Abstract
To track Aedes aegypti (L.) egg-laying behavior in the field in Iquitos, Peru, we developed methods for 1) sampling DNA from live mosquitoes and 2) high through-put parentage analysis using microsatellite markers. We were able to amplify DNA extracted from a single hind leg, but not from the pupal exuvia. Removal of a leg from teneral females caused no significant changes in female behavioral or life history traits (e.g., longevity, blood feeding frequency, fecundity, egg hatch rate, gonotrophic cycle length, or oviposition behavior). Using a panel of nine microsatellite markers and an exclusion-based software program, we matched offspring to parental pairs in 10 Ae. aegypti test families in which parents originated from natural development sites in Iquitos. By mating known individuals in the laboratory, retaining the male, sampling the female’s DNA before release, and collecting offspring in the field, the technique we developed can be used to genotype large numbers of Ae. aegypti, reconstruct family relationships, and track the egg-laying behavior of individual Ae. aegypti in nature.
Keywords: Aedes aegypti, DNA extraction, microsatellite genotyping, oviposition, individual behavior
The behavior of individual insects determines population processes such as dispersal, migration, mating success, and habitat selection. Thus, understanding behavior is critical to predicting the distribution and dynamics of populations (Krebs and Davies 1978, Stimac 1982). Insight into insect behavior is especially important when building models to predict the population-level outcomes for target species of conservation or control measures (Stimac 1982, Hamm et al. 2010). Parentage analysis using genetic markers has enhanced the field of behavioral ecology by allowing investigators to infer behaviors that are difficult to observe directly in nature (Tentelier et al. 2008). For example, sampling individuals from known habitats in the field and reconstructing parentage can provide insights into dispersal, oviposition-site choice, and reproductive success of parents of the progeny sampled. Our goal in this study was to use parentage analysis as a tool to better understand egg-laying behavior of the dengue virus vector, Aedes aegypti (L.), with the ultimate aim of improving strategies for its control.
Investigators of insect behavior have pioneered nonlethal methods to sample small amounts of genetic material, enabling behavioral and genetic analysis of released individuals and their offspring. These methods typically involve removing a leg (Fincke and Hadrys 2001, Starks and Peters 2002, Holehouse et al. 2003) or portion of the wing (Vila et al. 2009, Hamm et al. 2010), resulting in minimal effects on individual behavior and survival (Starks and Peters 2002, Holehouse et al. 2003, Vila et al. 2009, Hamm et al. 2010). Some investigators have developed completely non-invasive sampling techniques using the insect pupal exuvia as a source of DNA (Watts et al. 2005, Dhananjeyan et al. 2010).
In our current study, we compared the feasibility of extracting genomic DNA from ethanol-preserved female Ae. aegypti pupual exuvia versus a single leg. We also assessed the effect of leg removal on several Ae. aegypti behavioral and life history traits. Researchers have genotyped mosquitoes using a single leg from dead specimens (Scott et al. 1993, Ruiz et al. 2005), but there are no reports documenting the impact of leg removal on important life history traits (e.g., longevity or fecundity) or mosquito behavior. For Aedes species, physical damage to gravid females can induce “death stress oviposition,” during which females lay their eggs immediately without regard to appropriate substrate or offspring development site (DeCoursey and Webster 1952, Wallis and DeBishop 1957, Chadee and Ritchie 2010). If removal of a leg from live Ae. aegypti females caused sufficient stress to induce this behavior, leg removal would be unsuitable for studying natural dispersal or oviposition patterns of released females.
To study fine scale patterns in Ae. aegypti oviposition behavior, we developed and assessed a method for parentage analysis using microsatellite markers. Microsatellites are the most commonly used marker for parentage assignment because of their polymorphism, broad genome distribution, co-dominant expression, and abundance in most organisms (Jones and Ardren 2003, Jones et al. 2010). Until recently, few microsatellites were identified for studying Ae. aegypti, because of their propensity to be embedded in repetitive regions of the genome (Chambers et al. 2007). In previous genetic studies of Ae. aegypti behavior, markers such as randomly amplified polymorphic DNAs (RAPDs) (Apostol et al. 1994) and restriction fragment length polymorphisms (RFLPs) (Colton et al. 2003) were used in combination with statistical methods to cluster individuals into sibling families. These techniques, however, exhibited high misclassification rates (14.4% for RAPDs, Apostol et al. 1994; 20.5% for RFLPs, Colton et al. 2003), which limited their utility for establishing true relatedness.
Recent publication of the Ae. aegypti genome (Nene et al. 2007) has led to the discovery of numerous polymorphic microsatellites (Chambers et al. 2007, Slotman et al. 2007, Lovin et al. 2009) that are suitable for the clear delineation of parent-offspring relationships. In our study, we demonstrate that 1) extracting Ae. aegypti genomic DNA from a single leg provides sufficient DNA for reliable PCR amplification, 2) removal of a leg from live females does not alter behavioral or life history traits in ways that are detectable in the laboratory, and 3) a panel of nine microsatellite markers can be used to match Ae. aegypti offspring to parental pairs.
Materials and Methods
Nonlethal DNA Sampling
Mosquitoes
From April through June 2008, F2 generation Ae. aegypti (F0 individuals originated from natural development sites in Iquitos, Peru; 73.2° W, 3.7° S, 120 m above sea level) were reared in the University of California Davis (UCD) insectary (26–28°C, 30–50% RH, 12:12 light:dark photoperiod) following the protocol developed by Styer et al. (2007). F2 generation mosquitoes were used in this experiment to minimize variation in life history traits because of differences in parental rearing conditions in the field (e.g., Grech et al. 2007). Individual pupae were transferred to individual 20 ml Pyrex test tubes filled with 5 ml of de-ionized water until adult emergence. Within an hour of emergence, adult females were allocated to test or control groups (described below). Pupal exuviae from test females were transferred to individual 1.5 ml plastic vials filled with 96% ethanol and labeled.
