Abstract
The human 3q29 microdeletion syndrome is associated with mild facial dysmorphism, developmental delay and variable congenital malformations. We report three new unrelated patients with this syndrome. We also performed in silico RNA binding analysis in silico on the 3q29 critical region genes. Several genes within this genomic region including DLG1 and RNF168 are predicted to bind RNA. While recessive mutations in RNF168 cause RIDDLE syndrome, an immune deficiency and radiosensitivity disorder, the potential impact of heterozygous deletion of RNF168 on patients with the 3q29 deletion syndrome is still unknown.
Keywords: chromosome, 3q29, aCGH, in silico, RNA binding, deletion
INTRODUCTION
Following the advent of microarray based technology, several novel submicroscopic micro-deletion and duplication syndromes had been identified, including the 3q29 deletion (del 3q29) syndrome. Using subtelomeric fluorescent in situ hybridization (FISH), de novo del 3q29 syndrome was first identified in one young child with moderate intellectual disability, minor facial dysmorphism, horse shoe kidney, and hypospadias by Rossi et al. [2001]. Willatt et al. [2005] provided clinical and molecular characterization of the del 3q29 syndrome in six unrelated patients. Subsequently, several additional patients had been described. The syndrome is characterized by mild-moderate developmental delay, speech delay, mild nonspecific facial dysmorphic features, and variable structural abnormalities [Koolen et al., 2004; Baynam et al., 2006; Krepischi-Santos et al., 2006; Ballif et al., 2008; Shao et al., 2008; Digilio et al., 2009; Li et al., 2009; Mihçi et al., 2009; Pollazzon et al., 2009; Tyshchenko et al., 2009; Clayton-Smith et al., 2010; Quintero-Rivera et al., 2010; Wang et al., 2010]. Autistic traits have been noticed in a few patients [Willatt et al., 2005; Ballif et al., 2008; Quintero-Rivera et al., 2010]. All patients shared a small “sporadic” 1.6 Mb microdeletion except in three affected individuals with a familial deletion [Digilio et al., 2009; Li et al., 2009; Clayton-Smith et al., 2010]. In this report, we describe three new unrelated individuals with 3q29 heterozygous deletion which does not include the human micro-RNA (hsa-miRNA) 570 gene. We also used in silico RNA binding analysis to characterize the deleted genes.
CLINICAL DESCRIPTION
Patient 1
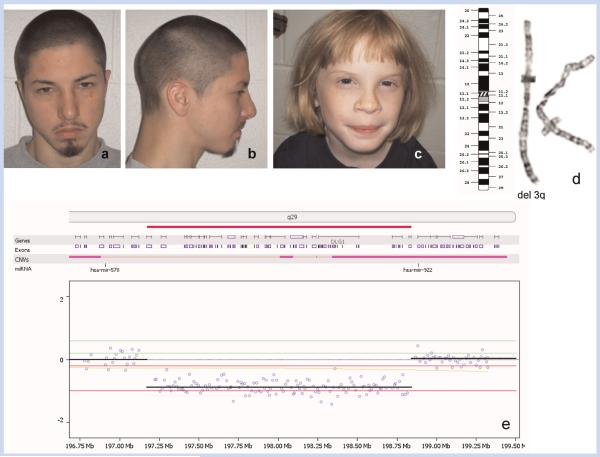
An 18.5-year-old Hispanic man (Fig. 1a,b) was the product of uncomplicated term gestation and delivery to a 25-year-old healthy mother. His growth measurements at birth were normal [weight 3,895 g (75th centile), length 52 cm (75th centile), head circumference 35.5 cm (25th centile)]. He had a right inguinal hernia and a heart murmur which was associated with a small uncomplicated secundum atrial septal defect. Severe expressive language and articulation delays were diagnosed at 31 months of age. He received speech therapy for several years which was helpful. Hearing and vision evaluations were normal. Because of recurrent otitis media, he had tympanostomy tubes. He was thought to have a submucous cleft palate but did not require surgical correction. More recently, he had an arteriovenous hemangioma resected from his left arm. Family history was unremarkable. Currently, his weight is 62.65 kg (25th centile), height 172 cm (25th centile). He has mild facial dysmorphism (narrow and long face with prominent chin), and a high palate with few indentations. He also has mild pectus excavatum. He is a shy and soft spoken adolescent. He had a high resolution karyotype and FISH studies for the 22q11 deletion which were interpreted as normal. Array comparative genomic hybridization (aCGH) analysis revealed a 1.66 Mb deletion at 3q29 (Fig. 1e). Using the same FISH probe set, his mother’s chromosomes demonstrated a normal hybridization pattern. His father was not available for analysis.
