Abstract
In axial organs of juvenile plants, the phytohormone auxin (indole-3-acetic acid, IAA) rapidly allows cell wall loosening and hence promotes turgor-driven elongation. In this study, we used rye (Secale cereale) coleoptile sections to investigate possible effects of IAA on the proteome of cells. In a first set of experiments, we document that IAA causes organ elongation via promotion of expansion of the rigid outer wall of the outer epidermis. A quantitative comparison of the proteome (membrane-associated proteins), using two-dimensional difference gel electrophoresis (2-D DIGE), revealed that, within 2 h of auxin treatment, at least 16 protein spots were up- or down-regulated by IAA. These proteins were identified using reverse-phase liquid chromatography electrospray tandem mass spectrometry. Four of these proteins were detected in the growth-controlling outer epidermis and were further analysed. One epidermal polypeptide, a small Ras-related GTP-binding protein, was rapidly down-regulated by IAA (after 0.5 h of incubation) by −35% compared to the control. Concomitantly, a subunit of the 26S proteasome was up-regulated by IAA (+30% within 1 h). In addition, this protein displayed IAA-mediated post-translational modification. The implications of these rapid auxin effects with respect to signal transduction and IAA-mediated secretion of glycoproteins (osmiophilic nano-particles) into the growth-controlling outer epidermal wall are discussed.
Keywords: Auxin, epidermis, cell elongation, cell wall, Golgi secretion, proteomics
INTRODUCTION
Eight decades ago, the botanist Frits Went (1903–1990) demonstrated the existence of a growth-promoting hormone, named auxin (indole-3-acetic acid, IAA), in developing oat coleoptiles. Went’s thesis of 1928, which was published 7 years later in an international journal (Went 1935), became a scientific classic, and his ‘oat coleoptile technique’ was soon accepted as the standard procedure for bioassay of auxins. In the 1930s it was shown that IAA increases the rate of elongation of excised, turgid coleoptile sections by altering the mechanical properties of the cell wall (Went 1935; Heyn 1940).
These studies led to the discovery that not all cell walls of the coleoptile display the same mechanical properties. Biophysical analyses of axial plant organs demonstrated that the thick, extension-limiting outer epidermal wall represents the structure of the organ that determines the rate of elongation, whereas the thin-walled, turgid inner tissues provide the driving force for growth (Kutschera 2001, 2003, 2008; Schopfer 2006). Hence, at least in maize (Zea mays), auxin-induced coleoptile elongation is mediated via IAA-regulated wall-loosening (and -stiffening) processes that are restricted to the peripheral organ wall (Kutschera 2001, 2003; Schopfer 2006).
The biophysical basis of auxin-induced coleoptile growth has been elucidated in some model plants such as maize. However, the biochemical process(es) that cause IAA-mediated changes in the mechanical properties of the extension-limiting peripheral organ wall are not yet clear (Schopfer et al. 2002; Cosgrove 2005; Kutschera 2006). The aims of this study were to re-investigate the general hypotheses that auxin may (a) primarily act in the epidermis of axial organs and (b) mediate wall expansion and thus stem elongation via the enhancement (or reduction) in the rate of biosynthesis of so-called ‘growth-related proteins’ in cells of the outer tissue layer (Edelmann & Schopfer 1989; Edelmann et al. 1989, 1995; Edelmann & Kutschera 1993; Schenck et al. 2010).
In order to detect possible hormone-induced changes, two-dimensional (2-D) difference gel electrophoresis (DIGE), combined with mass spectrometry, was employed (Deng et al. 2007; Tang et al. 2008 a,b). We selected coleoptiles of etiolated rye (Secale cereale) seedlings for our experimental analysis. This plant material has been used before to study the cytological basis of auxin action (Edelmann & Kutschera 2002; Kutschera 2003; Kutschera et al. 2010). However, the role of the outer epidermis with respect to IAA action has not yet been analysed in this cereal species.