Leg Removal
Teneral females were assigned in sequential order to one of three treatments: 1) test (chilled and one leg removed), 2) chilled control with no leg removal, or 3) nonchilled control with no leg removal. For test females, legs were removed soon after emergence to provide maximum recovery time before oviposition. Because our subsequent field studies involved releasing adult females to track reproductive behavior, assays on the effect of leg removal were conducted only on females. Test females were individually mouth-aspirated into a 20 ml glass scintillation vial (Wheaton Industries Inc., Millville, NJ) and immobilized by chilling the vial in ice water for 20 s. Females then were placed on a paper towelcovered Tissue-Tek cold plate (Sakura-Finetek USA Inc., Torrance, CA) where one hind leg was removed at the junction of the coxa and thorax using a pair of forceps. Because adult Ae. aegypti support themselves using the fore- and mid-legs (Christophers 1960), the hind leg was selected for removal to minimize interference with balance and resting behavior. The female’s hind leg is used during copulation (Roth 1948), so we later examined for effects of leg removal on insemination success (below). Total time for immobilization and leg removal was less than 1 min. Afterwards, females were aspirated into either individual plastic buckets or group cages (see below). Mosquito legs were placed in individual 1.5 ml plastic vials filled with 96% ethanol and labeled. Chilled control females were transferred to scintillation vials, submerged in ice water, and placed on the cold plate, but did not have legs removed. Nonchilled control females were transferred directly from test tubes to plastic buckets or cages by aspiration.
DNA Extraction and PCR Amplification
Legs and pupal exuviae were preserved in 96% ethanol at room temperature for 1.5–2.5 mo before DNA extraction to simulate transport time from our field site in Iquitos, Peru, to our UCD laboratory. Genomic DNA was extracted from the hind leg and pupal exuvia of 20 test females (chilled and one leg removed) following the potassium acetate/ethanol precipitation protocol described by Black and DuTeau (1997), with the exception that all reagents were used at half volume. Polymerase chain reactions (PCRs) were performed at the carboxypeptidase A locus (Fulton et al. 2001) using puReTaq Ready-To-Go PCR Beads (GE Healthcare, Piscataway, NJ). This protocol and locus were selected for testing PCR amplification of extracted DNA, because it had provided consistent amplification from hundreds of ethanol-preserved Ae. aegypti in a previous study (Wong et al. 2008). Each 25 μl reaction contained 1.5 μl of genomic DNA, 0.6 μmol/L of each primer, 10 mmol/L Tris-HCl, 50 mmol/L KCl, 1.5 mmol/L MgCl, 200 μmol/L of each dNTP, and 2.5 U pureTaq DNA polymerase. PCR conditions were: 95°C for 5 min; 30 or 35 cycles of 1) 95°C for 45 s, 2) Ta for 45 s, and 3) 72°C for 1 min; followed by a final extension at 72°C for 7 min. PCR products were visualized using 1% agarose gel electrophoresis.
Behavioral and Life History Observations
To assess whether chilling and/or leg removal induced a “death stress” response, test females were compared with chilled and nonchilled controls with respect to severalbehavioral and life history traits (Table 1). Individual females were housed in plastic buckets (22 cm diameter × 15.5 cm height; Container and Packaging Supply Inc., Eagle, ID) with two unmated males and observed for up to 30 d. In each bucket, five ovalshaped black plastic cups (6 × 4.5 × 3.5 cm height, Pactiv Corp., Lake Forest, IL) were filled with water to 50% capacity and lined with strips of brown paper towel to serve as potential oviposition sites. Females were offered blood daily from a human arm or leg placed on top of the bucket’s mesh lid for 10 min between 1500 and 1700 hours. Because wild female Ae. aegypti living in close association with humans take frequent bloodmeals and rarely feed on plant carbohydrates (Edman et al. 1992, Van Handel et al. 1994, Scott et al. 2000), no sugar was offered. Every day before feeding, plastic cups were inspected for eggs. If eggs were present, mosquitoes were temporarily transferred by mouth aspiration to an empty bucket and the paper liners changed. The number of eggs laid, number of oviposition containers used, blood feeding, and mortality were recorded daily. Two trials were conducted, with test and chilled control females observed during the first trial (April 2008, n = 8 females per treatment) and all three groups observed during the second trial (June 2008, n = 7 females per treatment).
Table 1.
Comparison of Ae. aegypti behavioral and life history traits among test, chilled control, and nonchilled control females
| Mean longevity up to 30 d (days ± SD) |
Mean blood feeding frequency (meals/d ± SD) |
Mean fecundity (eggs ± SD) |
Mean gonotrophic cycle length (days ± SD) |
Mean no. oviposition sites used (containers ± SD) |
|
|---|---|---|---|---|---|
| Trial 1 | |||||
| Test (n = 8) | 26.4 ± 7.2a | 0.33 ± 0.13b | 96.0 ± 26.5c | 4.0 ± 0.9e | 3.9 ± 1.0f |
| Chilled control (n = 8) | 29.0 ± 2.1a | 0.38 ± 0.09b | 100.8 ± 26.7c | 3.8 ± 1.0e | 3.7 ± 1.2f |
| Trial 2 | |||||
| Test (n = 7) | 24.6 ± 5.2a | 0.35 ± 0.08b | 74.0 ± 32.5d | 4.2 ± 1.8e | 4.1 ± 1.2f |
| Chilled control (n = 7) | 20.3 ± 9.9a | 0.28 ± 0.09b | 83.8 ± 32.4d | 3.8 ± 0.9e | 4.0 ± 1.0f |
| Nonchilled control (n = 7) | 24.7 ± 8.4a | 0.31 ± 0.09b | 77.1 ± 40.9d | 3.8 ± 1.7e | 3.8 ± 1.2f |
Observations were limited to the first five gonotrophic cycles for all females. Values denoted by the same letter are not significantly different after Bonferroni correction (P value ≥ 0.010).
