SUMMARY
Reversal of promoter DNA hypermethylation and associated gene silencing is an attractive cancer therapy approach. The DNA methylation inhibitors decitabine and azacitidine are efficacious for hematological neoplasms at lower, less toxic, doses. Experimentally, high doses induce rapid DNA damage and cytotoxicity, which do not explain the prolonged response observed in patients. We show that transient exposure of cultured and primary leukemic and epithelial tumor cells to clinically-relevant nanomolar doses, without causing immediate cytotoxicity, produce an anti-tumor “memory” response, including inhibition of subpopulations of cancer stem-like cells. These effects are accompanied by sustained decreases in genome-wide promoter DNA methylation, gene re-expression, and anti-tumor changes in key cellular regulatory pathways. Low dose decitabine and azacitidine may have broad applicability for cancer management.
INTRODUCTION
Decitabine (DAC) and its analog azacitidine (AZA), two major DNA de-methylating agents (Jones and Taylor, 1980), have recently emerged as potent therapies for the pre-leukemic hematological disease, myelodysplastic syndrome (MDS), and for established leukemias (Blum et al., 2007; Cashen et al., 2009; Issa et al., 2004), leading to FDA approval for patients with MDS (Kantarjian et al., 2006; Silverman et al., 2002). Remarkably, the improved clinical efficacy and safety profile have emerged only as doses of the drugs, given either alone (Issa et al., 2004; Kantarjian et al., 2006; Kantarjian et al., 2007) or in combination with histone deacetylase (HDAC) inhibitors (Gore et al., 2006), were significantly reduced. Despite the clinical efficacy observed in hematological neoplasms, these lower dosing regimens have not been thoroughly tested in patients with common solid tumors. Past trials with high doses have been plagued by extreme toxicities that have probably confounded the ability to document true clinical responses (Abele et al., 1987; Momparler et al., 1997). Even for the successes in hematologic neoplasms, it is still under debate whether epigenetic effects of the drugs account for all, or even some, of the therapeutic response (Issa and Kantarjian, 2009). In a recently completed clinical trial for advanced lung cancer using a low dose regimen which has efficacy in MDS, we have seen some very durable, complete, partial, and stable responses in a subset of patients who have failed multiple previous chemotherapy regimens (Juergens et al.). These results emphasize the importance of deciphering the mechanisms involved with therapeutic efficacy of DAC and AZA and understanding how low, nanomolar doses of DAC and AZA are effective at inducing sustained anti-tumor responses.
RESULTS
Transient, low dose DAC decreases tumorigenicity of cultured leukemia cells, with minimal acute DNA damage, cell cycle alterations, or apoptosis
DAC and AZA were originally designed as nucleoside analogues which, at high doses, clearly produce DNA damage and cytotoxicity (Karpf et al., 2001; Palii et al., 2008). However, these effects may not be the primary mechanisms responsible for the clinical efficacy in patients with MDS or leukemia. We, thus, first sought to separate low dose, from high dose effects of DAC on cultured leukemia cells. We employed the very low doses, indicated by pharmacokinetic studies to be in the nanomolar range for DAC (20 to 300 nM) (Cashen et al., 2008; Schrump et al., 2006), to which tumor cells in responding patients with MDS/AML are most likely exposed in settings of clinical efficacy. Kasumi-1 cells, an acute myelogenous leukemia (AML) line with a stem-cell like phenotype characterized by a high fraction of CD34+ early progenitor cells (Asou et al., 1991) (Figure S1A), are known to be sensitive to cytotoxic effects of high dose DAC (Berg et al., 2007). Indeed, daily doses of 500nM DAC for three days produced 50% apoptosis which reached over 90% by four days after drug withdrawal (Figure S1B), while 10nM produces little or no cell death at three days in Kasumi-1, KG-1, KG-1a AML cells, and histiocytic lymphoma U-937 cells (Figures 1A and S1C). Importantly, this lack of early cytotoxicity at 10 nM is subsequently followed, after drug withdrawal, by sustained rates of apoptosis leveling off at ~ 40% for Kasumi-1, and ~25% for KG-1 leukemia cells (Figure S1D). Consistent with these observations, the 3-day 10 nM DAC exposures, produce little cell cycle changes between mock and treated Kasumi-1 cells at day 3 (Figure 1B) and 4 and 11 days after drug withdrawal (Figure S1E) or significant increases in double-strand DNA breaks in CD34+ and CD34− Kasumi-1 cells at day 3 (Figure 1C). In contrast, 100 nM of cytarabine (Ara-C), a compound structurally similar to DAC and a standard cytotoxic chemo-therapeutic agent used for AML therapy, causes distinct prolongation of S-phase (Figure 1B).
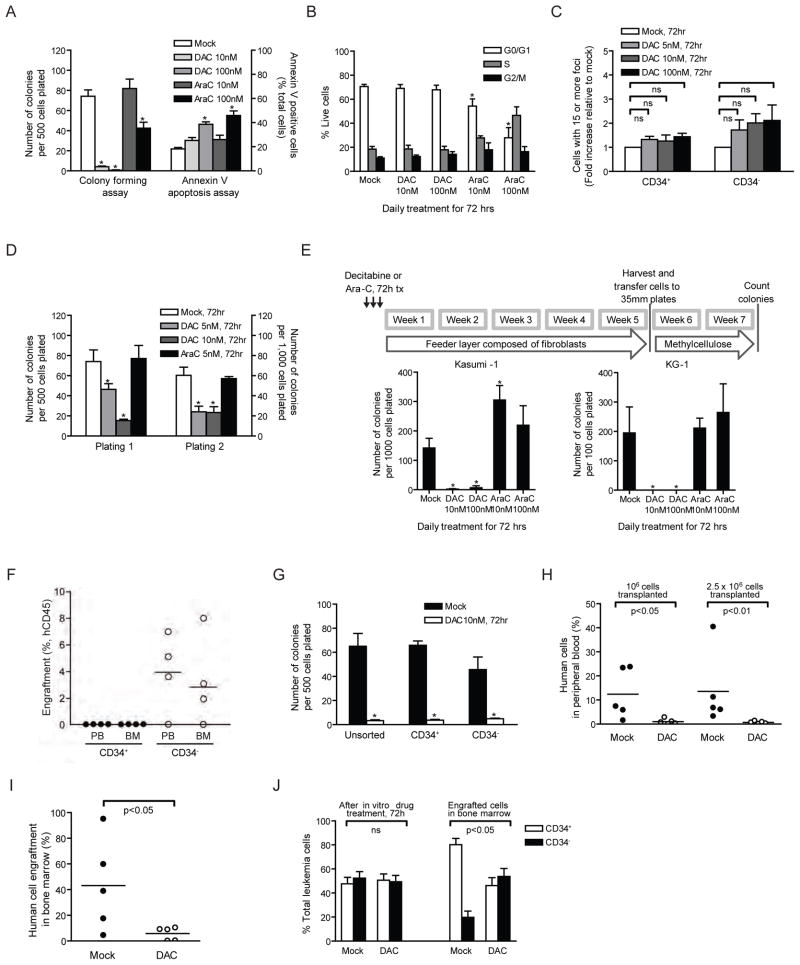
Figure 1. Low dose Decitabine (DAC) treatment diminishes self-renewing and leukemia-initiating capacities in cultured leukemia cells.
(A) Apoptosis and methylcellulose colony forming assays of Kasumi-1 cells following 72-hour daily treatment with DAC or cytarabine (Ara-C). *p<0.05 compared to mock by ANOVA and Dunnett’s multiple comparison test. (B) Cell cycle analysis of Kasumi-1 after DAC or Ara-C daily treatment for 72 hours. *p<0.05 by ANOVA and Bonferroni post-tests. (C) γH2AX foci formation in CD34+ and CD34− Kasumi-1 cells after 72-hour DAC treatment. For each treatment, cells containing more than 15 foci are counted and calculated as fold change relative to that of mock treatment. ns, not statistically significant by ANOVA and Bonferroni post-tests. (D) Serial methylcellulose replating assays of Kasumi-1 after 72-hour daily treatment of DAC or Ara-C. Equal numbers of viable cells were plated for each plating *p<0.01 compared to mock by ANOVA and Bonferroni post-tests. (E) Long-term culture-initiating cell assay of Kasumi-1 and KG-1 AML cells following 72-hour daily treatment of DAC or Ara-C. *p<0.05 compared to mock by Mann-Whitney test. (F) Engraftment assay in NOD/Shi-scid/IL-2Rγnull mice of CD34+ versus CD34− Kasumi-1 cells. Human CD45+ cells were analyzed in the peripheral blood (PB) and Bone Marrow (BM) of transplant mice 4 to 5 months post transplantation. No engraftment was observed in any mice receiving CD34− cells. (G) Methylcellulose colony forming assay of CD34+ and CD34− Kasumi-1 cells following 72-hour daily treatment with 10nM DAC. *p<0.001, compared to mock, by ANOVA with Bonferroni posttests. (H and I) Engraftment assay in NOD/Shi-scid/IL-2Rγnull mice of Kasumi-1 cells pretreated daily with 10nM DAC for 72 hours. Percentage of human leukemia cells in peripheral blood (H) and in bone marrow (I) is shown. p values are calculated by Mann-Whitney test. (J) Percentage of CD34+ and CD34− Kasumi-1 AML cells after daily treatment of 10nM DAC for 72h in vitro and after engraftment in mouse bone marrow. *p<0.05 by Mann-Whitney test. All error bars represent standard errors. See also Figure S1.