MATERIALS AND METHODS
Plant material and growth measurements
Caryopses of rye (Secale cereale L. cv. Picasso) were soaked for 2 h in water and thereafter germinated in moist standard potting soil (Pro-mix PGX, Premier Horticulture Ltd., Quakertown, PA, USA) in closed transparent plastic boxes (darkness, 25 ± 0.5 °C) for 3 d. Experiments were carried out on 15-mm segments excised from the region 2 mm below the tip of the coleoptile (Fig. 1A). After excision and removal of the enclosed primary leaf, the segments (40 per experiment) were allowed to equilibrate for 1 h in distilled water in darkness. Batches of 20 auxin-depleted segments were then transferred to Petri dishes containing 10 ml of either distilled water or IAA solution (concentration: 10 μM) and incubated on a shaker (50 rpm) for up to 4 h in the dark (25 ± 0.5 °C).
Fig. 1.
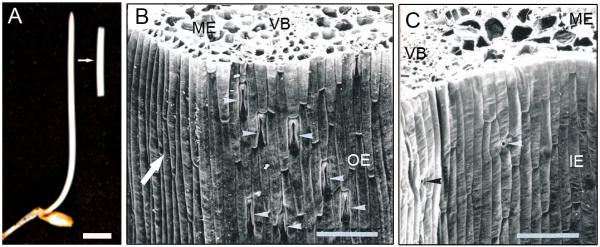
Photograph of a 3-d-old etiolated seedling of rye (Secale cereale) and an excised 15-mm coleoptile section, after removal of the enclosed primary leaf (A). In the scanning electron micrograph of cross-sections of the coleoptile, four different tissue systems are indicated (B, C). IE = inner epidermis, with small stomata (black and white arrowheads), OE = outer epidermis with large stomata in the region above the vascular bundles (white arrowheads), ME = mesophyll, VB = vascular bundle (phloem and xylem). Bars = 0.5 cm in A; 100 μm in B and C. The white arrow in B points to the flat region of the coleoptile from which epidermal peels were isolated for protein extraction.
Growth measurements on individual segments were performed as described in Kutschera et al. (2010), and the change in length (elongation response; unit: mm per segment) calculated. The ‘split section test’ was performed as described in Kutschera et al. (1987).
Electron microscopy
Scanning electron micrographs of transverse sections of 3-d-old rye coleoptiles were performed as described in Fröhlich & Kutschera (1994). For transmission electron microscopy, small pieces of epidermal tissue were cut from the middle of 15-mm sections that were treated for 1 h with IAA. Thereafter, the samples were fixed for 2 h in 2% glutaraldehyde/phosphate buffer (50 mM, pH 7.2), washed and post-fixed for 3 h in 2% OsO4 solution (w/v). Ultrathin sections were cut and electron microscopy performed as described in Fröhlich et al. (1994).
Sample preparation and extraction of microsomal proteins
Twenty auxin-depleted segments were incubated in IAA (10 μM) or distilled water (control). After 0.5 to 2.0 h, the segments were removed from the Petri dish, blotted dry, frozen in liquid nitrogen and stored at −80 °C. In a second set of experiments, 20 auxin-depleted segments were incubated for 1 h in IAA (10 μM) as described above. At the end of the incubation period, single segments were removed from the Petri dishes. Thereafter, one strip of epidermal tissue was cut with a razor blade from the flat side of each coleoptile (Fig. 1B, C) and frozen in liquid nitrogen. About 0.12 g of epidermal tissue was collected per experiment (± IAA). The frozen samples were stored at −80 °C. Microsomal proteins were extracted as described in detail in Kutschera et al. (2010). The protein extract (microsomal fraction) was dissolved in 2-D DIGE buffer (6 M urea, 2 M thiourea, 4% CHAPS) and quantified using a Biorad protein assay.
Two-dimensional difference gel electrophoresis (2-D DIGE)
The 2-D DIGE experiments were carried out as described in Li et al. (2011). In brief, 20 μg of microsomal protein (pH 8.5) from the samples (± IAA) was mixed with 80 pmol of Cy3 and Cy5 minimal dyes (± IAA, respectively), and incubated for 2 to 4 h in darkness on ice. The protein labelling reaction was terminated by adding 0.5 μl lysine (10 mM) to the mixtures. Thereafter, the Cy3- and Cy5-labelled microsomal proteins were combined and used for isoelectrofocusing (IEF), which was performed as described in Li et al. (2011). Cy3- and Cy5-labelled images were obtained using a Typhoon Trio scanner (GE Healthcare, Piscataway, NJ, USA). Analysis of DIGE images was performed as previously described (Li et al. 2011), based on four independent repeats for each auxin incubation experiment.