To assess whether leg removal negatively impacted insemination success, fecundity and egg-hatch rates were observed for females housed in cages. Ten females per treatment were each aspirated into a single cage (30 × 30 × 30 cm, BioQuip, Rancho Dominguez, CA) with 10 males and monitored for 10 d. A blue plastic cup (11 cm diameter × 7 cm height) filled with water to 50% capacity and lined with paper towel was provided for oviposition. Only one cup was provided per cage because we were unable to differentiate each females’ eggs, and hence could not distinguish individual egg-laying patterns. Caged females were offered blood from a human arm once per day and given no sugar. The daily number of eggs laid in each cage was recorded. We did not record when individual females fed or died. All three treatment groups were monitored during two trials (May and June 2008). Egg hatch rates were compared between the three treatments by attempting to hatch 100 eggs per cage per trial. Because of the tendency of Aedes eggs to hatch in installments (Gillett et al. 1977), we made three hatching attempts, each time submerging the eggs in hay infusion overnight.
The UCD Institutional Review Board (IRB) determined that feeding laboratory-reared mosquitoes on people in this experiment did not meet the requirements for human subject research, and thus, did not require IRB approval.
Data Analysis
Analysis of variance (ANOVA) was performed using R version 2.8.1 (R Development Core Team 2008) to examine the effects of treatment (leg removed, chilled control, and nonchilled control), trial, and treatment by trial interaction on female longevity (up to 30 d) and blood feeding frequency (number of bloodmeals taken per day alive). Longevity data were y3 transformed to meet ANOVA normality assumptions. Because fecundity, gonotrophic cycle length, and number of oviposition sites used were measured repeatedly for females over multiple gonotrophic cycles, the effects of treatment, trial, treatment by trial interaction, and gonotrophic cycle number on these variables were examined by repeated measures ANOVA using PROC MIXED in SAS version 9.2 (SAS Institute 2008). Normality assumptions for these models were met by applying y2 and log10 y transformations to fecundity and gonotrophic cycle length data, respectively.
Effects of treatment, trial, and treatment by trial interaction on the numbers of eggs laid per day were examined by repeated measures ANOVA using PROC MIXED in SAS version 9.2 (SAS Institute 2008). Egg hatch rates were compared among the three treatments by Fisher Exact test in R version 2.8. one (R Development Core Team 2008).
For the evaluation of insemination success, effects of treatment, trial, and treatment by trial interaction on the numbers of eggs laid per day were examined by repeated measures ANOVA using PROC MIXED in SAS version 9.2 (SAS Institute 2008). Egg hatch rates were compared among the three treatments by Fisher Exact test in R version 2.8. one (R Development Core Team 2008).
Parentage Analysis Using Microsatellites
Microsatellite Screening and Multiplexed-PCR
To screen for microsatellite polymorphisms in an Ae. aegypti population from Iquitos, we collected eggs naturally laid in 24 houses across the city. Individual mosquitoes from each house were randomly selected for inclusion in our study. Fifty-two individuals were selected from August 2008 collections and 20 individuals were selected from April 2010 collections to maximize coverage across Iquitos. Ae. aegypti collected in 2008 were reared to the adult stage under conditions previously described in Wong et al. (2011). Adults were killed after 1 wk and preserved in 96% ethanol for transport to UCD for genetic analysis. Ae. aegypti collected in 2010 were transported to UCD as eggs, and subsequently reared to the adult stage following the protocol by Styer et al. (2007).
Genomic DNA was purified from the 72 whole adult mosquitoes by potassium acetate/ethanol precipitation (Black and DuTeau 1997). We screened 18 microsatellite loci previously identified for Ae. aegypti (Chambers et al. 2007, Slotman et al. 2007) to assess polymorphism as well as fragment sizes in our population (Table 2). Microsatellites were initially divided into groups of no more than four loci per reaction for multiplexed PCR amplification. Because we used a set of four ßuorescent dyes, multiplex group size was limited to four loci to definitively identify PCR products corresponding to each microsatellite. PCR was carried out using ßuorescently labeled forward primers (Life Technology Corp., Carlsbad, CA) and the Qiagen Multiplex PCR Kit (Qiagen) (15 μl PCR reaction containing 1 μl of genomic DNA, 0.1–0.3 μmol/L of each primer, 1x PCR buffer, 3 mmol/L MgCl2, 0.2 mM of each dNTP, and 0.75 U HotStarTaq). PCR conditions were: 95°C for 15 min, followed by 35 cycles of 1) 94°C for 30 s, 2) 60°C for 90 s, and 3) 72°C for 1 min, followed by a final extension at 60°C for 30 min. PCR products were diluted 1:70 in ddH2O and submitted to the UCD College of Agriculture and Environmental Sciences Genomics Facility (http://cgf.ucdavis.edu/home/) for fragment analysis on an ABI 3730 XL capillary electrophoresis sequencer (Life Technology Corp.). Resulting chromatograms were analyzed using ABI Peak Scanner software (Applera Corp., Norwalk, CT). Genotypes were determined by comparing the relative position of ßuorescent peaks to GS600 LIZ size standard (Life Technology Corp.) peaks included with each sample.
Table 2.