Despite the above lack of acute cytotoxic effects, the 3 day, 10nM dose of DAC can fully, for Kasumi-1, KG-1 and KG-1a cells, and partially for U-937 cells, inhibit subsequent colony formation in methyl-cellulose assays performed over 20 days in drug free media (Figures 1A and S1C). Similarly, 100 nM DAC produces no initial apoptosis in chronic myelogenous leukemia, K-562 cells, but sharply reduces subsequent colony formation (Figure S1C). Acute promyelocytic leukemia, NB4 cells, are sensitive to both early apoptosis and diminishing of proliferation at 10 nM DAC (Figure S1C).
Importantly, these anti-cloning effects for sensitive cells such as Kasumi-1 are sustained and wane only slowly after drug removal. First, the 10 nM dose of DAC blunts clonogenic potential of the cells in repeated methylcellulose colony forming assays performed without subsequent drug exposure over 28 days (Figure S1F). Second, the transient 3-day exposure of 10 nM also inhibits, in serial methylcellulose replating assays, colony formation for Kasumi-1 (Figure 1D) and KG-1 (Figure S1G) cells, and markedly decreases this ability for U-937 and K-562 cells (Figure S1G). In contrast, 10 or 100 nM doses of Ara-C produce much less inhibition of colony growth (Figure 1A). Finally, 10 nM DAC for 3 days also markedly blunts cloning of Kasumi-1 and KG-1 cells, in a 5-week, drug-free, feeder layer assay which supports long-term culture-initiating cells (LTC-IC) (Figure 1E). Ara-C, again, fails to reduce LTC-IC (Figure 1E), consistent with known poor capability of this drug to target such leukemic cell populations (Guzman et al., 2001).
The ultimate test of tumor initiation for leukemia and solid tumor cells (Lapidot et al., 1994) (Al-Hajj et al., 2003; Li et al., 2007; O’Brien et al., 2007; Ricci-Vitiani et al., 2007; Schatton et al., 2008; Singh et al., 2004) is engraftment capability and tumor growth of the most tumorigenic or more “stem-like” cells in mice (Jordan et al., 2006). Kasumi-1 has a high percentage of stem-like CD34+ cells. Only purified CD34+, and not CD34−, Kasumi cells are tumorigenic in mice (Figure 1F). Importantly, our transient, low dose treatment with DAC inhibits colony formation of both CD34+ and CD34− Kasumi-1 cells (Figure 1G). Most strikingly, a 3 day treatment of Kasumi-1 cells to 10 nM DAC markedly delays, without any drug treatment of recipient animals, detectable engraftment of Kasumi-1 cells in the peripheral blood and the bone marrow of NOD/Shi-scid/IL-2Rγnull mice (Figures 1H and 1I) and decreases the percentage of CD34+ leukemic stem/progenitor cells that do appear in bone marrow (Figure 1J). Thus a 10 nM, transient DAC exposure markedly inhibits subsequent leukemia-initiating CD34+, in vivo.
Low dose DAC, without acute cytoxicity, blunts clonogenicity of primary leukemia cells but not primary normal bone marrow cells
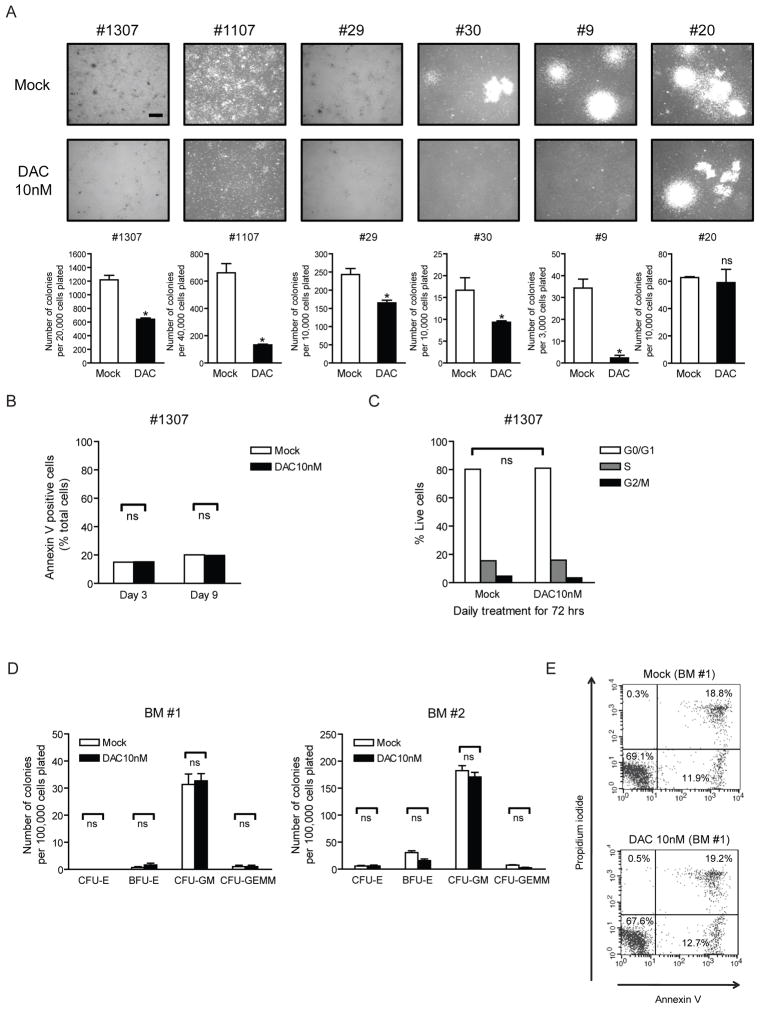
Low dose DAC can produce many of the same effects observed above on primary AML cells without the toxic effects on primary bone marrow cells. Thus, a 3 day, transient 10nM dose of DAC, markedly blunts the ability of five of six primary samples from patients with newly-diagnosed AML to subsequently clone in methyl-cellulose (Figure 2A) without causing early cell apoptosis or change in cell cycle (Figures 2B and 2C). This same 10nM dose of DAC fails to blunt the ability of a normal bone marrow sample to generate progenitor colonies for multiple marrow lineages and did not cause significant apoptosis (Figures 2D and 2E). All of the above results are consistent with clinical results for MDS/AML wherein delayed, robust anti-tumor responses can be produced while the normal bone marrow parameters are restored.
Figure 2. Decitabine (DAC) at non-acutely-cytotoxic doses blunts clonogenicity of primary human leukemia cells but not normal bone marrow cells.
(A) Methylcellulose colony forming assay of bone marrow mononuclear cells from 6 patients with newly diagnosed acute myelogenous leukemia (AML) following 72 hr daily DAC treatment and 4-day recovery period in vitro. Images and quantifications of colonies are shown in upper and lower panels, respectively. *p<0.05 compared to mock by ANOVA and Dunnett’s multiple comparison test. The scale bar represents 500μm. #1307 and #29: AML with FLT3-ITD mutation, #1107: AML FAB M2, #30: AML with mutated NPM1, #9: Secondary AML, #20: AML FAB M5. (B) Annexin V apoptosis assay of one representative AML patient sample (#1307) after 72hr daily DAC treatment in vitro. Bone marrow mononuclear cells were harvested for analysis at the end of 72 hr drug treatment (Day 3) and 6 days after drug removal (Day 9). ns, not statistically significant, by ANOVA with Bonferroni posttests. (C) Cell cycle analysis on one representative AML patient sample (#1307) was performed 4 days after drug removal (Day 7) with DNA content measured using propidium iodide staining. ns, not statistically significant, by ANOVA with Bonferroni posttests. (D) Methylcellulose colony forming assays of human normal bone marrow mononuclear cells (BM #1 and #2) following 72hr daily DAC treatment in vitro. CFU-E: Colony-Forming Unit-Erythroid, BFU-E: Burst-Forming Unit-Erythroid, CFU-GM: Colony-Forming Unit-Granulocyte, Macrophage, CFU-GEMM: Colony-Forming Unit-Granulocyte, Erythrocyte, Macrophage, Megakaryocyte. ns, not statistically significant, by ANOVA with Bonferroni posttests. (E) Annexin V apoptosis assay in one representative bone marrow sample (BM #1) after DAC treatment in vitro. Cells were double-stained with Annexin-V and propidium iodide, and percentage of cells relative to total cells is shown in each quadrant. All error bars represent standard errors.