Analysis of protein spots and mass spectrometry
The mechanical removal of individual, dye-labelled proteins (‘spot picking’), subsequent in-gel digestion by trypsin and reverse-phase liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS) analyses were performed as previously described (Chalkley et al. 2005; Kutschera et al. 2010).
The digests were separated by nanoflow liquid chromatography using a 100-μm × 150-mm reverse-phase Ultra 120-μm C18Q column (Peeke Scientific, Redwood City, CA, USA) at a flow rate of 350 nl min−1 in a Dionex/LC Packing UltiMate NanoFlow high-performance liquid chromatography system. Solvent A was water/0.1% formic acid, and solvent B was acetonitrile/0.1% formic acid; peptides were eluted in a linear gradient from 5% to 25% solvent over 30 min, followed by a short wash in 50% solvent B for 3 min. Half of each digest (5 μl) was injected. The liquid chromatography eluate was coupled to a LTQ mass spectrometer (LTQ, Thermo Scientific, San Jose, CA, USA) equipped with a nano-electrospray ion source.
Peptides were analysed in positive ion mode and in information-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. Acquisitions on the Thermo LTQ were done using a Triple Play method. This consisted of one survey scan at mass range 310–1400, using one 100 ms micro-scan with the AGC setting of 30 000. This was followed by three pairs of zoom scans and CID scans. The zoom scan was over a 10 Da range with one 50 ms micro-scan at an AGC setting of 3 000. The CID scans were one 100 ms micro-scan at an AGC setting of 10 000. The survey and zoom scan were in profile mode, and the CID scans in centroid mode. Peak lists were generated using an in-house script. Enzyme specificity was set to trypsin, and the maximum number of missed enzyme cleavages per peptide was set at one. The number of modifications was limited to two per peptide. Mass tolerance was 0.6 Da for precursor and 0.6 Da for fragment ions. The peaklists were searched using in-house Protein Prospector version 5.7.1 (http://prospector2.ucsf.edu/prospector/html/misc/revhist.htm) (public version at http://prospector.ucsf.edu) against a database that consisted of the Brachypodium protein database, (ftp://ftpmips.helmholtz-muenchen.de/plants/brachypodium/v1.0/) (http://files.brachypodium.org/Annotation/Bradi_1.0.pep.fa.gz), to which a randomised version was concatenated. The database contained all entries for Brachypodium (64510 entries searched) (Vogel et al. 2010). Carbamidomethylation of cysteine was included as a fixed modification; N-acetylation of the N-terminus of the protein, oxidation of methionine, formation of pyro-Glu from N-term-glutamine were all allowed as variable modifications (Tables 1, 2). CID peptide results were reported using a peptide false discovery rate of 0.45% according to concatenated database search results. In all protein identifications, a minimal protein score of 22, a peptide score of 15 and a minimal discriminant score threshold of zero were used for initial identification criteria. Maximum expectation values (number of different peptides with scores equivalent to or better than the result reported that are expected to occur in the database search by chance) for accepting individual spectra were set to 0.05. For identification of possible phosphorylation sites, a second search was performed allowing phosphorylation of serine, threonine or tyrosine as additional variable modifications.
Table 1.
Auxin-responsive proteins identified in the microsomal fraction of rye coleoptiles treated for 2.0 h (± IAA; concentration: 10 μM). In the expression ratio column, positive numbers indicate up-regulated, and negative values denote down-regulated, proteins with respect to IAA treatment (average values of four independent experiments).