Microsatellites initially screened for polymorphism in Ae. aegypti from Iquitos, Peru
| Locus | Reported size range (bp) |
Polymorphism level during screening |
|---|---|---|
| AG5a | 170–180 | High (≥3 alleles)b |
| AC1a | 193–209 | High (≥3 alleles)b |
| A10c | 233–239 | High (≥3 alleles)b |
| H08c | 199–205 | Low (<3 alleles)b |
| AG2a | 115–178 | High (≥3 alleles)b |
| AC5a | 149–163 | High (≥3 alleles)b |
| AT1a | 156–174 | High (≥3 alleles)b |
| B07c | 157–183 | High (≥3 alleles)b |
| AG3a | 164–178 | High (≥3 alleles)b |
| AG1a | 113–129 | High (≥3 alleles)b |
| M201c | 110–116 | Low (<3 alleles) |
| AG4a | 147–169 | Low (<3 alleles) |
| AC7a | 129–143 | Poor PCR amplification |
| AC2a | 176–190 | Low (<3 alleles) |
| M313c | 117–123 | Low (<3 alleles) |
| B19c | 156–186 | Low (<3 alleles) |
| G11c | 240–300 | Low (<3 alleles) |
| AG7a | 153–185 | Low (<3 alleles) |
Based on levels of polymorphism and fragment sizes observed, 10 microsatellites were selected for genotyping the 72 field-collected mosquitoes. By labeling primers that produced fragments in nonoverlapping size ranges with the same ßuorescent dye, we were able to combine amplification of these 10 microsatellites into two multiplexed PCR reactions (Table 3). PCR conditions and fragment analysis procedures were the same as described above. Arlequin version 3.5.1.2 (Excoffier and Lischer 2010) was used to calculate observed and expected heterozygosity and to test for deviations from Hardy–Weinberg equilibrium(1,000,000 Markov chains). Micro-Checker (van Oosterhout et al. 2004) was used to assess the presence of null alleles. Parentage exclusion probabilities were calculated according to the formula by Jamieson and Taylor (1997) (Table 3).
Table 3.
Ae. aegypti microsatellites included in multiplexed PCR for parentage assignment
| Locus | Multiplex PCR group |
Label | F and R primer conc. (μmol/L) |
Observed size range (bp)a |
No. alleles | HO | HE | HW P value | PEb |
|---|---|---|---|---|---|---|---|---|---|
| AG5c | A | VIC | 0.200 | 151–161 | 6 | 0.789 | 0.788 | 0.886 | 0.752 |
| AC1c | B | 6FAM | 0.200 | 176–191 | 4 | 0.652 | 0.601 | 0.827 | 0.501 |
| A10d | A | NED | 0.125 | 231–238 | 4 | 0.408 | 0.436 | 0.736 | 0.391 |
| H08d | A | VIC | 0.150 | 199–202 | 2 | 0.507 | 0.487 | 0.808 | 0.276 |
| AG2c | B | PET | 0.200 | 102–136 | 3 | 0.380 | 0.421 | 0.594 | 0.343 |
| AC5c | B | VIC | 0.200 | 131–145 | 5 | 0.606 | 0.742 | 0.042 | 0.676 |
| AT1c | A | NED | 0.100 | 141–155 | 6 | 0.609 | 0.771 | 0.011 | 0.728 |
| B07d | A | PET | 0.150 | 164–180 | 5 | 0.262 | 0.482 | <0.001 | 0.478 |
| AG3c | B | 6FAM | 0.200 | 147–153 | 4 | 0.088 | 0.534 | <0.001 | 0.471 |
| AG1c | A | PET | 0.300 | 102–108 | 4 | e | e | e | e |
Number of alleles and size ranges observed in 72 individuals collected across Iquitos, Peru. Loci deviating from Hardy–Weinberg equilibrium after correction for multiple tests are in bold (P value < 0.006).
Relative to GS600 LIZ size standard (Life Technology Corp.).
Exclusion probability for each locus (PEM3 from Jamieson and Taylor 1997): .
Not analyzed because of sex linkage.
Parentage Analysis Validation
A subset of the fieldcollected Ae. aegypti (20 individuals collected in 2010) were used to rear 10 test families for parentage analysis. At UCD, eggs were hatched and larvae reared according to the protocol by Styer et al. (2007). Throughout the rearing process, mosquitoes were separated by collection house. Pupae were identified to sex based on size and morphology (Christophers 1960). Ten male-female pairs of pupae were transferred to small plastic cups (4.5 × 3.5 cm height) and placed inside individual one pint, mesh-covered paper cartons. The paired individuals originated from the same house, but each mosquito pair was collected from a different part of the city. This was done to maximize our ability to resolve family relationships by increasing the number of shared alleles within a family while decreasing the number of shared alleles between families. Females were offered human blood daily and no sugar was provided. Mosquito pairs were allowed to mate for 5 d. Plastic cups were filled to 50% capacity with water and lined with white filter paper to collect eggs. After each female laid her first egg batch, parents were killed and preserved as described above. F1 eggs, separated according to family, were hatched and larvae reared in the laboratory as previously described. After adult emergence, 10 female and 10 male offspring were randomly selected from each family for parentage analysis. Genomic DNA was extracted from all selected mosquitoes and PCR amplified at the 10 microsatellite loci as described above. Exclusion-based parentage analysis was performed using PROBMAX version 1.2 (Danzmann 1997). During our analysis, parental genotypes were pooled together as 10 parental pairs and the 200 F1 genotypes were treated as potential offspring from any of the parental pairs.
Results
Nonlethal DNA Sampling
We successfully amplified genomic DNA extracted from each of the 20 hind legs, but not from any of the pupal exuviae (Fig. 1). No signs of “death stress oviposition” were observed when these females later became gravid. Data for female behavioral and life history traits were limited to the first five gonotrophic cycles because few females survived beyond that point (Table 1). After Bonferroni correction for multiple tests (P value cutoff = 0.01), females in the three treatments (chilled with leg removed, chilled control with no leg removal, and non-chilled control with no leg removal) showed no difference with respect to longevity (F(2, 32) = 0.01; P = 0.995), blood feeding frequency (F(2, 32) = 0.26; P = 0.772), fecundity (F(2, 31) = 1.30; P = 0.287), gonotrophic cycle length (F(2, 31) = 1.18; P = 0.322), or the number of oviposition sites used (F(2, 31) = 0.43; P = 0.656). Trial number had an effect on fecundity (F(1, 31) = 10.73; P = 0.003), with more eggs laid per female per gonotrophic cycle during the first trial (mean eggs = 98.5 ± 6.6 SE) than the second (mean eggs = 79.2 ± 7.8 SE). Trial number did not impact longevity (F(1, 32) = 4.89; P = 0.034), blood feeding frequency (F(1, 32) = 1.06; P = 0.310), gonotrophic cycle length (F(1, 31) = 0.22; P = 0.645), or the number of oviposition sites used (F(1, 31) = 1.78; P = 0.192). Gonotrophic cycle number did not affect fecundity (F(4, 109) = 2.44; P = 0.051), gonotrophic cycle length (F(4, 109) = 0.34; P = 0.848), or the number of oviposition containers used (F(4, 109) = 2.43; P = 0.052). No significant female treatment by trial interactions were observed during any of the ANOVA analyses (Table 1S, supplemental material available online only).
