Abstract
RUNX1 is a critical transcription factor during embryogenesis and neoplastic disease. To identify novel transcriptional targets of RUNX1 in the context of chromatin, we performed genome wide location analysis (ChIP-on-chip). Here we report that SERPINB13, a gene downregulated in head and neck cancers, is a novel RUNX1transcriptional target. RUNX1 binds the SERPINB13 promoter in chromatin to repress its transcription. Mutation of either RUNX1 binding site in the SERPINB13 promoter increased the activity of the promoter. Finally, overexpression of RUNX1 and concomitant decrease in SERPINB13 expression led to increased activity of cathepsin K, an enzyme inhibited by SERPINB13. These data demonstrate that RUNX1 is an important regulator of SERPINB13 and cathepsin K activity.
Keywords: RUNX, transcription, SERPINB13, chromatin immunoprecipitation
Introduction
RUNX1 is a transcription factor that binds to TGt/cGGT consensus site in gene regulatory elements [1; 2]. The last three nucleotides are critical for binding to RUNX1 [3] and 5 of the 6 nucleotides are important for RUNX proteins to bind the TCR enhancer [4]. RUNX1 is essential during development [5; 6] and conditional knockout of RUNX1 revealed that it is critical for the development of platelets and T-cells [7; 8].
RUNX1 is also involved in human malignancies. Sporadic and germline mutations and translocations in the RUNX1 gene are linked to leukemogenesis [9]. These alterations affect the DNA binding and transcriptional activities of RUNX1, indicating that RUNX1 transcriptional targets regulate proliferation and differentiation. Overexpression of RUNX1 in endometrial carcinoma has been reported [10] and in a subset of B-ALL patients and AML patients with trisomy 21 [reviewed by Blyth et al [11].
Consensus RUNX1 binding sites were confirmed by EMSA in several genes [12]. Reporter studies demonstrate that RUNX1 regulates transcription through these binding sites. In hematopoietic cells, it upregulates expression of M-CSFR [13], GM-CSF [14; 15], CD4 silencer [16], IL-3 [17], and BLK [18]. Little is known, however, about RUNX1 regulation of these genes the natural context of chromatin.
To identify novel transcriptional targets of RUNX1 in chromatin, we performed genome wide location or ChIP-on-chip. We identified novel RUNX1 genes including SERPINB13, an inhibitor of cathepsins K and L, which are associated with inflammation, survival, cancer development [19], and bone remodeling [20]. RUNX1 repressed SERPINB13 resulting in increased cathepsin K enzymatic activity. We propose that RUNX1 repression of SERPINB13 alters cathepsin activity in normal cells and perhaps in tumor cells.
Materials and Methods
Cell culture
HaCaT and K562 were maintained in RPMI 1640 medium (Invitrogen) and 293T cells were maintained in DMEM supplemented with 10% FBS (Omega), 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen).
Plasmids
pCAPP-HA-RUNX1b contains the HA-RUNX1b cDNA via SmaI. The pCAPP vector was created by digesting pCAGGS vector with HindIII and blunt-ending with T4 polymerase. The phosphoglycerate kinase promoter (PGK) regulates the puromycin resistance gene and was derived from the MSCVpac vector (Clontech).
MSCV-HA-RUNX1b-puro contains the HA-RUNX1b cDNA in the HpaI site of MSCV-puro (Clontech).
For control shRNA, primers used were: 5’-GATCCCCCCTCGAAGACATCGGCAGATTCAAGAGATCTGCCGATGTCTTCGAGGTTTTTA-3’ (top) and 5’-AGCTTAAAAACCTCGAAGACATCGGCAGATCTCTTGAATCTGCCGATGTCTTCGAGGGGG-3’ (bottom).
For RUNX1 shRNA, the primers used: 5’-GATCCCCACTTTCCAGTCGACTCTCATTCAAGAGATGAGAGTCGACTGGAAAGTTTTTTA-3’ (top) and 5’-AGCTTAAAAAACTTTCCAGTCGACTCTCATCTCTTGAATGAGAGTCGACTGGAAAGTGGG-3’ (bottom)
Oligos were inserted into pSuper.retro.puro (Oligoengine) via BglII-HindIII sites.
Wild-type SERPINB13 p romoter plasmids were provided by Dr. Clayman (MDACC) [21]. Mutagenesis of the wild-type promoter was performed with the primers (Quikchange XL, Stratagene):
For TGTGGT consensus site 1 mutation: 5'-GCTCTGTCCATCTGTTAGAAGTAAAGTCTGCAGTTAATGTATGTGTTCATCAGGC-3', 5’-GCCTGATGAACACATACATTAACTGCAGACTTTACTTCTAACAGATGGACAGAGC-3'
For the consensus TGCGGT site 2: 5' -TCCCAGCGTTGGCGGTACCTTGTAATCACCTGCTAGGATAAAAATTCTGATGCCTGG- 3', 5' - CCAGGCATCAGAATTTTTATCCTAGCAGGTGATTACAAGGTACCGCCAACGCTGGGA- 3'
The pRL-null (Promega) was co-transfected with reporter plasmids to normalize transfections with Renilla luciferase.
Generation of HA-RUNX1 K562 cells
Approximately 1 × 107 K562 cells were electroporated in 400 µl serum free media at 250V/950 µF with 50 µg of linearized pCAPP-HA-RUNX1b. Two days after electroporation, cells were selected in 4 µg/ml puromycin. Resistant cells were pooled and HA-RUNX1 expression was confirmed by immunoblotting with anti-HA antibody (Covance). Cell cycle analysis was performed as described [22].