Transient, low-doses of DAC and AZA inhibit cultured solid tumor stem-like cells and diminish tumorigenicity
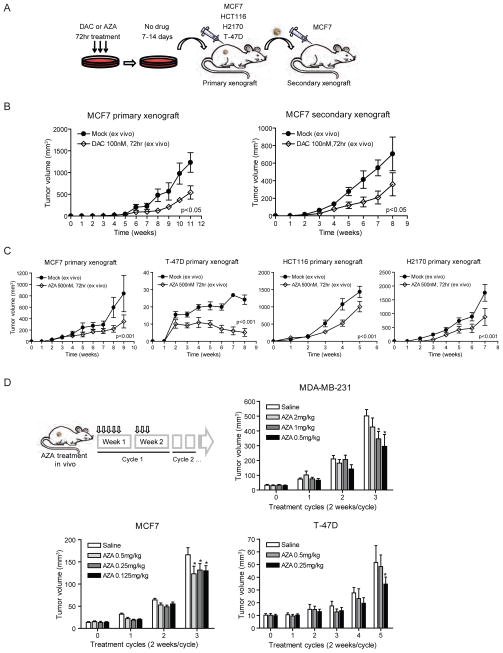
We next determined doses of DAC/AZA that were acutely non-toxic to cultured breast cancer cells but might affect their tumorigenic properties. We found that transient three day exposure of MCF7 cells to 100 nM DAC at day 3 of drug treatment or 4 days after drug withdrawal (Day 7), or an equivalent dose of 500 nM AZA at 7 days after drug withdrawal (Day 10), produced little apoptosis (Figure S2A). Similarly, 3 days of exposure to 100 nM DAC or 500 nM AZA led to minimal cell cycle changes overtime (Figure S2B). Significant decreases were observed for MCF7 cells in anchorage-independent growth in soft agar, a classic in-vitro transformation assay, for both 100nM DAC and 500nM AZA (Figure S2C). Similarly, three day treatments with10 nM DAC (Figure S2D), 100 nM DAC (Figure 3B), and 500nM AZA (Figure 3C) followed by subcutaneous injection into untreated NOD/SCID mice (Figure 3A) significantly reduced the size of MCF7 xenografts, and this diminished tumorigenicity persisted in serially transplanted secondary xenografts (Figures 3A and B). An even stronger decrease was produced in tumor growth for T-47D breast cancer cells. Growth was also suppressed for HCT116 colon and H2170 lung cancer lines (Figure 3C). Moreover, for clinical relevance, we tested several short cycles and therapeutic doses of AZA in mice bearing established xenografts of MDA-MB-231, MCF7 and T-47D breast cancer cells. In MDA-MB-231 and T-47D, the lowest dose of drug led to the largest reduction in tumor size whereas in MCF 7 xenografts, all doses were comparable (Figure 3D). There were, again, no changes in proliferative capacity of cells in the treated MCF7 tumors (Figure S2E).
Figure 3. Decitabine (DAC) and azacitidine (AZA) at non-cytotoxic doses decrease tumorigenicity in mouse tumor xenografts.
(A) Schematic outline of xenograft tumorigenicity assay in NOD/SCID mice for MCF7 (breast), T-47D (breast), HCT116 (colon) and H2170 (lung) cancer cells after 7–14 day recovery period following 100 nM DAC or 500 nM AZA daily treatment for 72 hrs. (B) Primary and secondary xenograft assays of MCF7 cells pretreated with 100 nM DAC. Equal numbers of viable cells were injected in each transplantation. p value is calculated by ANOVA test. n=6 for each treatment group. (C) Primary xenograft assays of MCF7, T-47D, HCT116 and H2170 cells pretreated with 500 nM AZA. p value is calculated by ANOVA test. n=10 for each treatment group. (D) AZA treatment of mice with pre-established MDA-MD-231, MCF7 or T-47D breast tumors. Tumor volumes are measured at the end of each cycle. *Statistical significance is determined by two-way ANOVA test (p<0.05). All error bars represent standard errors. See also Figure S2.
Transient, low-doses of AZA inhibit primary breast cancer stem-like and self-renewing cells
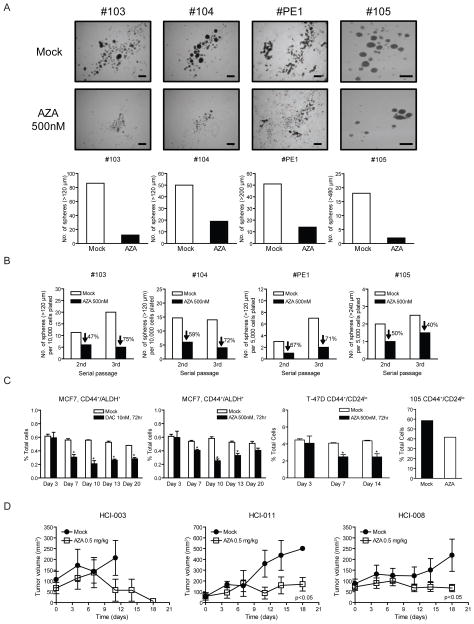
We further investigated the effects of transient, low dose AZA treatment on the self-renewal potential of primary cancer cells isolated from the pleural effusions of four patients with metastatic breast cancer of the luminal type. These cells were treated for 3 days with 500 nM AZA, followed by drug withdrawal and culture in suspension as tumor spheres and serial passaged to test for self-renewal capacity and stem-like cell activity (Figure 4A) (McDermott and Wicha, 2010, Korkaya, et al., 2009, Charafe-Jauffret et al., 2009; Dontu et al., 2003; Ginestier et al., 2007; Liu et al., 2006). There was dramatic inhibition of primary sphere formation and an inhibitory effect that persisted through two subsequent serial passages (Figure 4B), while spheres from mock treated cells showed a trend towards enrichment from passage two to three (See patients #103, PE1, #105, Figure 4B). Thus, three days of exposure to low dose AZA reduces the self-renewal capacity of primary breast cancer cells consistent with our observation that AZA treatment decreases a self-renewing population in breast cancer cell lines. Aldehyde dehydrogenase (ALDH1) and CD44+/CD24lo have emerged as markers for cancer stem-like cells in breast, and are predictive for metastasis and poor prognosis (Al-Hajj et al., 2003; Ginestier et al., 2007). We find, in agreement with others, that the MCF7 breast cancer line has ~ 0.5% ALDH1+ cells (Charafe-Jauffret et al., 2009) and T47Ds contain ~ 5% CD44+/CD24lo (data not shown). A three day treatment with 500 nM AZA and 10 nM DAC significantly reduced CD44+/ALDH1+ cells for up to 17 days after drug withdrawal (Figure 4C) with no changes observed in the larger CD44+/ALDH1− cell population (data not shown). Similar results were observed for 100nM DAC (data not shown). For T-47D cells, a similar 3 day treatment with 500 nM AZA reduced the CD44+/CD24lo population for up to11 days after drug withdrawal (Figure 4C). Likewise, 3 days of 500nM AZA decreased the CD44+/CD24lo population in mammospheres from patient #105 (Figure 4C).
Figure 4. Low dose Decitabine (DAC) and azacitidine (AZA) decrease self-renewal and tumorigenesis in primary breast cancer tissue from patients.