FIG. 1.
Facial profiles of patients 1 (a,b) and 2 (c) with the 3q29 deletion syndrome show narrow and long face (a,b), micrognathia (a), broad nose (c), and small mouth (a,d). Panel (d) ideogram and a partial karyotype of chromosome 3 in patient 1 shows a small deletion of 3q29. Panel (e) representative log2 ratio probe plot for patients 1, 2, and 3 showing the 3q29 deletion. Note the CNV which overlaps the hsa-mir-922 gene. [Color figure can be seen in the online version of this article, available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552-4833]
Patient 2
An 8-year-old Caucasian girl (Fig. 1c) was born at 36 weeks gestation which was complicated by maternal type 1 diabetes mellitus, significant hypertension, severe preeclampsia and polyhydramnios. Amniocentesis was normal (46,XX). Apgar scores were 6 and 8 and a patent ductus arteriosus closed by the second day of life. Failure to thrive, gastroesophageal reflux, urinary voiding dysfunction, ADHD and speech delay were diagnosed throughout early childhood. Family history was significant for attention deficit disorder in her sister while her mother had type 1 diabetes mellitus, hypothyroidism and multiple intestinal polyps. Routine chemistry studies including serum calcium, phosphorus and parathormone and 25-hydroxy vitamin D levels were unremarkable. Her bone age was normal. The patient’s weight was 21.2 kg (10th centile), height 118 cm (<5th centile), and head circumference 51 cm (25–50th centile). She had bilateral epicanthal folds, normal eye slanting and prominent central incisors. She also had an interrupted transverse left palmar crease. Blood aCGH also revealed a de novo 1.66 Mb deletion at 3q29.
Patient 3
A 1.13 Mb deletion at 3q29 (base position chr3:197085422–198217248) was found in a patient referred to CMDX (Irvine, CA) for clinical aCGH analysis. The child had developmental delay, a heart murmur in addition to a paternally inherited paracentric inversion of chromosome 21 (not shown) based on the data provided to the laboratory at the time of clinical testing. No formal clinical genetics evaluation was available for this patient. No other structural or numerical chromosomal abnormalities were reportedly seen on the routine karyotype.
MATERIALS AND METHODS
Chromosome and FISH Analysis
GTG-banded chromosomes and FISH analyses were performed on peripheral blood lymphocyte cultures using standard techniques. The BAC clones RP11-793A14 (chromosome 3p11.1) and RP11-652M22 (chromosome 3q29) were obtained from The Center for Applied Genomics (TCAG, Toronto, Canada) and were used for FISH based confirmation of array comparative genomic hybridization (aCGH) results in all three patients.
Array Comparative Genomic Hybridization
Diagnostic whole-genome array-based comparative genomic hybridization (array CGH) was performed using the CMDX Oligo HD Scan™ (Irvine, CA) designed to detect genome-wide copy number variations. The array contains 99,000+ probes (Agilent Technologies, Santa Clara, CA) covering coding and non-coding human genome sequences with content sourced from the UCSC hg18 human genome (NCBI build 36, March 2006) and an average probe spatial resolution of ~21 kb. DNA copy number in the patient sample is evaluated in relation to a reference diploid DNA sample, and targeted evaluation of copy number changes involving >6 probes is performed in all regions of the genome including pericentromeric, subtelomeric, and loci representative of microduplication/microdeletion syndromes. FISH and/or BAC array CGH analysis using probes targeted to the region delineated by oligonucleotide array CGH is used for confirmation. Non-diagnostic copy number changes are referenced to the Database of Genomic Variants [http://projects.tcag.ca/variation; Iafrate et al., 2004] and percentage of overlap with polymorphic regions is determined. Nexus Copy Number™ software (Biodiscovery, El Segundo, CA) was used for copy number analysis across the genome.
In Silico RNA Binding Analysis
The RNABindR [Terribilini et al., 2007] and Pprint [Kumar et al., 2008] methods for prediction of RNA-binding propensity in specific amino acids of structurally un-resolved proteins were used to predict the RNA binding ability of the genes included in the deleted interval in chromosome 3q29. The default support vector machine (SVM) threshold value of −0.2 was used with the Pprint method. Both of these models were constructed from a training set of crystallographically characterized protein-RNA complexes available from the Protein Databank, with RNA-interacting residues being defined according to spatial proximity to RNA fragments. Genes were classified as likely RNA-binders (at least 25 consensus amino acids), possible RNA-binders (between 10 and 24 consensus amino acids), likely RNA non-binders (1–9 consensus amino acids) and RNA non-binders (0 consensus amino acids).