Generation of retroviruses and HaCaT stable lines
Approximately 5 × 106 293T cells were co-transfected with 15 µg MSCV-puro vector (Clontech) or MSCV-HA-RUNX1b-puro and 10 µg of amphotrophic packaging pCL10A1 using calcium phosphate precipitation. Retrovirus was collected 48 hours after transfection and added to 5 × 105 HaCaT cells/well with 16 µg/ml polybrene (Sigma). Cells were centrifuged at 1400× g for 2.5 hours at 32°C followed by selection in 4 mg/ml puromycin.
For RUNX1 shRNA HaCaT pools, retroviruses were prepared from control vector pSuper (Oligoengine), pSuper-RUNX1 control shRNA, or pSuper-RUNX1shRNA. Reduction of endogenous RUNX1 was detected by immunoblotting with a RUNX1 specific antibody [23].
Chromatin Immunoprecipitation
Approximately 4 × 108 HA-RUNX1 K562 cells were crosslinked in PBS containing 1.1% formaldehyde for 10 minutes on ice followed by 0.125 M glycine treatment. Nuclei isolation was performed as described [24] and DNA was sonicated to an average length of 0.5 – 1 kb. Soluble chromatin was precleared with 100 µl sepharose beads (GE Healthcare) followed by dilution in 0.1% Triton X-100 and 0.1% sodium deoxycholate to 1 ml. Immunoprecipitations with 5 µg of anti-HA (Covance) or mouse IgG control (Sigma I5381) were performed at 4°C overnight. Immune complexes were obtained with 30 µl protein G sepharose (GE healthcare) including 1 µg/µl BSA and 1 µg/µl herring sperm DNA for 1 hour at 4°C. Immunoprecipitates were washed 7 times in 1 ml of RIPA buffer with 5 minute rotations between washes. A final wash with 1 ml of TE was performed. Crosslinks were reversed by incubation at 65°C overnight. DNA was purified by RNase and proteinase K, followed by phenol:chloroform extraction [24] and Qiaquick column (Qiagen).
Preparation of DNA for microarray
DNA from ChIP was used in a positive control PCR reaction for enrichment of human PU.1 URE [28]. PU.1 URE primer sequences were: forward 5’-GTTTCTCTGGGCCGCTGT-3’, reverse 5’-AGCTGCCCCTGTTTCCACATC-3’. DNA was prepared for microarray to the promoter microarray as described [25]. Statistical analysis was performed after scanning fluorescent microarray slides using GenePix Pro software (Molecular Devices). Putative target promoters were identified as enriched relative to total input (genomic DNA) with a P value of 0.001. Each promoter microarray was performed two independent times using independent ChIP.
SERPINB13 promoter PCR
Primers used for SERPINB13 promoter regions: Region 1 F: 5’-AAGCATTATGGCCGACTCAG-3’ Region 1R: 5’-AGGGCATATGGGTCATGTG-3’, Region 2F: 5’-GTGTGTGCGTGTGTGTGTG -3’ and Region 2R: 5’-CACCGCAGGTGATTACAATG-3’.
Approximately 0.1% of DNA from ChIP was used in amplification of each promoter region using quantitative real-time PCR reactions with (Platinum SYBR green QPCR, Invitrogen) and 0.5 µM of each primer. Duplicate reactions were analyzed by iCycler iQ (Bio-Rad Laboratories) with melt curve analysis. Products were analyzed by gel electrophoresis. Relative expression was calculated by correcting for Ct value differences between IgG control and HA immunoprecipitates. Promoter binding in IgG control was set to 1 and the relative binding of HA-RUNX1 is graphed.
Luciferase Reporter Assays
2 × 106 cells were electroporated with 12µg of DNA in 180 µl serum free RPMI at 250V/950 µF and assayed 24 hours later.
Ten µg of reporter plasmid were co-transfected with 1 µg of vector control or transcription factor indicated, and 0.2 µg of pRL-null. Electroporated cells were cultured for one day following transfection and then lysed for luciferase assays using the Dual Reporter Assay (Promega). Relative light units were measured using a luminometer (BD Monolight, BD Biosciences). Data from three independent experiments were averaged.
Quantitative RT-PCR
RNA was prepared using RNAzolB Reagent (TEL-TEST). cDNA was prepared (SuperScript II RT, Invitrogen) for quantitative real-time PCR reactions with Platinum SYBR green QPCR Supermix (Invitrogen) and 0.5µM of each primer. Duplicate reactions were analyzed using the iCycler iQ (Bio-Rad Laboratories) with melt curve analysis. Products were analyzed by agarose gel electrophoresis. For each gene analyzed, cDNA from at least three independent vector control and RUNX1 expressing pools were analyzed. Relative expression was calculated by correcting for Ct value differences for human GAPDH loading. Vector control was set to 1 and the relative fold change is graphed for SERPINB13 mRNA.
Primers used were hGAPDHF: 5’- GAGCTGAACGGGAAGCTCACTGG-3’, hGAPDHR: 5’- CAACTGTGAGGAGGGGAGATTCAG-3’, SERPINB13F: 5’- AGCCGATGAAAGTCGAAAGA-3’, SERPINB13R: 5’- CTGCAAGTCCTCCAGGAAAG- 3’.
Cathepsin activity assay
1 × 105 cells were plated on 22 mm square coverslips. 16–20 hours after plating, cathepsin substrates were added at 1× concentration (Cell Technology, CDK200-1, CDK300-1) to media for 45 minutes at 37°C, followed by Hoechst 33342 staining for 10 minutes at 37°C. Coverslips were washed twice in 1× PBS and mounted on glass slides. Fluorescence was detected using a BP 360–400 filter for Hoechst 33342 and BP 515–560 filter for the cathepsins, and images were acquired using a Spot Digital camera (Diagnostic Instruments). Multiple images were acquired from 3 independent vector control pools or HaCaT-HA-RUNX1 expressing pools.