Fig. 1.
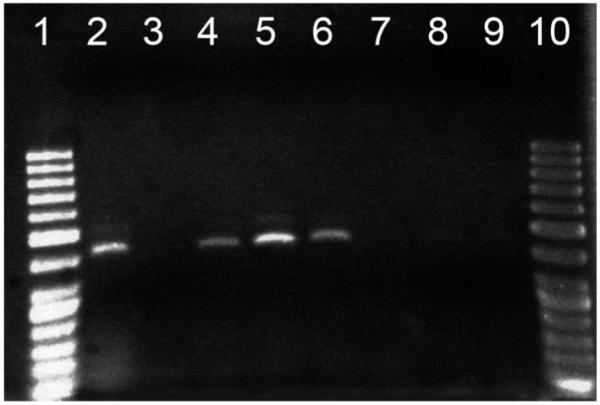
PCR products at the Carboxypeptidase A locus. Lanes: 1 (50 bp DNA ladder); 2 (positive control, DNA extracted from Ae. aegypti whole body); 3 (negative control, water); 4–6 (DNA extracted from single Ae. aegypti legs); 7–9 (DNA extracted from Ae. aegypti pupal exuviae), 10 (50 bp ladder).
For females housed as a group in cages (10 females per cage), the daily number of eggs laid per cage (mean 186 ± 13 SE) was not affected by treatment (F(2, 2) = 0.27; P = 0.790) or trial (F(1, 2) < 0.01; P = 0.956). No significant differences were observed for egg hatch rates among the different treatments during trial one (P = 0.625) or trial two (P = 0.776). Mean egg hatch rates were 94% (±1.7% SD) and 99% (±1.0% SD) during the first and second trials, respectively.
Parentage Analysis Using Microsatellites
From 18 microsatellites screened (Table 2), 10 of the most polymorphic loci were selected for parentage analysis, and genotyping was carried out for the 72 field-collected mosquitoes (Table 3). Among these individuals, two to six alleles were found per locus. Observed and expected heterozygosity for each locus is reported in Table 3. After Bonferroni correction for multiple tests, significant deviations from Hardy–Weinberg proportions because of heterozygote deficiency were detected at loci AG3 and B07. Microchecker identified evidence of null alleles at these two loci, as well as at locus AT1. When the Bonferroni correction was not applied (a more conservative approach), heterozygote deficiency was also noted for locus AC5, suggesting that in the Iquitos population, a null allele may exist at this locus as well.
After examination of parent and offspring genotypes from the 10 test families, the AG1 locus appeared to be sex-linked in some families, but not in others. Because of its unreliable behavior, AG1was eliminated from our subsequent analyses. Exclusion probabilities, reported for each locus in Table 3, describe the likelihood that a match at that locus is because of a true parent-offspring relationship rather than chance (Jamieson and Taylor 1997). When both maternal and paternal genotypes are known, the combined exclusion probability across all nine loci was 0.999 for Ae. aegypti from the Iquitos population. In other words, of 1,000 offspring matched to their parents, only one false match because of chance would be expected. Because the occurrence of null alleles makes parent-offspring relationships less clear, this probability may be an overestimate. We calculated overall exclusion probabilities for microsatellite panels in which one locus was progressively removed (Table 4), beginning with those exhibiting null alleles among our 10 test families (see below). When AG3, AT1, B07, and AG2 were removed, the overall exclusion probability remained high (0.982). Loci with null alleles still contributed some information toward parentage assignment, so we expect the true exclusion probability to lie between these two values.
Table 4.
Combined exclusion probabilities of microsatellites for parentage analysis
| No. markers | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 |
|---|---|---|---|---|---|---|---|---|---|
| Total PE | 0.999 | 0.998 | 0.997 | 0.988 | 0.982 | 0.946 | 0.925 | 0.876 | 0.752 |
| AG5 | AG5 | AG5 | AG5 | AG5 | AG5 | AG5 | AG5 | AG5 | |
| AC1 | AC1 | AC1 | AC1 | AC1 | AC1 | AC1 | AC1 | ||
| A10 | A10 | A10 | A10 | A10 | A10 | A10 | |||
| H08 | H08 | H08 | H08 | H08 | H08 | ||||
| AC5 | AC5 | AC5 | AC5 | AC5 | |||||
| AG2 | AG2 | AG2 | AG2 | ||||||
| AT1 | AT1 | AT1 | |||||||
| B07 | B07 | ||||||||
| AG3 |
Each column represents one panel of microsatellite markers. Exclusion probabilities decrease as one locus is successively removed. Loci showing evidence of null alleles (AG3, B07, AT1, and AG2) were removed first. Subsequently, loci were removed in order (lowest to highest) of locus-specific exclusion probabilities (Table 3).
Total exclusion probably across k loci (Jamieson and Taylor 1997): PE = 1 – (1 – P1) (1 – P2) (1 – P3) … (1 – Pk).
Of the 200 offspring tested, correct parentage was automatically assigned to 149 individuals. Parentage results were checked manually to confirm that assignments were correct. For 42 other individuals, a complete match with parents was identified after manual inspection of genotypes and consideration of null alleles. Among all 10 test families, null alleles were observed across four microsatellite loci (AG2, AG3, AT1, and B07). On an individual level, mosquitoes exhibited a maximum of two loci with null alleles.