(A) Tumor sphere assays of primary breast cancer cells from pleural effusions following 500nM AZA daily treatment for 3 days in vitro. Images and quantifications of tumor spheres are shown in upper and lower panels respectively. Scale bars represent 500μm. (B) Serial passages of tumor spheres formed by primary breast cancer cells following the initial 3-day AZA treatment. Equal numbers of viable cells were plated in each passage. The data shows a sustained decrease in self-renewal capacity of tumor spheres. (C) Flow cytometric analyses of CD44+/ALDH+ in MCF7 cells, CD44+/CD24lo cells in T-47D and primary breast cancer sample 105, following AZA or DAC daily treatment for 3 days and subsequent drug removal. Statistical significance was determined via two-way ANOVA test. (D) In vivo AZA treatment of immunodeficient mice bearing orthotopically-transplanted primary breast cancer cells from patients. 0.5mg/kg of AZA was administered intraperitoneally, 5 days a week. All error bars represent standard errors.
Lastly, we tested the effects of low dose AZA on primary tumor tissue from breast cancer patients. Therapeutic administration of AZA (0.5mg/kg) significantly inhibited the growth of three, pre-established, patient-derived tumors, engrafted orthotopically into immunodeficient mice (Figure 4D) (DeRose et al., 2011)
Low-dose DAC inhibits gene promoter DNA methylation, and causes re-expression of hypermethylated genes
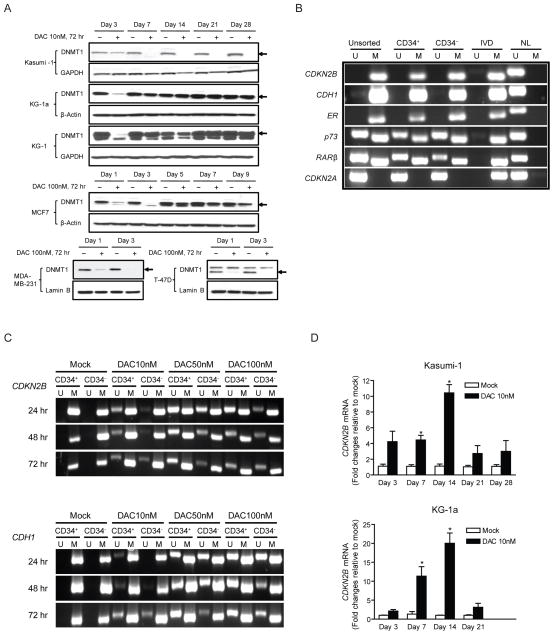
The ability of transient low doses of DAC to sustain long-term anti-tumor effects is associated, at least temporally, with the retained ability of this drug to target DNA methylation processes and alter gene expression, in each of the cell types under study. DAC and AZA, after their incorporation into DNA, are known to bind to and block the catalytic site of DNA methyltransferases (DNMT’s). This covalent binding effectively traps DNMTs as adducts to DNA (Ferguson et al., 1997; Gabbara and Bhagwat, 1995; Santi et al., 1984). Additionally DAC and AZA can trigger proteosomal degradation of the maintenance DNA methyltransferase DNMT1 (Ghoshal et al., 2005). Three days of 10 nM DAC leads to early depletion of DNMT1 protein in Kasumi-1, KG-1 and KG-1a leukemia cells and 100 nM gives a similar effect in three different breast cancer lines (Figure 5A). Despite these similar effects on early depletion of DNMT1 in all tested cell lines, the duration of the effect does not correlate with the later phenotypic consequences of drug treatment described above. Thus, while DNMT1 protein levels quickly recover in most of the lines, DNMT1 depletion lasts for at least 25 days after drug withdrawal in Kasumi-1 cells, although the phenotypic responses of these cells are similar to those of KG-1 cells (Figure 5A). Thus, transient or sustained inhibition of DNMT1 protein expression by 3 days of DAC treatment accompanies prolonged, subsequent, anti-tumor effects.
Figure 5. Low-dose decitabine (DAC) inhibits gene promoter DNA methylation, and causes sustained re-expression of hypermethylated genes.
(A) Western blot of DNMT1 expression levels in human leukemia cells (Kasumi-1, KG-1a and KG-1) and breast cancer cells (MCF7, MDA-MB-231 and T-47D) following 72 hr daily treatment of 10 nM (leukemia) or 100 nM (breast cancer) DAC. (B) Methylation-Specific PCR (MSP) analyses of gene promoter DNA methylation in unsorted, CD34+ and CD34− Kasumi-1 cells prior to DAC treatment. U: unmethylated sequence amplifications, M: methylated sequence amplifications, IVD: in-vitro methylated DNA, NL: normal lymphocyte DNA. (C) MSP analyses of CDKN2B and CDH1 promoters in CD34+ and CD34− Kasumi-1 cells at 24, 48 and 72 hr of DAC treatment. (D) Quantitative real-time PCR analyses of CDKN2B gene expression over time in Kasumi-1 and KG-1a cells following 72 hr daily treatment of 10nM DAC. Expression levels are adjusted to GAPDH for each sample and graphed as fold changes relative to mock. All error bars represent standard errors.
In keeping with the early depletion of DNMT1 protein levels, low-dose DAC causes demethylation and re-expression of DNA hypermethylated and epigenetically silenced genes. A three day exposure to 10 nM DAC produces, in both CD34+ and CD34− Kasumi-1 cells, significant de-methylation of the completely methylated and silenced tumor suppressors CDKN2B and CDH1 (Figures 5B and 5C). One particularly important finding with respect to the cellular phenotypes observed involves the expression of some initially silenced genes. In the treatment schema employed wherein cells are not acutely killed, we see in both Kasumi-1 and KG-1a cells increased transcription long after drug withdrawal. This is exemplified by CDKN2B, for which the peak expression is reached at day 14, or 11 days after cessation of drug administration (Figure 5D).
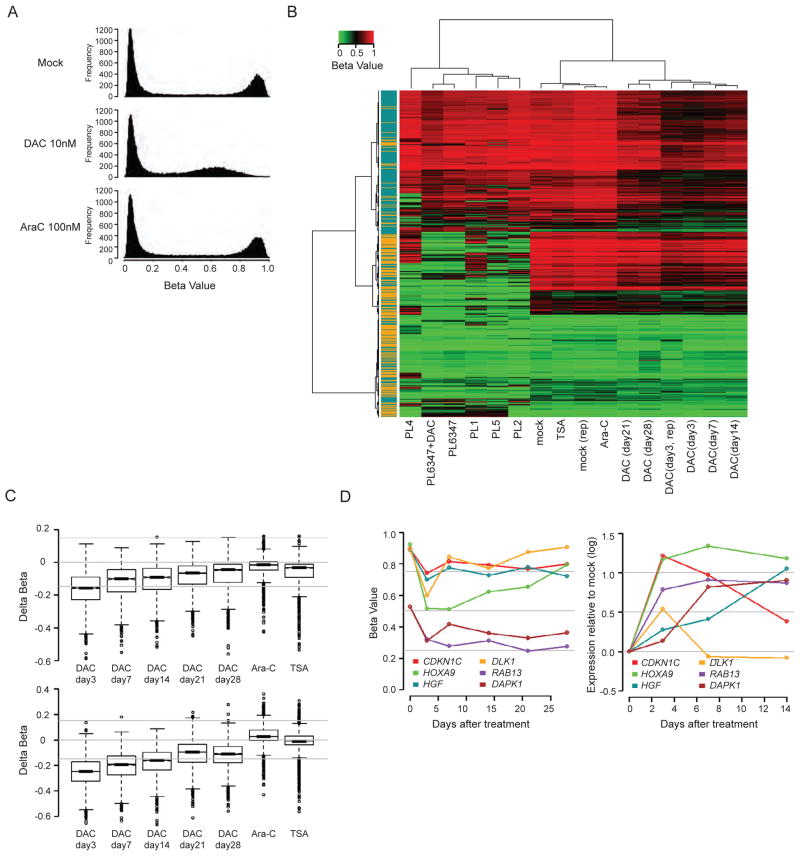
Importantly, these methylation effects are seen across the genome in analyses of DNA methylation by using the Methylation 27 BeadChip for cultured cells and the more probe-dense HumanMethylation450 BeadChip 450K for the treated 1107 and 1307 primary leukemia cells (Figure 2A). DAC (10 nM) for 72 hours causes a distinct decrease in methylation for essentially all probes and genes with high initial levels of DNA methylation in Kasumi-1 cells (Figures 6A and 6B), many of which are also heavily methylated in DNA from primary AML samples (Figure S3A). The decreases can be seen in promoter regions of both CpG island- and non-CpG island- containing genes (Figure 6C). This dose similarly reduces global methylation of highly methylated genes in KG-1, KG-1a, as does 100 nM in breast cancer MCF7 cells (Figure S3B). Treatment of a primary leukemia sample (PL6347) with 10 nM DAC, following proliferation stimulation with cytokines, similarly causes decreases in gene promoter region DNA methylation (Figures 6B and S3B). Virtually identical results have been seen for primary, treated leukemia samples 1107 and 1307 using the 450K platform (data not shown).