Results
RUNX1 Chromatin IP and promoter microarray
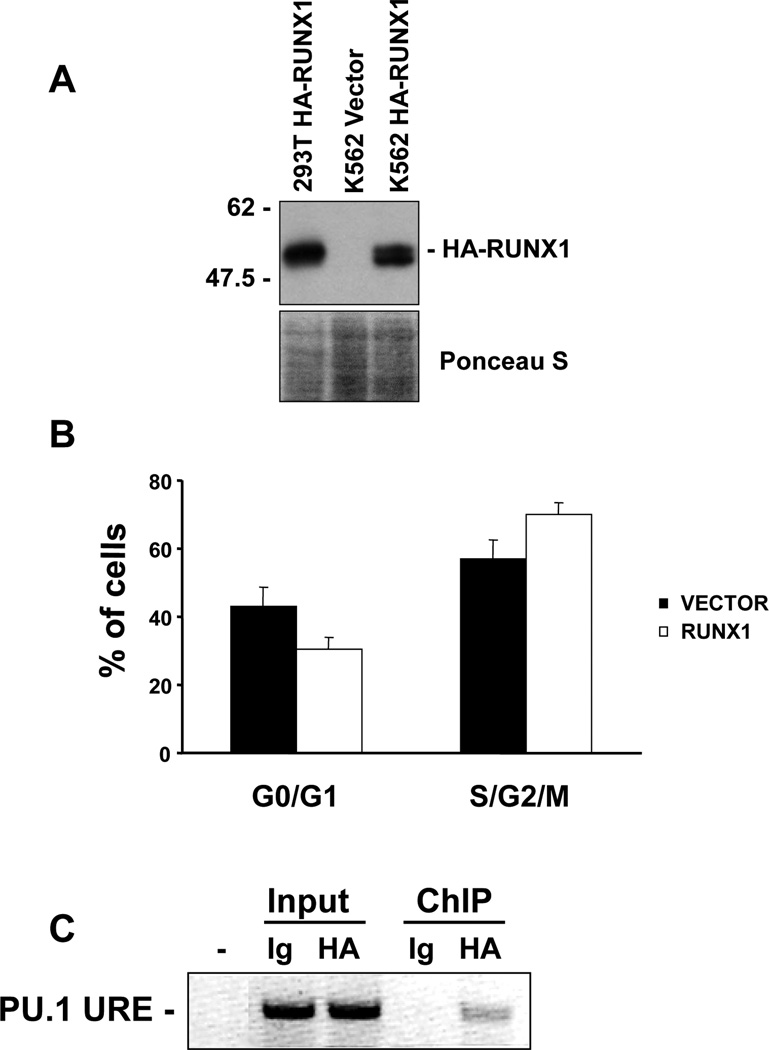
To identify novel RUNX1 target genes, we generated K562 cells expressing HA-tagged RUNX1 for chromatin immunoprecipitation assays (ChIP) (Fig. 1A). Expression of HA-RUNX1 in K562 cells did not inhibit cell proliferation compared to the vector control as reflected in the cell cycle distributions (Fig. 1B). The K562 HA-RUNX1 cells were expanded to perform ChIP with HA antibody or control mouse IgG. To validate the specificity of RUNX1 chromatin immunoprecipitation we analyzed binding to a known target gene, human PU.1 upstream regulatory region (URE) [25]. The PU.1 URE was enriched in HA-RUNX1 chromatin immunoprecipitates but not in the IgG control (Fig. 1C). This confirmed that the HA-RUNX1 ChIP was successful and this chromatin was used for hybridization to a promoter microarray.
Fig. 1.
Analysis of K562-HA-RUNX1b cells. (A) Immunoblot from K562 control pool or HA-RUNX1b expressing pool. 293T cells transfected with the HA-RUNX1b expression plasmid were used as a positive control. Ponceau S stain reflects relative protein loading. (B) Cell cycle distributions of vector control pools and HA-RUNX1b expressing pools. Cells were plated at the same density and stained with propidium iodide. The bar graph shows the average percentages of cells in G0/G1 or in S/G2/M from ModFit analysis. (C) ChIP of K562-HA-RUNX1b cells. Soluble chromatin was used for immunoprecipitation with either IgG control antibody (Ig) or HA antibody (HA). An inverted ethidium bromide gel is shown after PCR for the human PU.1 URE. Input lanes reflect the relative chromatin input for each immunoprecipitation and the ChIP lanes show the relative enrichment of the PU.1 URE in the HA-RUNX1 ChIP.
DNA that co-precipitated with either IgG or HA antibody was purified, amplified by ligation mediated PCR (LM-PCR) as described [24] and hybridized individually to human promoter microarrays containing approximately 8000 upstream regulatory regions. Following microarray analysis combined with statistics, genes enriched in HA-RUNX1 immunoprecipitates compared to genomic input with varying P values between 0 and .0088 were considered as putative RUNX1 target genes. Promoters enriched in control IgG micorarrays were considered false positives. Among the 27 novel RUNX1 target genes identified (Table 1), four genes were identified in two independent experiments, including ETF1, SERPINB13, HSD3B2, and cyclin A1.
Table 1. Putative RUNX1 targets from promoter microarray.