For the remaining nine mosquitoes (4.5%), a mismatch at a single allele prevented 100% agreement between offspring and parental genotypes. We suspect that these mismatches were because of genotyping error or mutation. We compensated by setting PROBMAX to allow for one mismatch between parents and offspring. Accepting a threshold of (typically one or two) mismatches is common during parentage exclusion analyses (Wang 2010). By accounting for null alleles and allowing a single mismatch, we were able to correctly assign all offspring to their true parents.
Discussion
To track the egg-laying behavior of free-ranging Ae. aegypti, we developed a protocol involving nonlethal DNA sampling and parentage analysis using microsatellite markers. This protocol requires mating known pairs of Ae. aegypti in the laboratory, removing a single hind leg from females before release, and genotyping parents and field-collected offspring. Removal of a single hind leg from adult females provided sufficient genomic DNA for PCR amplification and did not significantly alter the female behavioral or life history traits that we examined in the laboratory. In test families, we were able to assign 200 Ae. aegypti offspring to the correct parental pairs using nine polymorphic microsatellite markers and an exclusion-based software program.
We were unable to amplify genomic DNA from Ae. aegypti pupal exuviae. The extractions were tested using a PCR protocol known to be sensitive and consistent for Ae. aegypti genomic DNA. Therefore, lack of PCR amplification was likely because of low DNA quantity or quality. Interestingly, Dhananjeyan et al. (2010) reported PCR amplification of DNA from Ae. aegypti pupal exuviae. Their result is surprising because insect exoskeletons are composed of noncellular glycoproteins and contain no nucleic acid. The authors speculated that epithelial cells from the foregut and hindgut lining shed with the exoskeleton (Bertholf 1925) provided a source of DNA. Dhananjeyan et al. (2010) were unsuccessful, however, when using samples older than 24 h or collected from the field because the small number of epithelial cells may have degraded quickly. Although noninvasive genetic sampling techniques, such as using pupal exuviae, minimize chances of damaging the insect, these techniques frequently have been limited by poor DNA yield (Taberlet et al. 1999). We found the mosquito leg to be a reliable source of DNA, even when legs had been preserved in ethanol for over a month.
In gravid Aedes species, immediate forced egg-laying, or “death stress oviposition,” has been observed in response to physical and chemical insult (DeCoursey and Webster 1952, Wallis and DeBishop 1957, Chadee and Ritchie 2010). The methods we used to immobilize teneral females and remove a hind leg did not significantly affect survival, blood feeding, or oviposition behavior. We purposefully carried out this procedure on teneral females so they could recover before blood feeding and ovipositing. It is possible that more changes in egg-laying behavior would be seen if females were manipulated while gravid. Differences were observed in female fecundity between trials, with females laying more eggs per gonotrophic cycle during the first trial than the second (Table 1). Differences could be because of length of Ae. aegypti egg storage (eggs used in trial two were stored 2 mo longer than those used in trial 1) or small ßuctuations in temperature and/or humidity in our insectary. A limitation of our study was the relatively small sample size used. The time-intensive nature of feeding individually housed females and checking five oviposition sites per female limited our ability to conduct more replicates. Our data, however, are consistent with previous studies that detected no obvious detrimental effects of leg sampling on insects (Fincke and Hadrys 2001, Starks and Peters 2002, Holehouse et al. 2003). Despite limitations in our study design, we are confident that chilling and leg removal from teneral females did not induce a death stress response later in life. In-depth studies would be needed to determine whether these procedures have effects on other mosquito behaviors.
In previous genetic studies of Ae. aegypti behavior, markers such as RAPDs (Apostol et al. 1994) and RFLPs (Colton et al. 2003) were used to assign sibling relationships among field-collected larvae based on population allele frequencies. Although population genetic-based estimates of relatedness have the advantage of requiring no parental information, they can generate high misclassification rates. Because RAPDs segregate as dominant loci, such that individuals homozygous for the dominant allele cannot be distinguished from heterozygotes, they provide incomplete genotypic information (Lynch and Milligan 1994, Yan et al. 1999). Using RAPDs requires several important assumptions regarding Mendelian inheritance, identity in state of null (recessive) alleles, and Hardy– Weinberg equilibrium (Apostol et al. 1996, Yan et al. 1999). Any unmet assumptions will preclude accurate estimation of population allele frequencies, ultimately biasing calculations of relatedness (Yan et al. 1999). Apostol et al. (1993) tested the reliability of RAPD markers in resolving sibling families and found an average error rate of 14.4%. Codominant restriction fragment-length polymorphism markers are expected to yield more accurate estimates of population allele frequencies (Yan et al. 1999), but the study by Colton et al. (2003) revealed a 20.5% error rate in assignment of individuals to sibling groups. In particular, the statistical method chosen for estimating relatedness was unable to discriminate among closely related groups (Colton et al. 2003). A major drawback of restriction fragment-length polymorphism analysis is that it is not PCR-based and requires large starting amounts of DNA, resulting in a limited number of loci that can be analyzed per individual mosquito.
The exclusion-based parentage analysis technique we describe herein does not depend on estimates of population allele frequencies (Jones et al. 2010). Instead, it is based on the principles of Mendelian inheritance, in which parent-offspring relationships are rejected if incompatibilities exist between candidate parents and offspring. When both genotypes of parental pairs are known, exclusion-based parentage analysis can represent an extremely powerful method to identify candidate offspring (Jamieson and Taylor 1997). If only a single parental genotype is known (e.g., paternity analysis), the exclusion power of these markers is reduced. Details of their resolving power for paternity will depend on whether fathers can be narrowed down to a specific pool of candidates (see Jamieson and Taylor 1997 for formulae). We expect that a larger number of microsatellite loci would be required for a paternity study.