| Spot | Protein name | Abund -ance ratio |
P-value (t-test) |
Gene locus | Number of unique peptides |
Sequence coverage (%) |
Best expected value |
|---|---|---|---|---|---|---|---|
| 1 | putative HSP70 | − 1.34 | 0.0060 | Bradi3g53100 | 22 | 21.0 | 2.5 e-7 |
| 2 | alpha-glucosidase-like protein |
− 1.35 | 0.0000011 | Bradi1g69780 | 29 | 24.9 | 7.2 e-7 |
| 3* | phospholipase D | + 1.53 | 0.0029 | Bradi2g04480 | 12 | 18.1 | 1.1 e-8 |
| vacuolar proton- ATPase subunit A |
Bradi1g31690 | 11 | 19.1 | 4.2 e-8 | |||
| 4 | N-ethylmaleimide- sensitive fusion protein |
− 1.25 | 0.0019 | Bradi2g19570 | 28 | −38.1 | 5.1e-8 |
| 5 | N-ethylmaleimide- sensitive fusion protein |
− 1.27 | 0.0064 | Bradi2g19570 | 33 | 40.7 | 1.1e-8 |
| 6* | TCP-1/cpn60 chaperonin family |
− 1.46 | 0.0021 | Bradi5g02890 | 6 | 14.3 | 3.1e-8 |
| protein disulfide isomerase 1 pro-protein |
Bradi4g23180 | 5 | 8.7 | 7.0 e-7 | |||
| 7* | Protein phosphatase 2A A subunit |
− 1.46 | 0.00071 | Bradi4g08720 | 7 | 17.5 | 6.7 e-8 |
| protein disulfide isomerase 1 pro-protein |
Bradi4g23180 | 6 | 10.2 | 1.3e-7 | |||
| 8 | dihydrolipoamide dehydrogenase |
+ 1.59 | 0.0036 | Bradi2g34770 | 40 | 70 | 3.0 e-8 |
| 9 | dihydrolipoamide dehydrogenase |
+ 1.30 | 0.0032 | Bradi2g34770 | 44 | 69.2 | 1.2e-9 |
| 10 | 26S protease regulatory subunit T5 |
− 1.31 | 0.000024 | Bradi3g54490 | 24 | 54.7 | 7.3e-9 |
| 11 | 26S protease regulatory subunit S10B |
+ 1.33 | 0.010 | Bradi1g36400 | 26 | 60.8 | 9.3e-9 |
| 12 | 26S protease regulatory subunit S10B |
+ 1.48 | 0.018 | Bradi1g36400 | 20 | 54.1 | 1.7e-8 |
| 13* | 26S protease regulatory subunit S10B |
− 1.46 | 0.00018 | Bradi1g36400 | 11 | 33.2 | 1.1e-6 |
| large subunit ribosomal protein L4e |
Bradi1g55510 | 7 | 24.4 | 3.9e-8 | |||
| 14 | RNA helicase 2 | + 1.41 | 0.00050 | Bradi3g60590 | 10 | 17.3 | 3.3e-6 |
| 15 | putative protein | + 1.32 | 0.017 | Bradi2g43230 | 7 | 15.9 | 1.1e-5 |
| 16* | small Ras-related GTP- binding protein |
− 1.33 | 0.0000077 | Bradi2g15730 | 10 | 50.5 | 2.3e-6 |
| putative 40S ribosomal protein S3 |
Bradi3g16150 | 8 | 31.0 | 2.0e-9 |
Two proteins were identified in the same spots.
Table 2.
Auxin-responsive proteins identified in the microsomal fraction of the epidermal cells. Rye coleoptile segments were incubated for 1 h in the absence (−IAA) or presence (+IAA) of auxin (concentration: 10 μM) before the epidermal cell layer was isolated (average values of four independent experiments). See Table 1 for details.