The results from two independent genome wide location analyses are listed. The gene symbols and the P values and fold enrichment in HA ChIP relative to genomic input are indicated.
| Gene | Accession | P value | Array |
|---|---|---|---|
| CCKAR | NM_000730 | 1.00E-06 | 2.61 |
| SLC22A5 | NM_003060 | 0 | 3.98 |
| * ETF1 | NM_004730 | 0 | 3.25 |
| SF3B2 | NM_006842 | 0 | 2.51 |
| HSP105B | NM_006644 | 7.97E-04 | 2.06 |
| CCL19 | NM_006274 | 0 | 2.48 |
| RCC1 | NM_001048194 | 0 | 4.61 |
| RB | NM_000321 | 0 | 2.60 |
| TNFRSF21 | NM_014453 | 3.30E-05 | 2.26 |
| * SERPINB13 | NM_012397 | 0 | 10.15 |
| IL18 | NM_001562 | 1.50E-05 | 2.03 |
| * HSD3B2 | NM_000198 | 3.38E-03 | 10.90 |
| GALK2 | NM_002044 | 4.31E-03 | 9.39 |
| GHSR | NM_004122 | 6.00E-03 | 6.74 |
| MUSK | NM_005592 | 7.55E-03 | 6.34 |
| * CCNA1 | NM_003914 | 6.94E-03 | 7.98 |
| ZNF225 | NM_013362 | 8.81E-03 | 5.89 |
| UQCRB | NM_006294 | 9.36E-04 | 2.67 |
| ADRB2 | NM_000024 | 3.36E-04 | 3.12 |
| RNF19 | NM_183419 | 3.40E-05 | 9.46 |
| HIST1H3I | NM_003533 | 9.34E-04 | 2.67 |
| GPC6 | NM_005708 | 6.28E-04 | 2.82 |
| PFTK1 | NM_012395 | 8.59E-04 | 2.72 |
| RBM8A | NM_005105 | 7.68E-04 | 2.74 |
| RPS6 | NM_001010 | 4.73E-04 | 2.93 |
| HIST1H2BC | NM_003526 | 9.59E-04 | 2.67 |
| REG1A | NM_002909 | 5.80E-04 | 2.85 |
Genes identified in two independent microarrays
Eight putative RUNX1 targets from Table 1 were selected for confirmation. As shown in Table 2, ChIP assays with promoter specific primers revealed that the RUNX1 bound the promoters of SERPINB13, RCC1, RNF19, HSP 105B, and ZNF225. In contrast, we were unable to detect association with several targets including RB1, TNFRSF21, and ETF1, which may be due to transient association of RUNX1 with these promoters or insufficient sensitivity of our ChIP assay.
Table 2. Genes confirmed by promoter ChIP.
Genes listed with GenBank Accession numbers, known functions, enrichment in microarray and whether confirmed to interact with HA-RUNX1 in ChIP (YES) or not detected (ND).
| Gene | Accession | Function | Microarray | ChIP |
|---|---|---|---|---|
| ETF1 | NM_004730 | Translation | 3.3 | ND |
| HSP105B | NM_006644 | Chaperone | 2.1 | YES |
| RB | NM_000321 | Tumor Suppressor | 2.6 | ND |
| RCC1 | NM_001048194 | Ran GNEF | 4.6 | YES |
| RNF19 | NM_183419 | E3 ligase | 9.5 | YES |
| SERPINB13 | NM_012397 | Cathepsin inhibitor | 10 | YES |
| TNFRSF21 | NM_014453 | Apoptosis | 2.3 | ND |
| ZNF225 | NM_013362 | Transcription Factor | 5.9 | YES |
SERPINB13 is a novel RUNX1 target gene
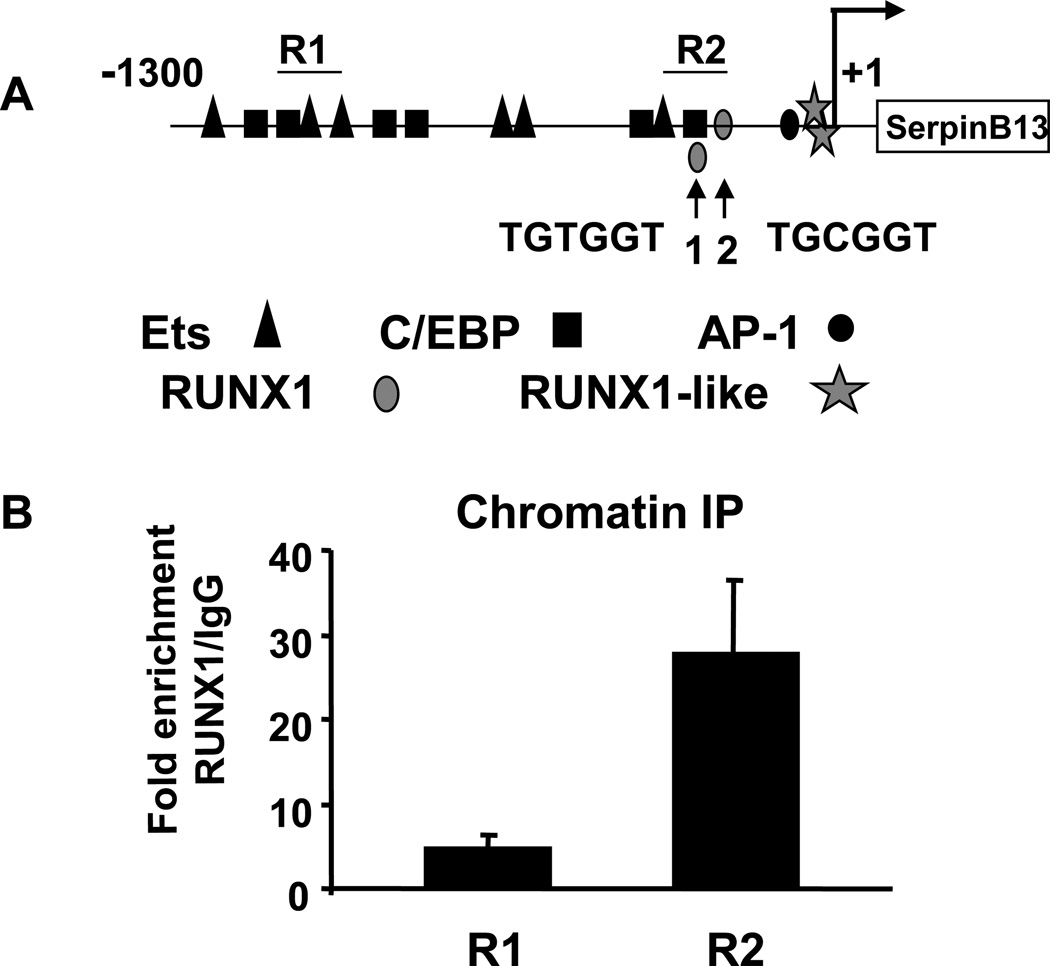
The SERPINB13 gene was selected for further study since it was one of the most highly enriched in RUNX1 ChIP. The SERPINB13 gene was identified as a gene modulated in squamous cell carcinomas and regulated by UV irradiation [26] [27]. The human SERPINB13 promoter [21] contains consensus RUNX, C/EBP and Ets binding sites (Fig. 2A). We hypothesized that RUNX1 may directly regulate the SERPINB13 promoter through a consensus TGt/cGGT binding site or instead indirectly occupy the promoter through known interacting partners Ets-1 [28; 29] or C/EBP [30].