With knowledge of both parental genotypes at nine polymorphic microsatellite loci, we were able to achieve complete exclusion of all but the true off-spring in our 10 test families. In other words, no ambiguities in parentage or false matches were observed. Because of the presence of null alleles at four loci, however, we found it necessary to relax our exclusion criteria by taking into account offspring genotypes that could result from parental null alleles at some loci appearing to be homozygous. We also set our matching criteria to accept one mismatch (because of mutation or genotyping error). Using this approach, we estimate that our power to exclude false parentage matches lies between 99.9 and 98.2%. Using the locus-specific exclusion probabilities listed in Table 3 and the formula published by Jamieson and Taylor (1997) (see Table 4), the total exclusion probability of any combination of the microsatellites we used can be estimated for Ae. aegypti from Iquitos, Peru. The design of our subsequent field studies, in which male–female pairs are mated in the laboratory and then the females released, allows us to know both the maternal and paternal genotypes for each set of parents. Our results support the use of this approach for genotyping large numbers of field-collected offspring to study oviposition behavior, components of reproductive success, dispersal patterns, and gene ßow among free-ranging Ae. aegypti in natural, field settings.
Supplementary Material
Acknowledgments
The authors thank H. Astete for providing field-collected Ae. aegypti eggs from Iquitos, Peru; R. Hemme and D. Severson for suggestions on which microsatellites to use; K. Glunt for help rearing mosquitoes; I. Baseer for laboratory assistance; and W. Reisen and A. Cornel for reviewing earlier versions of the manuscript. This research was supported by UC Davis Jastro–Shields, Hazeltine, and McBeth awards to JW; grants from the Innovative Vector Control Consortium, Regents of the University of California from the Foundationfor the National Institutes of Health through the Grand Challenges in Global Health Initiative, and National Institutes of Health grant R01 AI069341 to TWS.
Footnotes
The views expressed in this article are those of the author and do not necessarily reßect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
References Cited
- Apostol BL, Black WC, Miller BR, Reiter P, Beaty BJ. Estimation of the number of full sibling families at an oviposition site using RAPD-PCR markers: applications to the mosquito Aedes aegypti. Theor. Appl. Genet. 1993;86:991–1000. doi: 10.1007/BF00211052. [DOI] [PubMed] [Google Scholar]
- Apostol BL, Black WC, Reiter P, Miller BR. Use of randomly amplified polymorphic DNA amplified by polymerase chain reaction markers to estimate the number of Aedes aegypti families at oviposition sites in San Juan, Puerto Rico. Am. J. Trop Med. Hyg. 1994;51:89–97. doi: 10.4269/ajtmh.1994.51.89. [DOI] [PubMed] [Google Scholar]
- Apostol BL, Black WC, Reiter P, Miller BR. Population genetics with RAPD-PCR markers: the breeding structure of Aedes aegypti in Puerto Rico. Heredity. 1996;76:325–334. doi: 10.1038/hdy.1996.50. [DOI] [PubMed] [Google Scholar]
- Bertholf LM. The moults of the honeybee. J. Econ. Entomol. 1925;18:380–384. [Google Scholar]
- Black WC, DuTeau NM. RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton J, Beard CB, Louis C, editors. The molecular biology of disease vectors: a methods manual. Chapman & Hall; New York: 1997. pp. 361–171. [Google Scholar]
- Chadee DD, Ritchie SA. Oviposition behaviour and parity rates of Aedes aegypti collected in sticky traps in Trinidad, West Indies. Acta Trop. 2010;116:212–216. doi: 10.1016/j.actatropica.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Chambers EW, Meece JK, McGowan JA, Lovin DD, Hemme RR, Chadee DD, McAbee K, Brown SE, Knudson DL, Severson DW. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J. Hered. 2007;98:202–210. doi: 10.1093/jhered/esm015. [DOI] [PubMed] [Google Scholar]
- Christophers SR. Aedes aegytpi (L.) the yellow fever mosquito. Cambridge University Press; London, United Kingdom: 1960. [Google Scholar]
- Colton YM, Chadee DD, Severson DW. Natural skip oviposition of the mosquito Aedes aegypti indicated by codominant genetic markers. Med. Vet. Entomol. 2003;17:195–204. doi: 10.1046/j.1365-2915.2003.00424.x. [DOI] [PubMed] [Google Scholar]
- Danzmann RG. PROBMAX: a computer program for assigning unknown parentage in pedigree analysis from known genotypic pools of parents and progeny. J. Hered. 1997;88:333. [Google Scholar]
- DeCoursey JD, Webster AP. Effect of insecticides and other substances on oviposition by Aedes sollicitans. J. Econ. Entomol. 1952;45:1030–1034. [Google Scholar]
- Dhananjeyan KJ, Paramasivan R, Tewari SC, Rajendran R, Thenmozhi V, Leo SVJ, Venkatesh A, Tyagi BK. Molecular identification of mosquito vectors using genomic DNA isolated from eggshells, larval and pupal exuvium. Trop. Biomed. 2010;27:47–53. [PubMed] [Google Scholar]
- Edman JD, Strickman D, Kittayapong P, Scott TW. Female Aedes aegypti (Diptera: Culicidae) in Thailand rarely feed on sugar. J. Med. Entomol. 1992;29:1035–1038. doi: 10.1093/jmedent/29.6.1035. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molec. Ecol. Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fincke OM, Hadrys H. Unpredictable offspring survivorship in the damselßy, Megaloprepus coerulatus, shapes parental behavior, constrains sexual selection, and challenges traditional fitness estimates. Evolution. 2001;55:762–772. doi: 10.1554/0014-3820(2001)055[0762:uositd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Fulton RE, Salasek ML, DuTeau NM, Black WC, V. I. SSCP analysis of cDNA markers provides a dense linkage map of the Aedes aegypti genome. Genetics. 2001;158:715–726. doi: 10.1093/genetics/158.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett JD, Roman EA, Phillips V. Erratic hatching in Aedes eggs: a new interpretation. Proc. R. Soc. London B, Biol. Sci. 1977;196:223–232. doi: 10.1098/rspb.1977.0038. [DOI] [PubMed] [Google Scholar]