| Spot | Protein name | Abundance ratio |
P-value (t-test) |
Gene locus | Unique peptides |
Sequence coverage (%) |
Best expected value |
|---|---|---|---|---|---|---|---|
| 1 | 26S protease regulatory subunit S10B |
+ 1.30 | 0.0004 | Bradi1g36400 | 25 | 65.6 | 9.4e -8 |
| 2 | 26S protease regulatory subunit S10B |
+ 1.26 | 0.0008 | Bradi1g36400 | 9 | 30.9 | 1.1e -7 |
| 3 | Uncharacterised protein |
+ 1.25 | 0.0003 | Bradi2g43230 | 7 | 15.9 | 1.6e -5 |
| 4 | small Ras-related GTP-binding protein |
− 1.35 | 0.0022 | Bradi2g15730 | 2 | 9.5 | 6.4e-5 |
When several accession numbers in the database matched the same set of peptides identified, the entries with the most descriptive name were reported. Individual isoforms of proteins were reported according to the detection of peptides unique to their sequences. If several isoforms shared the same set of peptides identified, they were all reported. Only proteins with at least two peptides identified were further considered. In order to assign the modification site for peptides containing post-translational modifications, the MS/MS spectrum was reinterpreted manually by matching the observed fragment ions to a theoretical fragmentation obtained using MS Product (Protein Prospector).
RESULTS
Elongation of coleoptile segments in response to auxin
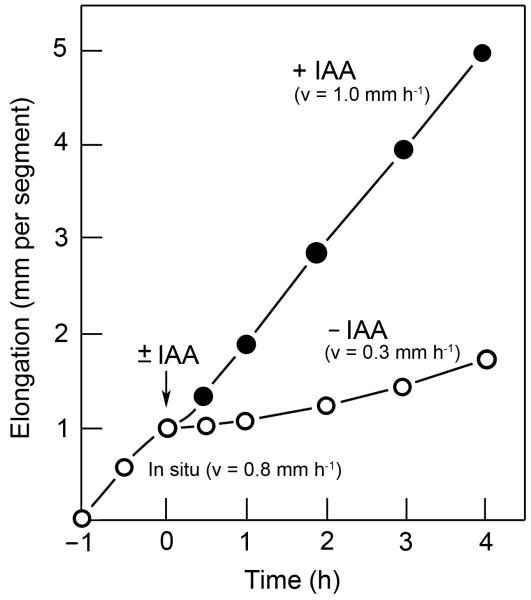
Isolated 15-mm segments cut from the sub-apical region of the coleoptiles of 3-d-old etiolated rye seedlings (Fig. 1A–C) continue to elongate in the absence of exogenous auxin at a rate of 0.8 mm h−1. About 30 min after excision, this endogenous growth response in water slows down and, within the subsequent 2 h, reaches a minimum. As Fig. 2 shows, these auxin-depleted coleoptile sections display a rapid elongation response upon the addition of IAA. We applied auxin at an optimal concentration of 10 μM. In response to this external provision of IAA, the endogenous growth of the organ was re-established within ca. 15 min after hormone treatment. In the presence of IAA, a steady-state growth rate of ca. 1.0 mm h−1 was recorded over the subsequent 4 h, which was about 20% higher than that measured in the intact, etiolated coleoptile (organ growth in situ).
Fig. 2.
The elongation response of excised 15-mm sections of etiolated rye coleoptiles to added auxin (IAA, concentration: 10 μM). One hour after cutting (= time zero), batches of ten coleoptile sections were either incubated in the absence (−IAA) or presence (+IAA) of auxin and the elongation response recorded. Data represent means of six independent experiments. The standard errors of the means (± SEM) are not shown because they are smaller than the size of the symbols. v = average rate of growth.
The epidermis as the target tissue for auxin action
Experiments with excised coleoptile and stem segments from etiolated maize and pea seedlings have shown that, when the turgid organs are split lengthwise through a medium cut, the halves bend outwards due to the release of tissue tension (Kutschera et al. 1987). A similar, tissue-specific response occurs when auxin-depleted rye coleoptile segments are incubated in water. In the presence of IAA, the outside elongates in response to the hormone, resulting in an inward bending of the split portions (Fig. 3A, B). This ‘split section curvature test’ documents that the thick, extension-limiting outer wall of the outer epidermis (Fig. 1B) displays a considerable elongation response to the added hormone, causing the conspicuous inward curvature of the organ halves. It should be noted that, at the outer epidermal wall of rye coleoptiles, IAA causes the appearance of osmiophilic nano-particles that are secreted via the Golgi system of the cells (Fig. 3C). These secretion products have been characterised in detail in previous reports (Fröhlich et al. 1994; Edelmann & Kutschera 2002).
Fig. 3.