Fig. 2.
RUNX1 binds the SERPINB13 promoter. (A) Schematic of the human SERPINB13 promoter with putative binding sites for C/EBP, Ets, RUNX1, and AP-1. Two different regions amplified in quantitative ChIP are indicated as R1 and R2. Two consensus RUNX1 sites are indicated and are located at −265 and −178 relative to the start site identified by Nakashima et al [24]. (B) ChIP of IgG and HA-RUNX1 from K562-HA-RUNX1 cells. Precipitated DNA was used in real-time PCR of the SERPINB13 promoter using primers that encompass either R1 or 2. The fold enrichment of DNA in HA-RUNX1 chromatin IP compared to control IgG was calculated from Ct values after correcting for input DNA.
To confirm promoter microarray results, chromatin immunoprecipitates of HA-RUNX1 or control IgG were used to amplify two different regions of the SERPINB13 promoter. The locations of promoter regions amplified are indicated as R1 and R2 in figure 2A. Region 1 does not contain consensus RUNX1 binding sites and showed an average of 5-fold enrichment in HA-RUNX1 ChIP compared to IgG control. Region 2 containing two consensus sites was enriched 28-fold in HA-RUNX1 immunoprecipitates compared to IgG (Fig. 2B). These data confirm that RUNX1 binds the human SERPINB13 promoter.
We next tested if RUNX1 regulates SERPINB13 transcription. RT-PCR analysis of SERPINB13 was performed in K562 vector control or HA-RUNX1 expressing cells that were used in genome wide location analysis. SERPINB13 mRNA levels in K562 cells were difficult to detect by RT-PCR (data not shown). We therefore examined RUNX1 regulation of SERPINB13 transcription in HaCaT cells, a cell line reported to express high levels of functional SERPINB13 [26; 27] which we confirmed by QPCR (data not shown).
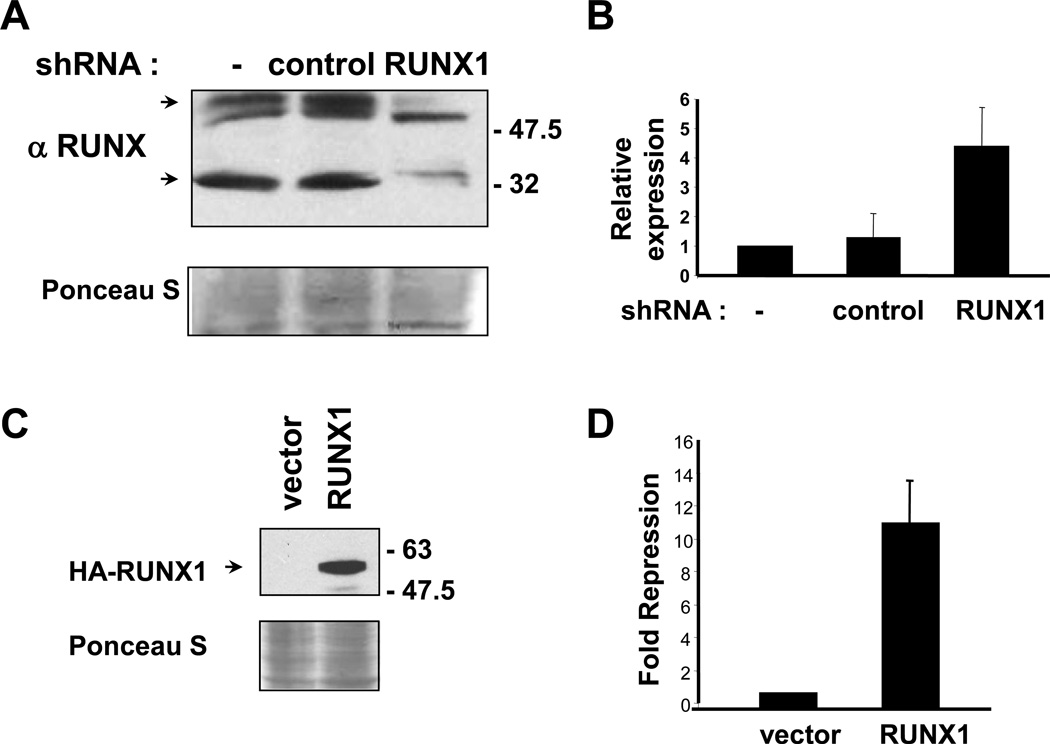
HaCaT cells express endogenous RUNX1 isoforms that were decreased by RUNX1 shRNA but not by a negative control shRNA (Fig. 3A). We examined three independent control pools or RUNX1 shRNA pools for changes in SERPINB13 expression by real-time RT-PCR. SERPINB13 mRNA levels increased in cells expressing reduced levels of RUNX1 (Fig. 3B), suggesting that RUNX1 may repress SERPINB13 expression. Stable overexpression of RUNX1 in HaCaT cells was also performed (Fig. 3C). Three independent control pools or HA-RUNX1 expressing pools were examined for SERPINB13 expression. RUNX1 overexpressing cells had significantly less SERPINB13 mRNA levels (11-fold repression) compared to vector control cells (Fig. 3D). These data indicate that RUNX1 is a negative regulator of SERPINB13 transcription.