RESULTS
Oligo-aCGH analysis in all three patients reported here showed three submicroscpic subtelomeric deletions in chromosome 3q29 as shown in the representative profile in panel 1e. The 1.66 Mb deletion [base position chr3:197174369–198842531] appeared to be similar in the first two patients. It was confirmed by FISH analysis in all three patients using the BAC clone RP11-652M22 (data not shown). The deletion was appreciated in Patient 1 on retrospective examination of his karyotype. The mother of Patient 1 and both parents of Patient 2 had normal FISH analysis using RP11-65M22. The father of Patient 1 was not available for testing. BAC-aCGH analysis in Patient 2 also showed deletion of the 3q29 region encompassing RP11-480A16→RP11-652M22 (data not shown).
In silico RNA binding analysis showed multiple genes with potential RNA binding. As shown in Table I, they are ranked by the number of consensus amino acids with RNF168 (74/571) predicted to have the strongest RNA binding ability.
TABLE I.
RNA Binding Analysis of the Genes Within the 3q29 Deletion Region
| Protein (number of consensus AA/total number of AA) | |
|---|---|
| Likely RNA-binders (>25 consensus AAs) | RNF168 (74/571); SENP5 (50/755); DLG1 (43/904); AX747828 (37/245); TNK2 (isoform 1) (34/1048); FBXO45 (31/286); NCBP2 (30/86); TNK2 (isoform 3) (26/1086); LOC152217 (25/99) |
| Possible RNA-binders (10–24 consensus AAs) | KIAA0226 (21/972), MFI2 (20/738), WDR53 (19/358), TNK2 (18/528), PCYT1A (14/367), AK124973 (10/101) |
| Likely RNA non-binders (1-9 consensus AAs) | UBXN7 (7/489), PAK2 (5/924), TFRC (2/760); BDH1 (2/343), C3orf43 (1/206), LRC33 (1/692) |
| RNA non-binders (0 consensus AAs) | ZDHHC19 (0/309), OSTalpha (0/340), TCTEX1D2 (0/142), KIAA0794 (0/341), TM4SF19 (0/209), OCTM4 (0/299), PIGX_I1 (0/258), PIGX_I2 (0/276), DKFZp686G18222 (0/100), PIGZ (0/86) |
Within each group, proteins are ranked based on the number of surface amino acids predicted to be available for RNA binding.
DISCUSSION
Large cytogenetically visible terminal deletions of chromosome 3 (3q27-ter) had been previously reported [Alvarez et al., 1984; Sargent et al., 1985; Brueton et al., 1989; Jokiaho et al., 1989; Chitayat et al., 1996; Senzaki et al., 2003; Pollazzon et al., 2009; Wang et al., 2010] in nine patients four of whom died in early childhood. As a group, they had significant structural abnormalities including cleft lip and palate, cerebral atrophy, Dandy-Walker malformation, meningocele and cardiomyopathy. Failure to thrive and short stature were frequent findings as well. More recently, Rossi et al. [2001] and then Willatt et al. [2005] identified and characterized a much smaller distal deletion limited to the terminal 3q29 band. Our three unrelated patients also have a very small terminal deletion (1.13–1.66 Mb) involving 3q29 which was easily identified and characterized using oligo-aCGH and confirmed by BAC-aCGH or fluorescent in situ hybridization. The relatively mild phenotype we report in these patients is consistent with the initial observations of Willatt et al. [2005] and others. While these patients have mild facial dysmorphic features, these features are not specific or clinically diagnostic. In the recent review by Quintero-Rivera et al. [2010], mild/moderate developmental delay (96%), speech delay (65%), microcephaly (58%) were the most commonly reported findings in patients with this syndrome. Also, unlike children who were born with larger “3q27-ter” deletions, children with the 3q29 deletion syndrome seem to have milder and fewer structural malformations including cleft lip/palate (4%), congenital heart disease (12%) and renal anomalies (8%) [Quintero-Rivera et al., 2010]. Congenital heart defects were reported in a father–son pair who also had 3q29 deletion [Li et al., 2009]. The father had pulmonic stenosis and PDA, while his son had subvalvular aortic stenosis and PDA. Patient 1 in this series had a hemodynamically insignificant secundum ASD without known family history of congenital heart disease while Patient 2 had prematurity-related PDA. C3orf43 (base position chr3:197718147–197726634, hg18) is a gene located within the critical region for this syndrome, and it encodes a hypothetical protein LOC255798 which is expressed in heart, connective tissue and skeletal muscle [G2SBC database]. Given the complex inheritance of congenital heart disease and the unknown genotype–phenotype correlation for the deleted region on chromosome 3q29, it is not possible to confirm a direct causal relationship. The report of congenital microophthalmia and cataract in the patient reported by Tyshchenko et al. [2009] expands the phenotypic spectrum the 3q29 microdeletion syndrome. It is interesting to note that the breakpoints reported by Li et al. [2009] and Tyshchenko et al. [2009] overlap with the breakpoints we report here in these three patients.