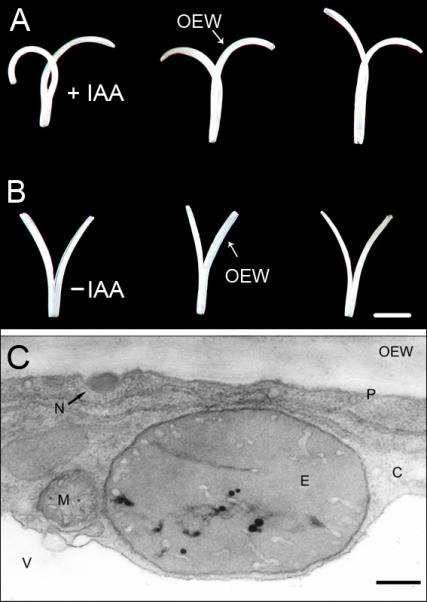
Effect of auxin (IAA, concentration: 10 μM) on the growth of split coleoptile sections, 15 mm in length, that were cut from 3-d-old etiolated rye seedlings (see Fig. 1). (A, B). After 24 h of incubation in the presence (+IAA) or absence (−IAA) of auxin, large differences are apparent in the three representative organ segments depicted here. IAA causes an inward curvature of the split halves via a selective promotion of the elongation of the growth-limiting outer epidermal wall. Transmission electron micrograph of a cross-section of an epidermal cell from an IAA-treated rye coleoptile segment (incubation time: 1 h; auxin concentration: 10 μM). (C). Note the electron-dense osmiophilic nano-particle in the periplasmic space between the plasma membrane and the cytoplasm. C = cytoplasm, E = etioplast, M = mitochondrion, N = osmiophilic nano-particle, OEW = outer epidermal wall, P = plasma membrane, V = vacuole. Bars = 1 cm (A, B); 0.5 μm (C).
Taken together, the data of Fig. 3A–C document that the outer epidermis appears to be the primary target tissue of IAA action in the coleoptile of etiolated rye seedlings. Hence, in the proteomic studies described below, we first analysed the microsomal (i.e. membrane-associated) proteins extracted from entire coleoptile sections and, in the second step, those obtained from the outer epidermis.
Quantitative analysis of proteomic changes
The coleoptile of the rye seedling is composed of four tissue systems: outer epidermis, mesophyll, vascular bundles (phloem, xylem) and inner epidermis (Fig. 1B, C). In the regions above the vascular bundles, large dumbbell-shaped stomata are present (length 40–50 μm; Fig. 1B). These specialised two-cell systems for the regulation of gas exchange are lacking in the epidermal layer that covers the flat sides of the organ. On the inner epidermis of the coleoptile, much smaller stomata were observed (length ca. 20 μm) and documented using scanning electron micrographs (Fig. 1C).
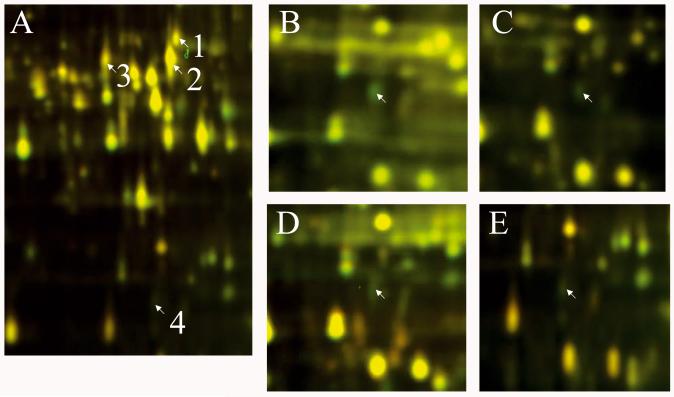
In a first set of experiments, we incubated auxin-depleted segments for 0.5, 1.0 and 2.0 h in IAA (10 μM) and, at the end of the corresponding time period, extracted microsomal proteins for quantitative 2-D DIGE analyses. A comparison of the two-dimensional DIGE maps (2 h ± IAA) revealed at least 16 protein spots that were up- or down-regulated by IAA (Fig. 4). As Table 1 shows, the abundance ratios of these proteins varied between −1.35 and +1.59, with positive values denoting up-regulated and negative values down-regulated proteins (corresponding to red and green spots in Fig. 4).