Figure 6. Genome-wide methylation and expression analyses after short-term exporsure to low dose decitabine (DAC).
(A) Histogram of Infinium results for Kasumi-1 methylation profiles following daily DAC or ARA-C treatment for 72 hrs. Y-axis: frequency of probes. X-axis: CpG probe beta scores (lowest to highest = increasing DNA methylation). (B) Heat map composed of minimal variation beta probes located at −1,000 to +200 bp surrounding transcription start site (~ 5,500 genes). Blue and orange colors on the left denote non-CpG and CpG island promoters, respectively. PL6347, PL1, PL2, PL4, PL5: primary leukemia samples. PL6347+DAC: primary leukemia after 72hr 10 nM DAC treatment. mock and mock (rep): untreated Kasumi-1 cells and its replicate. Ara-C: Kasumi-1 cells treated with 100nM Ara-C daily for 72 hrs. TSA: Kasumi-1 cells treated with 300 nM trichostatin A for 9 hrs. DAC (day 3, day 3 rep, 7, 14, 21, 28): Kasumi-1 cells harvested at various time points post 72hr daily treatment of 10 nM DAC. (C) Box plots showing beta value changes over time in CpG island (upper panel) and non-CpG island gene promoters (lower panel) in Kasumi-1 cells following 72hr daily treatment of 10nM DAC. Ara-C: 100nM AraC treatment for 72 hrs. TSA: 300nM TSA treatment for 9 hrs. (D) Left panel: beta value changes after 72 hr, 10 nM DAC treatment for 6 genes in Kasumi-1 with basal beta values ≥ 0.5. Right panel: corresponding Agilent expression changes normalized to day 0. Gray horizontal lines: bottom to top, no change (log2 scale = 0), 1.4 fold (0.5) and 2.0 fold (1.0). See also Figure S3.
The above genomic promoter demethylation induced by low dose DAC is accompanied by widespread increases in gene expression (scatter plot for Kasumi-1 cells – Figures S3C and S3D). In fact, up to 50% of genes whose expression increase following transient low dose exposure, remain increased for up to 14 days (Figure S3E). Among such genes are those having decreases in their cancer specific promoter hypermethylation after drug treatment, such as CDKN2B in Kasumi-1 (Figure 5C) and KG-1a AML cells (Figure S5), CDKN2A in KG-1a cells, and six hypermethylated genes (Figure 6D) studied by Lubbert and colleagues, or our lab, in Kasumi-1 cells, DAPK1, CDKN1C (also known as p57KIP2), HOXA9, HGF, DLK1 and RAB13 (Flotho et al., 2009). All these eight genes have increased expression by day 3 of low dose decitabine treatment, and seven have sustained increases at least 11 days after treatment cessation (Figure 6D).
Transient low doses of DAC and AZA induce sustained alterations in major cancer cell signaling pathways
Cancer is driven by alterations of gene function in key cellular pathways (Hanahan and Weinberg, 2011). We find, using Metacore analyses of gene expression, changes in many key anti-tumor pathways (Figures 7A,B, S4A,B, Tables S1 and S2) as described below.
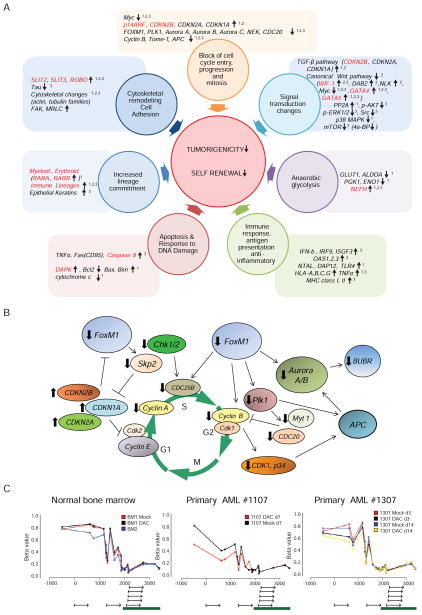
Figure 7. Metacore pathway analyses of Agilent expression changes following decitabine (DAC) or azacitidine (AZA) treatment.
(A) Key pathways and/or master genes, shown here for studies in cultured cells. Genes in black = genes with expression changes not directly linked to promoter DNA methylation. Genes in red = genes with basal promoter hypermethylation, drug induced demethylation, and corresponding increases in expression. Superscripts by genes or pathways indicate: 1, Kasumi-1 AML cells, 2, KG-1a AML cells, and 3, MCF7 breast cancer cells with expression changes concordant in the indicated cell line. (B) An example of pathway diagram summarizes gene changes for cultured and primary AML and breast cancer samples after transient in vitro treatment of DAC or AZA. Events depicted illustrate key expression increases in cyclin dependent kinase inhibitors (CDKi’s – CDKN2B, CDKN2A and CDKN1A) which are known to trigger decreased activity of the FOXM1 pathway and all of the decreases shown above for FOXM1 itself and the other key participants for cycle entry and progression and including the oncogene, Skp2. (C) Analyses of DNA methylation by HumanMethylation450 BeadChip for multiple CpG sites in the CDKN1A proximal promoter region. Note that a normally heavily methylated (two normal bone marrow samples) region 5′ to the unmethylated CpG island (large green bar), surrounding an alternate gene start site (small arrow), is distinctly demethylated after 10 nM DAC treatment in 1107 primary AML cells which show all of the pathway changes but not to the same degree in 1307 AML cells where CDKN1A expression is not increased and the pathway changes seen are not present. See also Figure S4, Tables S1 and S2.
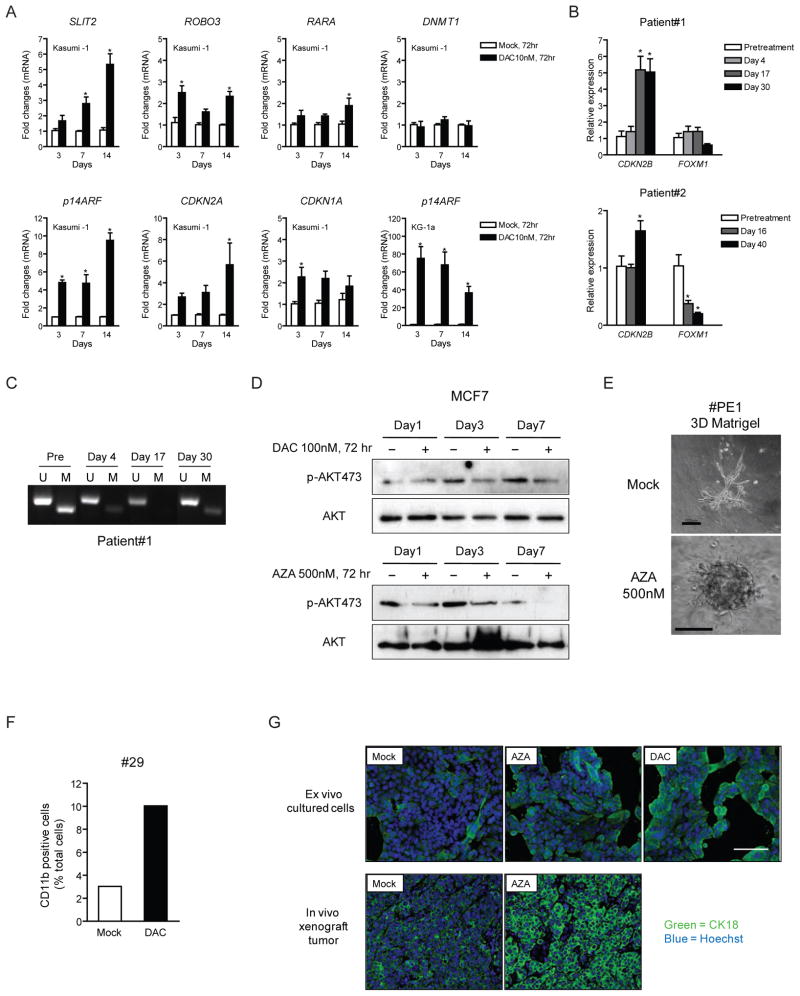
Changes in gene expression, which would block cell proliferation and decrease tumor self-renewing populations, were often correlated with increases in expression of more than one cyclin-dependent kinase inhibitor CDKN2B, CDKN2A, CDKN1A, and were associated with CpG island DNA demethylation for CDKN2B in leukemia cells (Figures 7A and 7B). The upregulation of p14ARF, CDKN2A, CDKN1A, and other key hypermethylated pathway genes, such as SLIT2 and ROBO3, were validated in cultured leukemia cells (Figure 8A). The FOXM1-polo-kinase (PLK) -Aurora kinase pathway would be predicted to be down-regulated secondary to these above gene increases (Koo et al., 2011). We observed this in both cultured (KG-1a AML) and primary leukemia cells (#1107 in Figure 2A), and in primary breast cancer, mammosphere samples (#103 and #104 in Figure 4A) (Figure 7B). In each case, CDKN1A expression was increased and could be linked to DNA demethylation in a DNA methylated region (Figure 7C) just upstream from a non-hypermethylated CpG island. This pathway, required for cells to enter and perform cell division and to maintain DNA damage-triggered cell cycle check points, is increased in tumors with poor prognosis, and has been linked to progenitor cell renewal and increased cellular invasion, motility, and metastatic potential (Koo et al., 2011; Raychaudhuri and Park, 2011). Loss of G2M cell cycle check point control and the down regulation of FOXM1, Aurora kinases, polo kinases, CHK1, CHK2, MYT1, represent proteins currently targeted by the pharmaceutical industry to induce cancer cell apoptosis and/or sensitization to cytotoxic drugs (Carrassa and Damia, 2011; Merry et al., 2011). In bone marrow samples from two leukemia patients who responded to DAC treatment in a clinical trial, sustained DNA demethylation and increased expression of CDKN2B is detected at early time points before reduction of the leukemic tumor clone and co-incident with decreases in FOXM1 RNA (Figures 8B and 8C).
