narbus CM, Kratovac Z, Israelow B, Sourisseau M, Evans MJ. Species-specific regions of occludin required by hepatitis C virus for cell entry. J Virol. 2010;84:11696–11708. doi: 10.1128/JVI.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **58.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. This study demonstrates for the first time that a step in the HCV life-cycle can be recapitulated in an inbred mouse strain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Long G, Hiet MS, Windisch MP, Lee JY, Lohmann V, Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.06.010. The authors demonstrate that HCV viral assembly can occur in murine hepatoma cell line if sufficient apolipoproteins are present. This is an important observation aiding the development of an inbred mouse model that can sustain the entire HCV life-cycle. [DOI] [PubMed] [Google Scholar]
- 60.Ploss A, Rice CM. Towards a small animal model for hepatitis C. EMBO Rep. 2009 doi: 10.1038/embor.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]