Fig. 4.
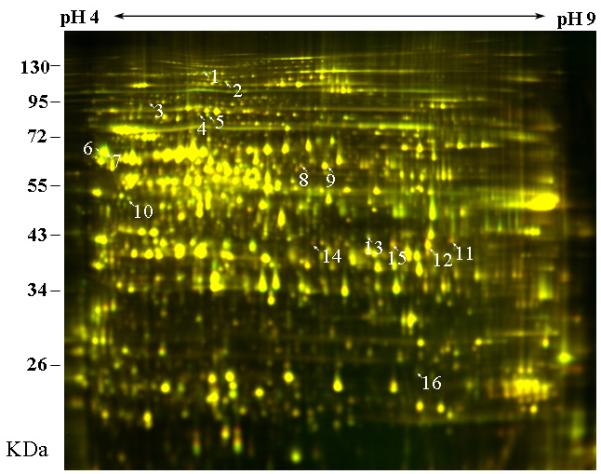
Effects of auxin (IAA) on the proteome of rye coleoptile, as revealed with two-dimensional DIGE analysis of microsomal proteins. Segments, 15 mm in length, were incubated for 2 h in the presence or absence of auxin (± IAA, concentration: 10 μM). Thereafter, membrane-associated proteins were extracted and labelled with Cy3 dye (−IAA) or C5 dye (+IAA). The labelled microsomal proteins were mixed and separated on the first dimension with 24-cm, pH 3 to pH 10, NL IPG strips, and thereafter on a second dimension with a 10% polyacrylamide SDS-PAGE gel. The images were analysed with a Typhoon Trio scanner and superimposed. Protein spots up-regulated in the presence of IAA appear in red, and those down-regulated in green. Spots that are unaffected by the hormone are yellow.
In a second series of experiments we extracted microsomal proteins from epidermal tissue isolated from 15-mm coleoptile segments incubated for 0.5 and 1.0 h in IAA. The corresponding 2-D DIGE maps (± IAA) revealed four protein spots that were up- or down-regulated by the hormone (Fig. 5A). The expression (i.e. abundance) ratios of these 16 and four proteomic changes, respectively, are listed in Tables 1 and 2. All of these changes were statistically significant (P-values < 0.05; n = 4 independent experiments).
Fig. 5.
Effects of auxin (IAA) on the proteome of the outer epidermis in rye coleoptile segments, as revealed by two-dimensional DIGE analysis of microsomal proteins (A). Coleoptile segments were incubated for 1 h in the presence or absence of auxin (± IAA, concentration: 10 μM). At the end of the incubation period, epidermal strips were cut from the flat side of the organ segments, as indicated in Fig. 1B. The down-regulated protein spot no. 4 occurs in intact coleoptile segments incubated for 0.5 (B), 1.0 (C) and 2.0 h (D) in IAA and, in an independent second experiment, after 1.0 h of auxin treatment in epidermal cells (E). As in Fig. 4, protein spots that are up-regulated in the presence of IAA appear in red, and those down-regulated in green. Proteins that are not affected by the hormone are yellow.
Protein identification with tandem mass spectrometry and quantitative analysis
The 16 and four protein spots, respectively, that were up- or down-regulated by IAA, as revealed and quantified by two-dimensional DIGE (Figs 4, 5), were subjected to protein identification with tandem mass spectrometry (LC-MS/MS) analysis (Tang et al. 2008a,b). A representative mass spectrum and the corresponding interpretation are shown in Fig. 6. Based on the published protein database for the model grass Brachypodium distrachyon (Vogel et al. 2010), the 16 spots differentially up- or down-regulated in the entire coleoptile by IAA were identified (Table 1). In five of the 16 spots (Nos. 3, 6, 7, 13 and 16), two different proteins were identified and listed in the corresponding column. It should be noted that no up-regulated glycoproteins related to the extracellular osmiophilic nano-particles (Fig. 3C) were discovered.
Fig. 6.