Figure 8. Validation of changes in multiple signaling pathways following decitabine (DAC) or azacitidine (AZA) treatment.
(A) Quantitative real-time PCR verification for selected genes with expression changes indicated by the Metacore analyses. *p<0.05 determined by ANOVA and Bonferroni post-tests. (B) Quantitative real-time PCR analyses of gene expression changes in bone marrow samples taken from patients #1 (partial responder with AML) and #2 (complete responder with CMML) at pretreatment and various time points during or after administration of DAC (20mg/m2 IV over 1 hour daily × 5 days every 4 weeks). mRNA levels of two genes, CDKN2B and FOXM1, at each time point were ploted relative to the baseline level after adjusted by a control house-keeping gene, GAPDH. (C) Methylation-specific PCR analysis of CDKN2B promoter on the bone marrow sample of patient #1 at pretreatment, day 4, day 17 and day 30 during the treatment cycle. U: unmethylated sequence amplifications, M: methylated sequence amplifications. (D) Western blot analysis of total and phosphorylated AKT in MCF7 breast cancer cells post 100 nM DAC (upper panel) and 500 nM AZA (lower panel) treatment. (E) 3D assay in Matrigel of a primary breast cancer sample PE1 following 3 day treatment with 500 nM AZA and subsequent drug removal. Scale bars represent 100μm. (F) Flow cytometric analysis of surface CD11b expression in primary AML sample #29 at 14 days after 72 hr 10nM DAC treatment. (G) Immunofluoresence staining of cytokeratin 18 in cultured MCF7 cells 4 days after 72 hr 100nM DAC or 500 nM AZA daily treatment (upper panel), and in MCF7 xenograft tumors removed from mock and AZA-treated mice (lower panel) at 6 weeks. The scale bars represents 100μm. All error bars represent standard errors. See also Figure S5.
We also saw decreased AKT mRNA, and down regulation of key regulatory genes, TWIST, SLUG, and SNAIL (Hanahan and Weinberg, 2011), which activate epithelial to mesenchymal transition (EMT) (Figure 7A). EMT has been closely linked to stem-like populations in breast cancer and we observe regulation of EMT in treated, self-renewing primary mammosphere cells from patients 103 and 104 (Figure 4A and Table S2). Correlative studies in the laboratory reveal decreased phosphorylation of AKT in cultured MCF7 breast cancer cells (Figure 8D).
In the self-renewing, primary mammosphere cultures we saw changes predictive of diminished cell motility, invasiveness, induction of angiogenesis, and metastatic potential including decreased expression for genes encoding the integrins, metalloproteinases, and the key cytoskeletal remodeling protein ezrin (Figure S4A and Table S2). These genes can be over-expressed in breast cancer, and their down-regulation is matched by upregulation for genes encoding proteins which suppress these cell processes, such as the often hypermethylated tissue inhibitors of metalloproteinases (TIMP’s), thrombospondin, and the complement receptor, C5AR (Table S2). Importantly, breast cancer sample PE1, following 3 day treatment with 500 nM AZA, appears less invasive and forms a more compact sphere in matrigel (Figure 8E).
Finally, both cultured and primary cells harbored gene expression changes that predict cell maturation events. Kasumi-1 AML, and primary AML sample 1107, had decreased expression of the G-CSF receptor and of the c-myc gene and increased expression, with decreased DNA methylation of the transcription region, for the transcription factor, RARA (Figures 8A and S5). Coordinately, myeloid commitment at the progenitor cell level was indicated by DNA demethylation and increased expression of the normally methylated early myeloid lineage genes, LYZ and ELANE (Figures S4B, S5 and Table S1), increases in CD13 (Figure S4B), a marker of cells with bipotential maturation to erythroid and granulocyte cells (Chen et al., 2007) and mature granulocyte formation (Levy et al., 1990) by increased p47-phox, p67-phox, and Gp91-phox (Figure S4B and Table S1). All of these changes were observed both in DAC treated total AML cells and tumorigenic CD34+ cells from Kasumi-1 AML(Figure S4B). Also, increased TGF-beta signaling (Figures 7A), promotes maturation and inhibition of leukemic progenitor cell renewal (Lewis et al., 2001; Watabe and Miyazono, 2009). Concomitantly, there is decreased canonical WNT pathway signaling associated with reduced methylation, and increased expression of WIF1 (Figures 7A and S5), a Wnt antagonist gene, and downstream decrease in expression of c-myc (Figures 7A), a canonical target for Wnt pathway activation (He et al., 1998). Importantly, we validated myelocyte differentiation by flow cytometric analysis of a surface marker for mature granulocyte, CD11b, in a treated primary AML sample 29 (Figure 8F). Sustained increase of cytokeratin 18 (CK18) in DAC- or AZA- treated MCF7 cells, as well as in tumor xenografts from mice treated for 6 weeks with AZA (Figure 8G) was also observed. CK18 expression reflects maturation of breast luminal cells, and loss of CK18 is associated with poor prognosis in breast cancer patients (Woelfle et al., 2004).
DISCUSSION
Our present studies indicate that treatment of cancer cells with clinically-relevant, low doses of DAC and AZA can exert sustained changes in gene expression, and critical signaling pathways involved in tumorigenesis, without inducing immediate cytotoxic effects such as DNA damage, apoptosis, and cell cycle arrest. Our preclinical use of transient, 3 day exposure to such doses ex vivo produces a “memory” type of anti-tumor response in mice bearing transplanted tumor xenografts, which may resemble the prolonged time to response seen in patients with hematologic neoplasms (Kantarjian et al., 2006; Oki et al., 2008; Silverman et al., 2002). In patient tumors and mammospheres, the anti-tumor response occurred more quickly and appeared to be more sensitive to AZA than that observed for cancer cell line generated tumors. This may be explained by the fact that the human tumors were exposed to a longer, more sustained treatment and that primary tissues may be initially more sensitive to AZA. All of these above dynamics following transient drug exposure are accompanied by genome-wide, prolonged gene promoter DNA de-methylation and sustained increases in gene expression, some which occur for key tumor suppressor genes in leukemia and breast cancer cells.
There are several reasons to suggest that at least one key mechanism underlying the anti-tumor responses we demonstrate may involve a resetting of abnormal epigenetic states in treated cancer cells. First, we have achieved a sustained change in the pattern of gene expression without changing primary DNA sequence. Second, as defined for an epigenetic change, these sustained changes persist for significant periods of time after a transient, subsequently withdrawn, signal - ie, in this case, drug treatment. Third, the new expression patterns are accompanied by a new cellular phenotype, anti-tumor effects. Importantly, we probably have brought out these aspects of DAC and AZA treatment by using doses that do not acutely kill cells and, thus, allow the sustained alterations in both gene expression patterns and appearance of a new phenotype to emerge. Importantly, these changes include anti-tumor events in multiple key pathways, such as apoptosis, increased lineage commitment, down-regulation of cell cycling, and others, which continue well after drug removal. Reprogramming might, then, be considered a very desirable type of targeted therapy that can blunt multiple tumor signaling pathways simultaneously.