The deleted 3q29 region contains a relatively small number of genes. While there is no known strong genotype–phenotype correlation, haploinsufficiency of PAK2 and DLG1 (also known as SAP97, synapse-associated protein 97) was thought to be responsible for the developmental delays seen in these patients. These two genes represent the autosomal homologues of known X-linked mental retardation genes, PAK3 (OMIM No. 300142) and DLG3 (OMIM No. 300189). DLG1 and CASK (calcium/calmodulin-dependent serine protein kinase) are members of the membrane-associated guanylate kinase (MAGUK) protein family which represent scaffolding proteins associated with intercellular junctions. Dlg is expressed in murine epithelial, mesenchymal, neuronal, endothelial, and hematopoietic cells during embryogenesis and is required in normal craniofacial development, possibly in a subset of neural crest cells or their derivatives [Caruana and Bernstein, 2001]. Mice homozygous for the dlg mutation [Caruana and Bernstein, 2001] as well as CASK 22 insertional mutants exhibit prenatal growth retardation and secondary cleft palate [Laverty and Wilson, 1998]. Through its L27 domain mediated homo and heteromulti-merization with other proteins, DLG1/SAP97 regulates synaptic delivery of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) in hippocampal neurons [Nakagawa et al., 2004]. RNAi knockdown of endogenous SAP97 reduced surface expression of both GluR1 and GluR2 and inhibited both AMPA and NMDA excitatory postsynaptic currents. PAK2 and PAK3 are members of group (1) p21-activated kinases which have diverse cellular functions. Loss of function of PAK3 is responsible for X-linked non-syndromic mental retardation whereas gain of PAK3 function is associated with cancer [Kreis et al., 2008].
MicroRNAs (miRNAs) are a large family of small “17–25 nucleotides long” noncoding RNAs that posttranscriptionally extensively regulate gene expression in animals, plants, and protozoa. They interact with proteins and mRNA (by base paring with the 3′ untranslated region) causing translational repression and mRNA decay [Fabian et al., 2010]. Small RNAs including miRNAs are involved in a wide range of physiologic functions, such as developmental transitions and neuronal patterning, apoptosis, fat metabolism, and regulation of hematopoietic lineage differentiation [Jin et al., 2004; Qurashi et al., 2007; Stefani and Slack, 2008; Kim et al., 2009]. Their dysregulation is likely linked to the pathogenesis of neurological disorders [Cao et al., 2006; Stefani and Slack, 2008; Chang et al., 2009; Qurashi and Jin, 2010]. The roles of specific miRNAs have been explored in several neurodevelopmental disorders such as fragile X syndrome, Rett syndrome, velocardiofacial/ DiGeorge syndrome and Down syndrome. Altered miRNA transcription and biogenesis, dosage of miRNA genes associated with segmental duplications, SNPs in the target sites throughout the human genome as well as variations in the target mRNA sequences could modulate the activity of specific miRNAs and alter phenotype accordingly [Clop et al., 2006; Georges et al., 2006]. Two miRNAs (570 and 922, base positions chr3 195,424,272–195,428,368 and 197,399,367–197,403,447, respectively) are encoded within the 3q29 region. Each of these two miRNAs is known to have many predicted downstream targets [miRBase database]. While miRNA 922 was deleted in Patients 1 and 2 and deletion involving this gene is a known CNV, miRNA577 was not deleted in any of our three patients. Given the large size deletions in patients with cytogenetically detectable 3q27-ter reported in the literature, we suspect that both miRNAs 570 and 922 were deleted as well which may explain their more severe phenotype and clinical heterogeneity. C3orf43, a gene consistently deleted in this syndrome is expressed in the heart and interacts with several downstream microRNAs [G2SBC database]. Based on the recent evidence which implicated DGCR8 (a gene within 22q11.2) in microRNA biogenesis and silencing of embryonic stem cell self-renewal [Wang et al., 2007], and in altered brain miRNA biogenesis leading to phenotypic abnormalities [Stark et al., 2008], we hypothesized that one or more of the deleted genes within 3q29 region may be an RNA binding gene and may exert a similar effect. We examined this possibility using the RNABindR and Pprint methods. The number of consensus amino acids predicted by both methods provides a reasonable measure of a protein’s realistic likelihood of binding RNA, since a successful binding event is contingent upon favorable RNA-interaction with a significant number of surface amino acids. Surprisingly, several potential RNA binding genes within 3q29 were identified including DLG1 (Tables I and II).