Fig. 3.
RUNX1 regulates SERPINB13 expression. (A) Immunoblotting for RUNX1 from HaCaT cells stably expressing control or RUNX1 shRNA or vector control (−). Ponceau S staining shows the relative protein loading. Arrows indi cate RUNX1 isoforms decreased by shRNA. (B) Quantitative RT-PCR analysis of SERPINB13 mRNA using RNA prepared from 3 independent vector control (−) pools, or 3 control or RUNX1 shRNA pools. The relative expression was calculated after correcting for cDNA input using human GAPDH primers. Error bars indicate the differences for data averaged from 3 pools. (C) HA immunoblot from HaCaT stably expressing HA-RUNX1 or vector control. (D) Quantitative RT-PCR analysis of SERPINB13 mRNA using RNA prepared from 3 independent vector control pools or 3 HA-RUNX1 expressing pools. The fold repression was calculated after correcting for cDNA input as described in B.
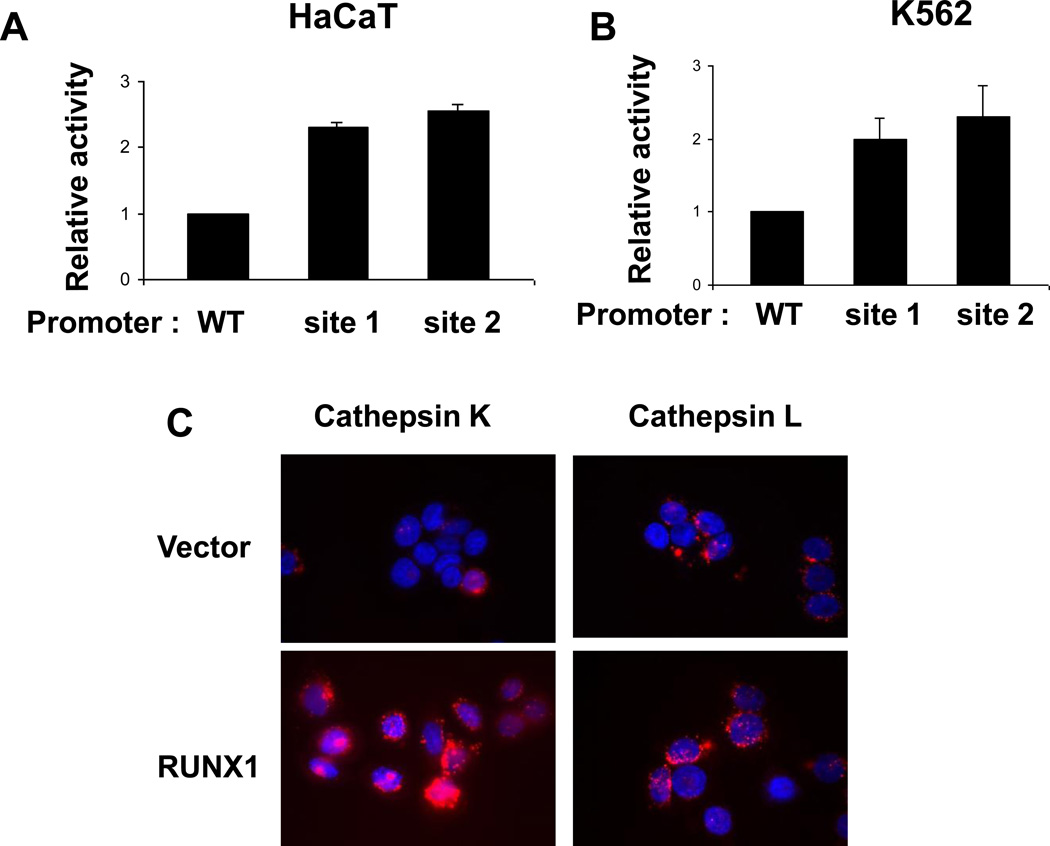
We next examined the mechanism of RUNX1 repression of SERPINB13 levels. As diagrammed in figure 2A, the SERPINB13 promoter contains two consensus RUNX1 sites, a TGTGGT site 1 and TGCGGT site 2 in Region 2. The consensus site 1 and the consensus site 2 were mutated to TGTTAG or TGCTAG, respectively, which eliminates binding of RUNX1 to target sites in EMSA analysis [3]. HaCaT cells were transiently transfected with either the wild-type SERPINB13 reporter (WT), the site 1, or the site 2 mutant reporters. Interestingly, mutation of the consensus site 1 or the site 2 consistently led to increased reporter activity in HaCaT cells (Fig. 4A) and in K562 cells (Fig. 4B). Additionally, transfection of HaCaT cells with the DNA biding domain of RUNX1 was sufficient to inhibit promoter activity (data not shown). These data suggest that RUNX1 represses SERPINB13 transcription by binding to consensus sites in the promoter.
Fig. 4.