Perhaps, one of the most striking effects observed in our study concerns the fact that nanomolar doses of both DAC and AZA appear to target, in both cell lines and primary samples of leukemia and breast cancers, self-renewing and/or tumorigenic cell subpopulations. This involves stem-like CD34+ cells in leukemia and CD44+/ALDH1+ and mammosphere forming cells in breast cancer. These findings should be considered in the context that one of the most important problems in cancer therapy is the failure of most therapies to target such subpopulations that are most responsible for sustained tumor cell renewal (Jordan et al., 2006). Our findings suggest perhaps that the often prolonged time course to response in patients with myelodysplasia or frank AML might involve a progressive exhaustion of such cell populations. Such a hypothesis is supported by recent reports that depleting DNMT1, by non-pharmacologic means, in normal mouse hematopoietic and human epithelial cells blocks self-renewal and proper cell maturation leading to cellular depletion of progenitor cells (Broske et al., 2009; Sen et al., 2010; Trowbridge et al., 2009). DAC and AZA do deplete DNMT1 in leukemia and breast cancer cells for variable time periods after transient exposure (Figure 4A). However, the mechanisms underlying our pharmacologically induced responses are certainly more complex since both DAC and AZA inhibit the catalytic sites of DNMT3a and 3b as well. Also, all the DNMT’s assuredly participate in complex protein interactions where they may exert scaffolding functions with effects on other chromatin regulatory features (Robertson et al., 2000; Rountree et al., 2000). They also, experimentally, have effects for repressing gene expression which may not require their directly catalyzing DNA methylation (Bachman et al., 2001). This complexity may actually give DAC and AZA an important advantage in treating cancer cells where they may target all of these functions and their role in governing the epigenetic aberrancies present in cancer. If so, the activities we find for low nanomolar doses, may be very important in rendering these drugs as “gold standards” for the development of agents to target DNA methylation, and other epigenetic abnormalities, as an anti-cancer strategy to inhibit the most tumorigenic subpopulations of cells.
In summary, our findings have much relevance for strategies to use DAC and AZA more widely for the management of cancer. These drugs are already making an impact for patients with hematological malignancies. Our findings now suggest ways and biomarkers that might be used to predict and/or monitor clinical efficacy. The similar “memory” type of responses we find for primary and cultured breast cancer cells, and the key pathway changes seen, indicate that the treatment of many cancers may be considered and with doses that will be not only efficacious but also minimally toxic to patients. Such possibilities are emerging in our recently completed Lung Cancer SPORE and Stand-up to Cancer (SU2C) sponsored trial for patients with advanced non-small cell lung carcinoma (Juergens et al.). SU2C trials have now begun in breast cancer which might be well informed by the pre-clinical studies we now report. Moreover, it is especially appealing to consider that DAC and AZA might sensitize tumor cells to other drugs, as is suggested in Juergens et al., that target the oncogenic pathways we have shown to be altered and allow use of reduced, less toxic, doses for these other agents.
EXPERIMENTAL PROCEDURES
Methylcellulose colony forming assay and serial re-plating assay
After a 3 day drug treatment, equal numbers of viable cells were plated in triplicate onto MethoCult® H4434 Classic or MethoCult® H4435 Enriched (StemCell Technologies, Vancouver, BC, Canada). Colonies containing more than 40 cells were quantified at 10–16 days under an inverted microscope. For serial re-plating assays, equal numbers of viable cells from the first plating were plated in triplicate in the second plating. Colonies were again scored after 16–21 days.
Long-term culture-initiating cell assay
After a 3 day drug treatment, equal numbers of viable leukemia cells (trypan blue negative) were plated onto a feeder layer of irradiated M2-10B4 mouse fibroblasts (StemCell Technologies Vancouver, BC, Canada) and maintained via bi-weekly one half medium changes in MyeloCult™ H5100 according to manufacturers’ instructions (StemCell Technologies Vancouver, BC, Canada). After 5 weeks, cells were harvested, plated onto MethoCult® H4435 Enriched (StemCell Technologies Vancouver, BC, Canada) and colonies were counted after 14–18 days.
Immunophenotypic staining and fluorescence-activated cell sorting (FACS)
Human anti-CD34-FITC, anti-CD34-APC, anti-CD38-PE, anti-CD33-APC, anti-CD15-FITC, anti-CD14-PE, anti-CD11b-APC, anti-CD45-perCP, anti-CD45-APC, anti-CD44-APC, anti-CD24-PE and corresponding isotype controls were obtained from BD Biosciences, San Jose, CA. ALDEFLUOR® Assay kit (StemCell Technologies Vancouver, BC, Canada) was used to identify cells expressing high levels of aldehyde dehydrogenase (ALDH). Cells were stained according to manufacturers’ instructions and analyzed on FACSCalibur™ Flow Cytometer (BD Biosciences, San Jose, CA) using BD CellQuest Pro v.5.2 (BD Biosciences, San Jose, CA). For sorting, CD34 cells with upper or lower 10% fluorescent intensity were collected (FACSVantage Cell sorter or BD FACSAria™ II cell sorter, BD Biosciences, San Jose, CA).
Leukemia engraftment assay
Cells were tail vein injected into sub-lethally irradiated (3.0 cG) female NOD/Shi-scid/IL-2Rγnull mice 6–8 weeks of age. Peripheral blood was sampled every 4 weeks and bone marrow was harvested from each mouse at 13–15 weeks. Engraftment was determined by immunophenotypic staining using human anti-CD45-perCP and anti-CD34-APC (BD Biosciences, San Jose, CA).
Solid tumor xenograft tumorigenecity assay
MCF7, T-47D, HCT116, and H2170 cells were pre-treated with 100nM decitabine, 500nM azacytidine or PBS (Mock) for 72 hours followed by another 4–7 days in culture without drug. Harvested cells were injected (1×106) subcutaneously into both flanks of 4–6-week-old NOD/SCID mice. Female mice receiving MCF7 cells were implanted with estrogen pellets (0.72mg/60 days, Innovative Research of America). Secondary xenografts were generated by collagenase dissociation of primary MCF7 tumors at 8 weeks, followed by re-injection of 1×106 viable cells into mice.
Therapeutic administration of AZA to NOD-SCID mice with pre-established tumors
Mice with pre-established MDA-MB-231, MCF7 or T-47D xenotumors (0.3 – 0.5 cm) received subcutaneous daily injections with saline (mock) or azacitidine (2, 1, 0.5, 0.25, 0.125 mg/kg) for 8 days over a 14-day period per cycle, as outlined in Figure 2D. Five mice (10 tumors) were used per treatment group.
For xenograft experiments using cell lines, tumors were measured weekly and volume was calculated as: 0.5*(L×W2) (mm3). Protocols for all animal experiments conducted at Johns Hopkins were approved by the John Hopkins University Animal Care and Use Committee and guidelines were strictly enforced. Experiments using human tumor xenografts and conducted at the Huntsman Cancer Institute were reviewed and approved by the University of Utah Institutional Animal Care and Use Committee.
In vitro culture of primary human leukemia and normal bone marrow cells
Frozen bone marrow mononuclear cells from patients with freshly diagnosed acute myelgenous leukemia (#1107 and #1307) or healthy controls (BM #1 and #2) were thawed and cultured in Poietics™ HPGM (Lonza, Inc.) supplemented with multiple cytokines described in Supplemental Experimental Procedures. Provision of all primary bone marrow and leukemia samples was through IRB approved protocols at the University of Maryland School of Medicine, The Johns Hopkins Medical Institutions, and The University of Texas M. D. Anderson Cancer Center.
Tumor sphere assays of primary breast cancer cells
Malignant plueral effusions were collected, under an IRB approved protocol at the Johns Hopkins Medical Institutions, from women with metastatic breast cancer undergoing thoracentesis as part of clinical care and who provided a written consent for use of leftover fluid. The fluid was enriched for malignant epithelial cells via centrifugation and lysing of red blood cells. Tumorspheres were maintained on ultra-low adherent plates (Corning Inc., Lowell, MA) in MammoCult® (StemCell Technologies Vancouver, BC, Canada) or MCF10A media. (See Supplemental Experimental Procedures for details) Tumorspheres were treated daily for 3 days with 500 nM AZA and were cultured 4 more days following drug withdrawal. Primary spheres were counted and digested with 0.05% trypsin (Invitrogen, Carlsbad, CA) into single cells. Equal numbers of live cells were plated in ultra-low attachment plates to generate the 2nd spheres. Again, spheres were counted on day 7 and digested to generate the 3rd spheres.