TABLE II.
Examples of RNA Binding Prediction Analysis
| RNF168: uc003fwq.1 (RNF168) length=571 |
| 74 consensus residues / likely RNA binding |
| MALPKDAIPSLSECQCGICMEILVEPVTLPCNHTLCKPCFQSTVEKASLCCPFCRRRVSSWTRYHTRRNSL VNVELWTIIQKHYPRECKLRASGQESEEVADDYQPVRLLSKPGELRREYEEEISKVAAERRASEEEENKAS EEYIQRLLAEEEEEEKRQAEKRRRAMEEQLKSDEELARKLSIDINNFCEGSISASPLNSRKSDPVTPKSEK KSKNKQRNTGDIQKYLTPKSQFGSASHSEAVQEVRKDSVSKDIDSSDRKSPTGQDTEIEDMPTLSPQISLG VGEQGADSSIESPMPWLCACGAEWYHEGNVKTRPSNHGKELCVLSHERPKTRVPYSKETAVMPCGRTESGC APTSGVTQTNGNNTGETENEESCLLISKEISKRKNQESSFEAVKDPCFSAKRRKVSPESSPDQEETEINFT QKLIDLEHLLFERHKQEEQDRLLALQLQKEVDKEQMVPNRQKGSPDEYHLRATSSPPDKVLNGQRKNPKDG NFKRQTHTKHPTPERGSRDKNRQVSLKMQLKQSVNRRKMPNSTRDHCKVSKSAHSLQPSISQKSVFQMFQR CTK |
| SENP5: uc003fwz.2 (SENP5) length=755 |
| 50 consensus residues / likely RNA binding |
| MKKQRKILWRKGIHLAFSEKWNTGFGGFKKFYFHQHLCILKAKLGRPVTWNRQLRHFQGRKKALQIQKTWI KDEPLCAKTKFNVATQNVSTLSSKVKRKDAKHFISSSKTLLRLQAEKLLSSAKNSDHEYCREKNLLKAVTD FPSNSALGQANGHRPRTDPQPSDFPMKFNGESQSPGESGTIVVTLNNHKRKGFCYGCCQGPEHHRNGGPLI PKKFQLNQHRRIKLSPLMMYEKLSMIRFRYRILRSQHFRTKSKVCKLRKAQRSWVQKVTGDHQETRRENGE GGSCSPFPSPEPKDPSCRHQPYFPDMDSSAVVKGTNSHVPDCHTKGSSFLGKELSLDEAFPDQQNGSATNA WDQSSCSSPKWECTELIHDIPLPEHRSNTMFISETEREIMTLGQENQTSSVSDDRVKLSVSGADTSVSSVD GPVSQKAVQNENSYQMEEDGSLKQSILSSELLDHPYCKSPLEAPLVCSGLKLENQVGGGKNSQKASPVDDE QLSVCLSGFLDEVMKKYGSLVPLSEKEVLGRLKDVFNEDFSNRKPFINREITNYRARHQKCNFRIFYNKHM LDMDDLATLDGQNWLNDQVINMYGELIMDAVPDKVHFFNSFFHRQLVTKGYNGVKRWTKKVDLFKKSLLLI PIHLEVHWSLITVTLSNRIISFYDSQGIHFKFCVENIRKYLLTEAREKNRPEFLQGWQTAVTKCIPQQKND SDCGVFVLQYCKCLALEQPFQFSQEDMPRVRKRIYKELCECRLMD |
| DLG1: uc003fxo.2 (DLG1) length=904 |
| 43 consensus residues / likely RNA binding |
| MPVRKQDTQRALHLLEEYRSKLSQTEDRQLRSSIERVINIFQSNLFQALIDIQEFYEVTLLDNPKCIDRSK PSEPIQPVNTWEISSLPSSTVTSETLPSSLSPSVEKYRYQDEDTPPQEHISPQITNEVIGPELVHVSEKNL SEIENVHGFVSHSHISPIKPTEAVLPSPPTVPVIPVLPVPAENTVILPTIPQANPPPVLVNTDSLETPTYV NGTDADYEYEEITLERGNSGLGFSIAGGTDNPHIGDDSSIFITKIITGGAAAQDGRLRVNDCILRVNEVDV RDVTHSKAVEALKEAGSIVRLYVKRRKPVSEKIMEIKLIKGPKGLGFSIAGGVGNQHIPGDNSIYVTKIIE GGAAHKDGKLQIGDKLLAVNNVCLEEVTHEEAVTALKNTSDFVYLKVAKPTSMYMNDGYAPPDITNSSSQP VDNHVSPSSFLGQTPASPARYSPVSKAVLGDDEITREPRKWLHRGSTGLGFNIVGGEDGEGIFISFILAG GPADLSGELRKGDRIISVNSVDLRAASHEQAAAALKNAGQAVTIVAQYRPEEYSRFEAKIHDLREQMMNSS ISSGSGSLRTSQKRSLYVRALFDYDKTKDSGLPSQGLNFKFGDILHVINASDDEWWQARQVTPDGESDEVG VIPSKRRVEKKERARLKTVKFNSKTRDKGEIPDDMGSKGLKHVTSNASDSESSYRGQEEYVLSYEPVNQQE VNYTRPVIILGPMKDRINDDLISEFPDKFGSCVPHTTRPKRDYEVDGRDYHFVTSREQMEKDIQEHKFIEA GQYNNHLYGTSVQSVREVAEKGKHCILDVSGNAIKRLQIAQLYPISIFIKPKSMENIMEMNKRLTEEQARK TFERAMKLEQEFTEHFTAIVQGDTLEDIYNQVKQIIEEQSGSYIWVPAKEKL |