Regulation of SERPINB13 promoter activity. (A) Relative activities of the WT, RUNX1 site 1 mutant, or RUNX1 site 2 mutant SERPINB13 reporters in HaCaT cells. The activity of the WT promoter was set to 1. The average of three independent experiments is indicated and error bars show the standard deviations. (B) Same as A except the luciSferase activity was measured in K562 cells. (C) Micrographs of HaCaT vector control cells or HA-RUNX1 cells incubated with cathepsin K or cathepsin L substrates. Hoechst staining shows nuclei (blue) and red fluorescence in lysosomes reflects the cathepsin enzymatic activity.
RUNX1 upregulates cathepsin K activity
SERPINB13 protein inhibits lysosomal cathepsins K and L in vitro [31; 32]. Therefore, we tested whether repression of SERPINB13 by RUNX1 could increase cathepsin K or L enzymatic activity in cells. Control or RUNX1 expressing HaCaT cells from three independent pools were incubated with a cell permeable substrate specific for either cathepsin K or L. Following cleavage of the cathepsin substrate by active enzyme, fluorescence was visualized by microscopy. RUNX1 expressing cells contained higher cathepsin K activity compared to vector control cells (Fig. 4C), however no significant increase in cathepsin L activity was detected (Fig. 4C). These data further support that RUNX1 regulates SERPINB13 expression and enzymatic activity in cells.
Discussion
Cellular proliferation and differentiation involves the activity of transcription factors that activate or repress critical target genes. Knowledge of RUNX1 target gene regulation in the context of chromatin is lacking for many genes. With the combination of chromatin crosslinking of RUNX1 and the high throughput microarray technique, we have identified several new RUNX1 target genes and characterized one novel target gene, SERPINB13.
SERPINB13 is an inhibitor of lysosomal cathepsin enzymes K and L [31; 32] and is downregulated in head and neck cancers [21] suggesting that these tumors may have elevated activity of cathepsins K and L. Cathepsin K facilitates invasion of tumor cells due to degradation of the ECM and is expressed in breast [33] and prostate cancer [34], indicating that SERPINB13 may be repressed in some of these tumors. Future studies should explore if RUNX1 is overexpressed in these cancers, disrupting the ratio of SERPINB13 and cathepsin activity. The advantage of downregulating SERPINB13 in tumors may be linked to its anti-angiogenic activity [35].
We demonstrate for the first time that reduction of SERPINB13 levels alters cathepsin K activity in cells, supporting previous in vitro enzymatic studies [31; 32]. Cathepsin K is the major proteinase secreted by osteoclasts during bone remodeling. Bone remodeling by osteoclasts that line the bone marrow facilitates mobilization of hematopoietic progenitor cells [36]. RUNX1 regulates the number of hematopoietic stem cells in the bone marrow, and cathepsin K knockout mice have alterations in the number of hematopoietic cells in the bone marrow [37]. Therefore, RUNX1 may influence cathepsin K activity in the bone marrow by altering SERPINB13 levels.
Cyclin A1 was also identified in this study. Overexpression of cyclin A1 leads to abnormal myelopoiesis and leukemia development [38], similar to mutation of the RUNX1 gene in patients with FPD or sporadic gene mutations. Therefore, it is possible that RUNX1 may negatively regulate cyclin A1 expression in hematopoietic cells. Other microarray targets worthy of further investigation include IL-18, which is important for angiogenesis [39] and immune modulation [40], two processes regulated by RUNX1 in vivo. Thus, genome wide location of RUNX1 has provided a platform to identify novel functions associated with differentiation and transformation.
Highlights.
Highlights of Boyapati et al BBRC submission 5–2011
Genome wide location analysis identifies novel RUNX1 targets
SERPINB13 is repressed by RUNX1
RUNX1 increases cathepsin K activity
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants CA096735 and CA104509 (DEZ), and an NRSA fellowship 5F32HL079900 (AB).
We wish to thank the Zhang Laboratory for discussions, Drs. Z. Li and C. Wu for microarray advice, Dr. Clayman for SERPINB13 plasmids, This is manuscript 18614 from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M, Ito Y, Shigesada K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J.Virol. 1990;64:4808–4819. doi: 10.1128/jvi.64.10.4808-4819.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melnikova IN, Crute BE, Wang S, Speck NA. Sequence specificity of the core-binding factor. J.Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML- 1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol.Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornell A, Holm M, Grundstrom T. Purification of SEF1 proteins binding to transcriptional enhancer elements active in T lymphocytes. J.Biol.Chem. 1993;268:21946–21954. [PubMed] [Google Scholar]
- 5.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. PNAS. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 7.Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Hirai H, Ogawa S, Kurokawa M. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat.Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 8.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, kashi KA, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamagata T, Maki K, Mitani K. Runx1/AML1 in normal and abnormal hematopoiesis. Int.J Hematol. 2005;82:1–8. doi: 10.1532/IJH97.05075. [DOI] [PubMed] [Google Scholar]
- 10.Planaguma J, az-Fuertes M, Gil-Moreno A, Abal M, Monge M, Garcia A, Baro T, Thomson TM, Xercavins J, Alameda F, Reventos J. A differential gene expression profile reveals overexpression of RUNX1/AML1 in invasive endometrioid carcinoma. Cancer Res. 2004;64:8846–8853. doi: 10.1158/0008-5472.CAN-04-2066. [DOI] [PubMed] [Google Scholar]