Global gene expression and methylation analysis
Gene expression profiles for queried cells were analyzed using Agilent Human 44K expression arrays (Agilent Techonologies, Santa Clara, CA. Global methylation analysis was performed using the Illumina Infinium Human Methylation27 BeadChip (Illumina, Inc. San Diego, CA) ), and for primary leukemia samples, with the more recent HumanMethylation450 BeadChip (Illumina, Inc. San Diego, CA). Data analyses are described in Supplemental Experimental Procedures.
Metacore (GeneGo, Inc.) pathway analysis
All probes from the Agilent Human 44K expression arrays with a change of at least.5 (log2 scale) up or down at any of the investigated time points were selected for further analysis and used as input for the Metacore pathway analysis. Details are described in Supplemental Experimental Procedures.
Supplementary Material
SIGNIFICANCE.
The mechanisms underlying clinical efficacies of the DNA methylation inhibitors decitabine and azacitidine are unclear. Understanding how these drugs work would be a pivotal step in furthering epigenetic therapy. Clinical clues suggest that these drugs work in myelodysplastic syndrome because low doses provide anti-tumor effects over time rather than acutely exerting cytotoxic effects. We show that nanomolar doses of both drugs have, without acute cytotoxic effects, anti-tumor effects on both cultured and primary human leukemic and epithelial tumor cells, including the most tumorigenic, self-renewing and drug resistant cell populations. These effects are accompanied by sustained, genome-wide changes in promoter DNA methylation and gene expression which affect multiple key regulatory pathways that are high priority targets for pharmacologic anti-cancer strategies.
HIGHLIGHTS.
Transient DAC or AZA sustains anti-tumor responses and reduces self-renewal.
Acute cytotoxicity does not appear to account for the lasting effects of the drugs.
Low-dose DAC causes sustained global DNA demethylation and gene re-expression.
Low dose DAC or AZA sustains anti-tumor responses through key signaling pathways.
Acknowledgments
We thank Drs. Peter A. Jones and Charles M. Rudin and multiple other members of our Stand Up To Cancer Team, charged with bringing epigenetic therapy to the clinical management of all types of cancer, for critically reviewing the paper, and for making helpful suggestions throughout. We thank Dr. Ying Ye for providing part of primary leukemia samples used in this study. We thank Ada J. Tam from the flow cytometry core facility and Wayne Yu from the microarray core facility at Sidney Kimmel Comprehensive Cancer Center for their technical assistance. We also thank Stacie Jeter and Shannon Slater for regulatory support and recruitment of patients to the clinical study.
Funding: This work was, in part, supported by Grant CA 116160 from the National Institutes of Health and by funds from the Entertainment Industry Foundation and Lee Jeans, Stand up to Cancer (SU2C), the Samuel Waxman Cancer Research Foundation, the Department of Defense Breast Cancer Research Program (A.L.W., BC075015) and The Huntsman Cancer Foundation (A. L.W.). Dr. Hsing-Chen Tsai is the David Workman student scholar of the Samuel Waxman Cancer Research Foundation and Dr. Cynthia Zahnow is the Evelyn Grollman Glick Scholar. The clinical study was supported by the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research.
Footnotes
Authors Contribution: H-C.T., C.A.Z and S.B.B designed the study, analyzed data and wrote the paper. L.V.N performed bioinformatic analyses. C.A.Z. designed and conducted animal studies for solid tumors. Y-Y J. and S.J.S helped design animal studies for leukemia. H-C.T., H-L.L., J.J.S, K.H., E.P., J.H., I.M. conducted solid tumor xenograft mouse experiments. H-C.T., R-W.C.Y. and H-L.L. performed all in vitro assays for leukemia and breast cancer cells. Y.C. and H-L.L. performed western assays. C.R. and F.V.R conducted DNA damage assays and provide primary leukemia samples. H-C.T. and R-W.C.Y performed methylation and gene expression studies. W.M. provided primary leukemia samples and helped with study design. V.S. was responsible for all regulatory aspects of patient recruitment to the clinical study which provided primary effusion samples and D.F-K. and L.B.Y. assisted with patient recruitment and conducted thoracentesis. A.L.W. and Y-C. L. provided and treated primary human xenografts with AZA. J-P. I. provided DNA and RNA samples from DAC-treated leukemia patients. S.B.B, C.A.Z, L.V.N and H-C.T. contributed to pathway anaylses.
Competing Interests: The authors declare no competing financial interests.
Accession numbers
All microarray data are deposited in the GEO database under accession number GSE20945.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abele R, Clavel M, Dodion P, Bruntsch U, Gundersen S, Smyth J, Renard J, van Glabbeke M, Pinedo HM. The EORTC Early Clinical Trials Cooperative Group experience with 5-aza-2′-deoxycytidine (NSC 127716) in patients with colo-rectal, head and neck, renal carcinomas and malignant melanomas. Eur J Cancer Clin Oncol. 1987;23:1921–1924. doi: 10.1016/0277-5379(87)90060-5. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Berg T, Guo Y, Abdelkarim M, Fliegauf M, Lubbert M. Reversal of p15/INK4b hypermethylation in AML1/ETO-positive and -negative myeloid leukemia cell lines. Leuk Res. 2007;31:497–506. doi: 10.1016/j.leukres.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Blum W, Klisovic RB, Hackanson B, Liu Z, Liu S, Devine H, Vukosavljevic T, Huynh L, Lozanski G, Kefauver C, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Carrassa L, Damia G. Unleashing Chk1 in cancer therapy. Cell Cycle. 2011;10:2121–2128. doi: 10.4161/cc.10.13.16398. [DOI] [PubMed] [Google Scholar]
- Cashen AF, Schiller GJ, O’Donnell MR, Dipersio JF. Multicenter, Phase II Study of Decitabine for the First-Line Treatment of Older Patients With Acute Myeloid Leukemia. J Clin Oncol. 2009 doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61:759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gao Z, Zhu J, Rodgers GP. Identification of CD13+CD36+ cells as a common progenitor for erythroid and myeloid lineages in human bone marrow. Exp Hematol. 2007;35:1047–1055. doi: 10.1016/j.exphem.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MTW, Factor R, Matsen C, Milash BA, Nelson E, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AT, Vertino PM, Spitzner JR, Baylin SB, Muller MT, Davidson NE. Role of estrogen receptor gene demethylation and DNA methyltransferase. DNA adduct formation in 5-aza-2′deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- Flotho C, Claus R, Batz C, Schneider M, Sandrock I, Ihde S, Plass C, Niemeyer CM, Lubbert M. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- Gabbara S, Bhagwat AS. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem J. 1995;307(Pt 1):87–92. doi: 10.1042/bj3070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, Luger SM, Jordan CT. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, Kantarjian HM. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination Epigenetic Therapy Has Efficacy in Patients with Refractory Advanced Non-Small Cell Lung Cancer. Cancer Discovery. 1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O’Brien S, Cortes J, Faderl S, Bueso-Ramos C, Ravandi F, Estrov Z, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]
- Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Levy R, Rotrosen D, Nagauker O, Leto TL, Malech HL. Induction of the respiratory burst in HL-60 cells. Correlation of function and protein expression. J Immunol. 1990;145:2595–2601. [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Merry C, Fu K, Wang J, Yeh IJ, Zhang Y. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2011;9:279–283. doi: 10.4161/cc.9.2.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momparler RL, Bouffard DY, Momparler LF, Dionne J, Belanger K, Ayoub J. Pilot phase I-II study on 5-aza-2′-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anticancer Drugs. 1997;8:358–368. doi: 10.1097/00001813-199704000-00008. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Oki Y, Jelinek J, Shen L, Kantarjian HM, Issa JP. Induction of hypomethylation and molecular response after decitabine therapy in patients with chronic myelomonocytic leukemia. Blood. 2008;111:2382–2384. doi: 10.1182/blood-2007-07-103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrump DS, Fischette MR, Nguyen DM, Zhao M, Li X, Kunst TF, Hancox A, Hong JA, Chen GA, Pishchik V, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12:5777–5785. doi: 10.1158/1078-0432.CCR-06-0669. [DOI] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.