| AX747828: uc003fwo.l (AX747828) length=245 |
| 37 consensus residues / likely RNA binding |
| MGRWETPPWILPGLPYSAWGFPLILPSKPQSLRSSSAPPASGGLSPVKCKGTAEPTGLAVKAGGPEPARAT AMAPRGPERGRAGRGPGQNDLAGEERKKGGEEMHRTAGLHREGKMMKEEEEGGGPGAAPYSSRPRVPGPAG ADGLGPFPGLGWRGVGGSPSTLLGDRERGYRERGGAVSRLNPETPTPSSPLPQEGGRQLWVKPEGGMPEGK GKPGADPLPAPDPTLHLPVTASLTGNGGELLN |
RNABindR predictions are highlighted in red, while underlined amino acids are predicted by Pprint.
The RING finger protein RNF168 is a new ubiquitin ligase that functions as a chromatin modifier, through ubiquitination of histones H2A and H2AX [Pinato et al., 2009]. Its function is critical for DNA double-strand breaks repair (DSB) where it co-localizes with γH2AX and 53BP1 which then increases the local concentration of ubiquitinated proteins. Recessive mutations in RNF168 cause RIDDLE syndrome, a recently discovered syndrome which is characterized by Radiosensitivity, ImmunoDeficiency, Dysmorphic features, and LEarning difficulties [Stewart et al., 2007; Stewart et al., 2009; Stewart, 2009]. The father of this patient with RIDDLE syndrome had chronic B-cell leukemia, suggesting that RNF168 may also act as a tumor suppressor gene. In patients with radiosensitivity disorders, the underlying DNA double strand break repair defects have considerable impact on V(D)J gene recombination, class switching and lymphocyte maturation, leading to increased infections and cancer risk [Nahas and Gatti, 2009]. So far, none of the patients with the 3q29 deletion syndrome who are haploinsufficient for the RNF168 gene was reported to have immunodeficiency or malignancy. DNA analysis of the RNF168 intact allele and cellular radiosensitivity studies will be necessary to identify recessive mutations in these patients.
In conclusion, the phenotype seen in patients with the 3q29 microdeletion syndrome is somewhat non-specific and the structural malformations are relatively mild. The study of more patients and long term follow up of such individuals will help better define this syndrome and potential future risk of immunodeficiency and malignancy. This region of the genome encodes several putative RNA binding proteins which are predicted to play very important role in gene regulation and expression. The roles of these predicted RNA binding proteins and their interaction partners in the pathogenesis of this syndrome should be validated through direct experimental studies.