- 11.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat.Rev.Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 12.Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DE, Fujioka K, Hetherington CJ, Shapiro LH, Chen HM, Look AT, Tenen DG. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1) Mol.Cell Biol. 1994;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank R, Zhang J, Uchida H, Meyers S, Hiebert SW, Nimer SD. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 15.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae SC, Lu J, Maruyama M, Zhang YW, Oka H, Arai N, Arai K. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor. PEBP2 Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 16.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 17.Uchida H, Zhang J, Nimer SD. AML1A and AML1B can transactivate the human IL-3 promoter. J.Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- 18.Libermann TA, Pan Z, Akbarali Y, Hetherington CJ, Boltax J, Yergeau DA, Zhang DE. AML1 (CBFalpha 2) Cooperates with B Cell-specific Activating Protein (BSAP/PAX5) in Activation of the B Cell-specific BLK Gene Promoter. J.Biol.Chem. 1999;274:24671–24676. doi: 10.1074/jbc.274.35.24671. [DOI] [PubMed] [Google Scholar]
- 19.Nomura T, Katunuma N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J.Med.Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- 20.Goto T, Yamaza T, Tanaka T. Cathepsins in the osteoclast. J.Electron Microsc.(Tokyo) 2003;52:551–558. doi: 10.1093/jmicro/52.6.551. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima T, Pak SC, Silverman GA, Spring PM, Frederick MJ, Clayman GL. Genomic cloning, mapping, structure and promoter analysis of HEADPIN, a serpin which is down-regulated in head and neck cancer cells. Biochim.Biophys.Acta. 2000;1492:441–446. doi: 10.1016/s0167-4781(00)00100-7. [DOI] [PubMed] [Google Scholar]
- 22.Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc.Natl.Acad.Sci.U.S.A. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, Zhang DE. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Van CS, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc.Natl.Acad.Sci.U.S.A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat.Genet. 2007 doi: 10.1038/ng.2007.7. Epub. [DOI] [PubMed] [Google Scholar]
- 26.Abts HF, Welss T, Mirmohammadsadegh A, Kohrer K, Michel G, Ruzicka T. Cloning and characterization of hurpin (protease inhibitor 13): A new skin-specific, UV- repressible serine proteinase inhibitor of the ovalbumin serpin family. J.Mol.Biol. 1999;293:29–39. doi: 10.1006/jmbi.1999.3159. [DOI] [PubMed] [Google Scholar]
- 27.Spring P, Nakashima T, Frederick M, Henderson Y, Clayman G. Identification and cDNA cloning of headpin, a novel differentially expressed serpin that maps to chromosome 18q. Biochem.Biophys.Res.Commun. 1999;264:299–304. doi: 10.1006/bbrc.1999.1453. [DOI] [PubMed] [Google Scholar]
- 28.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 29.Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999;18:1609–1620. doi: 10.1093/emboj/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang DE, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen HM, Hiebert SW, Tenen DG. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol.Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayakumar A, Kang Y, Frederick MJ, Pak SC, Henderson Y, Holton PR, Mitsudo K, Silverman GA, EL-Naggar AK, Bromme D, Clayman GL. Inhibition of the cysteine proteinases cathepsins K and L by the serpin headpin (SERPINB13): a kinetic analysis. Arch.Biochem.Biophys. 2003;409:367–374. doi: 10.1016/s0003-9861(02)00635-5. [DOI] [PubMed] [Google Scholar]
- 32.Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von MA, Ruzicka T, Bird PI, Abts HF. Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry. 2003;42:7381–7389. doi: 10.1021/bi027307q. [DOI] [PubMed] [Google Scholar]
- 33.Littlewood-Evans AJ, Bilbe G, Bowler WB, Farley D, Wlodarski B, Kokubo T, Inaoka T, Sloane J, Evans DB, Gallagher JA. The osteoclast-associated protease cathepsin K is expressed in human breast carcinoma. Cancer Res. 1997;57:5386–5390. [PubMed] [Google Scholar]
- 34.Brubaker KD, Vessella RL, True LD, Thomas R, Corey E. Cathepsin K mRNA and protein expression in prostate cancer progression. J.Bone Miner.Res. 2003;18:222–230. doi: 10.1359/jbmr.2003.18.2.222. [DOI] [PubMed] [Google Scholar]
- 35.Shellenberger TD, Mazumdar A, Henderson H, Briggs K, Wang M, Chattopadhyay C, Jayakumar A, Frederick M, Clayman GL. Headpin: a serpin with endogenous and exogenous suppression of angiogenesis. Cancer Res. 2005;65:11501–11509. doi: 10.1158/0008-5472.CAN-05-2262. [DOI] [PubMed] [Google Scholar]
- 36.Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, Elson A, Lapidot T. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat.Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 37.Gowen M, Lazner F, Dodds R, Kapadia R, Feild J, Tavaria M, Bertoncello I, Drake F, Zavarselk S, Tellis I, Hertzog P, Debouck C, Kola I. Cathepsin K knockout mice develop osteopetrosis due to a deficit in matrix degradation but not demineralization. J.Bone Miner.Res. 1999;14:1654–1663. doi: 10.1359/jbmr.1999.14.10.1654. [DOI] [PubMed] [Google Scholar]
- 38.Liao C, Wang XY, Wei HQ, Li SQ, Merghoub T, Pandolfi PP, Wolgemuth DJ. Altered myelopoiesis and the development of acute myeloid leukemia in transgenic mice overexpressing cyclin. A1 Proc.Natl.Acad.Sci.U.S.A. 2001;98:6853–6858. doi: 10.1073/pnas.121540098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim KE, Song H, Kim TS, Yoon D, Kim CW, Bang SI, Hur DY, Park H, Cho DH. Interleukin-18 is a critical factor for vascular endothelial growth factor-enhanced migration in human gastric cancer cell lines. Oncogene. 2006 doi: 10.1038/sj.onc.1209926. [DOI] [PubMed] [Google Scholar]
- 40.Muhl H, Pfeilschifter J. Interleukin-18 bioactivity: a novel target for immunopharmacological anti-inflammatory intervention. Eur.J.Pharmacol. 2004;500:63–71. doi: 10.1016/j.ejphar.2004.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.