- Grech K, Maung LA, Read AF. The effect of parental rearing conditions on offspring life history in Anopheles stephensi. Malaria J. 2007;6:130. doi: 10.1186/1475-2875-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm CA, Aggarwal D, Landis DA. Evaluating the impact of non-lethal DNA sampling on two butterßies, Vanessa cardui and Satyrodes eurydice. J. Insect Conserv. 2010;14:11–18. [Google Scholar]
- Holehouse KA, Hammond RL, Bourke AFG. Non-lethal sampling of DNA from bumble bees for conservation genetics. Insectes Soc. 2003;50:277–285. [Google Scholar]
- Jamieson A, Taylor CS. Comparisons of three probability formulae for parentage exclusion. Anim. Genet. 1997;28:397–400. doi: 10.1111/j.1365-2052.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- Jones AG, Ardren WR. Methods of parentage analysis in natural populations. Molec. Ecol. 2003;12:2511–2523. doi: 10.1046/j.1365-294x.2003.01928.x. [DOI] [PubMed] [Google Scholar]
- Jones AG, Small CM, Paczolt KA, Ratterman NL. A practical guide to methods of parentage analysis. Molec. Ecol. Res. 2010;10:6–30. doi: 10.1111/j.1755-0998.2009.02778.x. [DOI] [PubMed] [Google Scholar]
- Krebs JR, Davies NB. Behavioral ecology: an evolutionary approach. Sinauer; Sunderland, MA: 1978. [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Milligan BG. Analysis of population genetic-structure with RAPD markers. Molec. Ecol. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi ZY, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. version 2.8.1 R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Roth LM. A study of mosquito behavior: an experimental laboratory study of the sexual behavior of Aedes aegypti (L.) Am. Midland Naturalist. 1948;40:265–352. [Google Scholar]
- Ruiz F, Quinones ML, Erazo HF, Calle DA, Alzate JF, Linton YM. Molecular differentiation of Anopheles (Nyssorhynchus) benarrochi and An. (N.) oswaldoi from Southern Colombia. Memorias Do Instituto Oswaldo Cruz. 2005;100:155–160. doi: 10.1590/s0074-02762005000200008. [DOI] [PubMed] [Google Scholar]
- SAS Institute . SAS user’s guide. version 9.2 SAS Institute; Cary, NC: 2008. [Google Scholar]
- Scott TW, Amerasinghe PH, Morrison AC, Lorenz LH, Clark GG, Strickman D, Kittayapong P, Edman JD. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J. Med. Entomol. 2000;37:89–101. doi: 10.1603/0022-2585-37.1.89. [DOI] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Slotman MA, Kelly NB, Harrington LC, Kitthawee S, Jones JW, Scott TW, Caccone A, Powell JR. Polymorphic microsatellite markers for studies of Aedes aegypti (Diptera: Culicidae), the vector of dengue and yellow fever. Molec. Ecol. Notes. 2007;7:168–171. [Google Scholar]
- Starks PT, Peters JM. Semi-nondestructive genetic sampling from live eusocial wasps, Polistes dominulus and Polistes fuscatus. Insectes Soc. 2002;49:20–22. [Google Scholar]
- Stimac JL. History and relevance of behavioral ecology in models of insect population dynamics. Fla. Entomol. 1982;65:9–16. [Google Scholar]
- Styer LM, Minnick SL, Sun AK, Scott TW. Mortality and reproductive dynamics of Aedes aegypti (Diptera: Culicidae) fed human blood. Vector-borne Zoon. Dis. 2007;7:86–98. doi: 10.1089/vbz.2007.0216. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Waits L, Luikart G. Noninvasive genetic sampling: look before you leap. Trends Ecol. Evol. 1999;14:323–327. doi: 10.1016/s0169-5347(99)01637-7. [DOI] [PubMed] [Google Scholar]
- Tentelier C, Guillemaud T, Ferry S, Fauvergue X. Microsatellite-based parentage analysis reveals non-ideal free distribution in a parasitoid population. Molec. Ecol. 2008;17:2300–2309. doi: 10.1111/j.1365-294X.2008.03743.x. [DOI] [PubMed] [Google Scholar]
- Van Handel E, Edman JD, Day JF, Scott TW, Clark GG, Reiter P, Lynn HC. Plant sugar, glycogen, and lipid assay of Aedes aegypti collected in urban Puerto Rico and rural Florida. J. Am. Mosq. Control Assoc. 1994;10:149–153. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley PF. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molec. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Vila M, Auger-Rozenberg MA, Goussard F, Lopez-Vaamonde C. Effect of non-lethal sampling on lifehistory traits of the protected moth Graellsia isabelae (Lepidoptera: Saturniidae) Ecol. Entomol. 2009;34:356–363. [Google Scholar]
- Wallis RC, DeBishop RJ. Death-stress oviposition by Aedes canadensis. J. Econ. Entomol. 1957;50:112. [Google Scholar]
- Wang JL. Effects of genotyping errors on parentage exclusion analysis. Molec. Ecol. 2010;19:5061–5078. doi: 10.1111/j.1365-294X.2010.04865.x. [DOI] [PubMed] [Google Scholar]
- Watts PC, Thompson DJ, Daguet C, Kemp SJ. Exuviae as a reliable source of DNA for population-genetic analysis of odonates. Odonatologica. 2005;34:183–187. [Google Scholar]
- Wong J, Astete H, Morrison AC, Scott TW. Sampling considerations for designing Aedes aegypti (Diptera: Culicidae) oviposition studies in Iquitos, Peru: substrate preference, diurnal periodicity, and gonotrophic cycle length. J. Med. Entomol. 2011;48:45–52. doi: 10.1603/me10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Tripet F, Rasgon JL, Lanzaro GC, Scott TW. SSCP analysis of scnDNA for genetic profiling of Aedes aegypti. Am. J. Trop. Med. Hyg. 2008;79:511–517. [PubMed] [Google Scholar]
- Yan G, Romero-Severson J, Walton M, Chadee DD, Severson DW. Population genetics of the yellow fever mosquito in Trinidad: comparisons of amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) markers. Molec. Ecol. 1999;8:951–963. doi: 10.1046/j.1365-294x.1999.00647.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