REFERENCES
- Alvarez AMC, Rivera H, Moller M, Valdivia A, Vigueras A, Cantu JM. De novo del(3)(q2800) Ann Genet. 1984;27:109–111. [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, McDonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynam G, Goldblatt J, Townshend S. A case of 3q29 microdeletion with novel features and a review of cytogenetically visible terminal 3q deletions. Clin Dysmorphol. 2006;15:145–148. doi: 10.1097/01.mcd.0000198934.55071.ee. [DOI] [PubMed] [Google Scholar]
- Brueton LA, Barber JC, Huson SM, Winter RM. Partial monosomy 3q in a boy with short stature, developmental delay, and mild dysmorphic features. J Med Genet. 1989;26:729–730. doi: 10.1136/jmg.26.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Caruana G, Bernstein A. Craniofacial dysmorphogenesis including cleft palate in mice with an insertional mutation in the discs large gene. Mol Cell Biol. 2001;21:1475–1483. doi: 10.1128/MCB.21.5.1475-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Wen S, Chen D, Jin P. Small regulatory RNAs in neurodevelopmental disorders. Hum Mol Genet. 2009;18:R18–R26. doi: 10.1093/hmg/ddp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitayat D, Babul R, Silver MM, Jay V, Teshima IE, Babyn P, Becker LE. Terminal deletion of the long arm of chromosome 3 [46,XX,del(3)(q27-qter)] Am J Med Genet. 1996;61:45–48. doi: 10.1002/(SICI)1096-8628(19960102)61:1<45::AID-AJMG9>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Giblin C, Smith RA, Dunn C, Willatt L. Familial 3q29 microdeletion syndrome providing further evidence of involvement of the 3q29 region in bipolar disorder. Clin Dysmorphol. 2010;19:128–132. doi: 10.1097/MCD.0b013e32833a1e3c. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bib eB, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Digilio M, Bernardini L, Mingarelli R, Capolino R, Capalbo A, Giuffrida M, Versacci P, Novelli A, Dallapiccola B. 3q29 Microdeletion: A mental retardation disorder unassociated with a recognizable phenotype in two mother-daughter Pairs. Am J Med Genet Part A. 2009;149A:1777–1781. doi: 10.1002/ajmg.a.32965. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Georges M, Clop A, Marcq F, Takeda H, Pirottin D, Hiard S, Tordoir X, Caiment F, Meish F, Bib e B, Bouix J, Elsen JM, Eychenne F, Laville E, Larzul C, Milenkovic D, Tobin J, Charlier AC. Polymorphic microRNA target interactions: A novel source of phenotypic variation. Cold Spring Harb Symp Quant Biol. 2006;71:343–350. doi: 10.1101/sqb.2006.71.056. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:351–949. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Jokiaho I, Salo A, Niemi KM, Blomstedt GC, Pihkala J. Deletion 3q27-3qter in an infant with mild dysmorphism, parietal meningocele, and neonatal miliaria rubra-like lesions. Hum Genet. 1989;83:302–304. doi: 10.1007/BF00285180. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Nillesen WM, Versteeg MH, Merkx GF, Knoers NV, Kets M, Vermeer S, van Ravenswaaij CM, de Kovel CG, Brunner HG, Smeets D, de Vries BB, Sistermans EA. Screening for subtelomeric rearrangements in 210 patients with unexplained mental retardation using multiplex ligation dependent probe amplification (MLPA) J Med Genet. 2004;41:892–899. doi: 10.1136/jmg.2004.023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P, Rousseau V, Thévenot E, Combeau G, Barnier JV. The four mammalian splice variants encoded by the p21-activated kinase 3 gene have different biological properties. J Neurochem. 2008;106:1184–1197. doi: 10.1111/j.1471-4159.2008.05474.x. [DOI] [PubMed] [Google Scholar]
- Krepischi-Santos AC, Vianna-Morgante AM, Jehee FS, Passos-Bueno MR, Knijnenburg J, Szuhai K, Sloos W, Mazzeu JF, Kok F, Cheroki C, Otto PA, Mingroni-Netto RC, Varela M, Koiffmann C, Kim CA, Bertola DR, Pearson PL, Rosenberg C. Whole-genome array-CGH screening in undiagnosed syndromic patients: Old syndromes revisited and new alterations. Cytogenet Genome Res. 2006;115:254–261. doi: 10.1159/000095922. [DOI] [PubMed] [Google Scholar]
- Kumar M, Gromiha MM, Raghava GPS. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins. 2008;71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- Laverty HG, Wilson JB. Murine CASK is disrupted in a sex-linked cleft palate mouse mutant. Genomics. 1998;53:29–41. doi: 10.1006/geno.1998.5479. [DOI] [PubMed] [Google Scholar]
- Li F, Lisi E, Wohler E, Hamosh A, Batista D. 3q29 interstitial microdeletion syndrome: An inherited case associated with cardiac defect and normal cognition. Eur J Med Genet. 2009;52:349–352. doi: 10.1016/j.ejmg.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Mihçi E, Ozcan M, Berker-Karaüzüm S, Keser I, Taçoy S, Hapsolat S, Lüleci G. Subtelomeric rearrangements of dysmorphic children with idiopathic mental retardation reveal 8 different chromosomal anomalies. Turk J Pediatr. 2009;51:453–459. [PubMed] [Google Scholar]
- Nahas SA, Gatti RA. DNA double strand break repair defects, primary immunodeficiency disorders, and ‘radiosensitivity’. Curr Opin Allergy Clin Immunol. 2009;9:510–516. doi: 10.1097/ACI.0b013e328332be17. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel H, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44:453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol. 2009;10:55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollazzon M, Grosso S, Papaa F, Katzaki E, Marozza A, Mencarelli M, Uliana V, Balestri P, Mari F, Renieri A. A 9.3 Mb microdeletion of 3q27.3q29 associated with psychomotor and growth delay, tricuspid valve dysplasia and bifid thumb. Eur J Med Genet. 2009;52:131–133. doi: 10.1016/j.ejmg.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: Case report and review. Am J Med Genet Part A. 2010;152A:2459–2467. doi: 10.1002/ajmg.a.33573. [DOI] [PubMed] [Google Scholar]
- Qurashi A, Chang S, Peng J. Role of microRNA pathway in mental retardation. Sci WorldJ. 2007;7:146–154. doi: 10.1100/tsw.2007.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurashi A, Jin P. Small RNA-mediated gene regulation in neuro-developmental disorders. Curr Psychiatry Rep. 2010;12:154–161. doi: 10.1007/s11920-010-0102-1. [DOI] [PubMed] [Google Scholar]
- Rossi E, Piccini F, Zollino M, Neri G, Caselli D, Tenconi R, Castellan C, Carrozzo R, Danesino C, Zuffardi O, Ragusa A, Castiglia L, Galesi O, Greco D, Romano C, Pierluigi M, Perfumo C, Di Rocco M, Faravelli F, Bricarelli F Dagna, Bonaglia M, Bedeschi M, Borgatti R. Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations. J Med Genet. 2001;38:417–420. doi: 10.1136/jmg.38.6.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent C, Burn J, Baraitser M, Pembrey ME. Trigonocephaly and the Opitz C syndrome. J Med Genet. 1985;22:39–45. doi: 10.1136/jmg.22.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzaki H, Inui M, Ban S, Masutani S, Morsy M, Kobayashi T, Nagasaka H, Sasaki N, Kyo S, Yokote Y. Dilated cardiomyopathy in a 3-year-old girl with a terminal deletion, 46, XX,del(3)(q27-qter), of the long arm of chromosome 3. Eur J Pediatr. 2003;162:403–405. doi: 10.1007/s00431-003-1160-8. [DOI] [PubMed] [Google Scholar]
- Shao L, Shaw CA, Lu XY, Sahoo T, Bacino CA, Lalani SR, Stankiewicz P, Yatsenko SA, Li Y, Neill S, Pursley AN, Chinault AC, Patel A, Beaudet AL, Lupski JR, Cheung SW. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: A study of 5,380 cases. Am J Med Genet Part A. 2008;146:2242–2251. doi: 10.1002/ajmg.a.32399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ, Taylor AM. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci. 2007;104:16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Stewart GS. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 2009;8:1532–1538. doi: 10.4161/cc.8.10.8351. [DOI] [PubMed] [Google Scholar]
- Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: A server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyshchenko N, Hackmann K, Gerlach E, Neuhann T, Schrock E, Tinschert S. 1.6 Mb deletion in chromosome band 3q29 associated with eye abnormalities. Eur J Med Genet. 2009;52:128–130. doi: 10.1016/j.ejmg.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Naik H, Khan A, Nowaczyk MJ. An uncommon 3.4-Mb interstitial deletion at 3q29. Clin Dysmorphol. 2010;19:133–136. doi: 10.1097/MCD.0b013e3283387b21. [DOI] [PubMed] [Google Scholar]
- Willatt L, Cox J, Barber J, Cabanas E Dachs, Collins A, Donnai D, FitzPatrick DR, Maher E, Martin H, Parnau J, Pindar L, Ramsay J, Shaw-Smith C, Sistermans EA, Tettenborn M, Trump D, de Vries BBA, Walker K, Raymond FL. 3q29 microdeletion syndrome: Clinical and molecular characterization of a new syndrome. Am J Hum Genet. 2005;77:154–160. doi: 10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB RESOURCES
- The Center for Applied Genomics (TCAG) http://www.cag.icph.org/
- Database of Chromosome Genomic Variants. http://projects.tcag.ca/variation
- Genes-to-Systems Breast Cancer (G2SBC) Database. http://www.itb.cnr.it/breastcancer/
- miRBase: the microRNA database. http://www.mirbase.org/