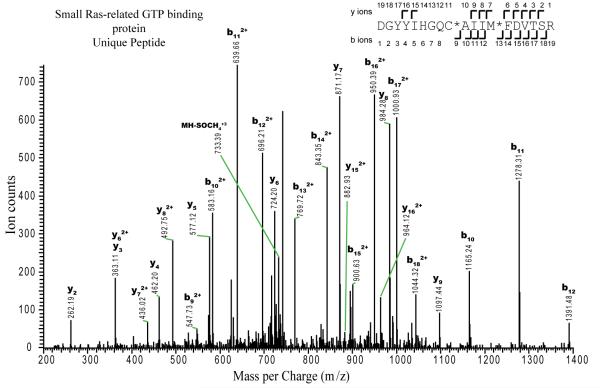
Tandem mass spectrum of a triple-charged precursor ion with mass per charge ratio (m/z) of 754.5048, observed during the LC-MS/MS analysis of the tryptic digest of spot 16 (see Fig. 4). Database searching identified this spectrum as corresponding to the peptide spanning residues 80–98 of the small Ras-related GTP binding protein of Brachypodium (theoretical mass of the triple protonated peptide would be 754.6821; measured mass error −0.18 Da). Fragment ions labelled as yn are C-terminal fragment ions, where the number corresponds to the number of amino acids present. Ions labelled as bn are N-terminal ions containing the indicated number of amino acids. The fragments observed are annotated on the peptide sequence as angled marks. Amino acids in the sequence are indicated using their one-letter abbreviations: C*, carbamidomethyl cysteine; M*, oxidised methionine; MH2-SOCH42+, side-chain loss of oxidised methionine from precursor ion.
In a second series of experiments, four IAA-modulated, epidermal membrane-associated proteins were identified and found to be identical to four of those detected in the entire, IAA-treated coleoptile (Table 2). Two of these four epidermal proteins, the up-regulated 26S protease regulatory subunit S10B (spots 11, 12, and 1, 2 in Figs 4 and 5 A, respectively) and the down-regulated small Ras-related GTP-binding protein (spots 16 and 4 in Figs 4 and 5 A, respectively), were analysed in more detail. The promotive effect of IAA on the up-regulated 26S-protease subunit (+30%) was detected after 1 and 2 h. In addition, our proteomic data indicate that this protein was modified post-translationally by the hormone, since its basic forms increased while its acetic form decreased (data not shown). In intact coleoptile segments, the inhibitory action of auxin on the down-regulated GTP-binding protein (−35%) occurred 0.5, 1.0 and 2.0 h after the addition of IAA (Fig. 5B, C, D). The same effect was detected in the epidermal cells removed from segments that were treated for 1 h with auxin (Fig. 5E) (n = 4 independent experiments each).
DISCUSSION
Inhibitor studies have shown that auxin-mediated cell wall loosening and turgor-driven coleoptile elongation are dependent on (1) the steady biosynthesis of certain ‘growth-relate’” polypeptides and (2) the secretion of not yet identified glycoproteins that are incorporated into the extension-limiting outer epidermal wall (Kutschera et al. 1987; Edelmann & Schopfer 1989; Edelmann et al. 1989, 1995; Fröhlich et al. 1994). The basic tenets of this so-called ‘gene expression hypothesis’ of primary IAA action (1) have recently been re-investigated by Schenck et al. (2010). Using hypocotyl sections cut from Arabidopsis thaliana seedlings, these authors monitored IAA-mediated gene expression and organ elongation during the early phase of hormone action. Although IAA-induced gene expression (i.e. protein biosynthesis) is clearly imperative for sustained hormone-mediated organ growth, the authors concluded that the initiation of cell elongation is not dependent on ‘global gene expression’ (Schenck et al. 2010).
In this study we focussed on the second aspect of the ‘auxin protein biosynthesis’ question outlined above (2). Specifically, we addressed the question of whether IAA rapidly up- or down-regulates certain proteins in the growth-controlling outer epidermis of the rye coleoptile. These proteins may be related to the secretion of specific glycoproteins, as visualised in the osmiophilic nano-particles (Fig. 3C), which are incorporated into the peripheral organ wall (Kutschera et al. 1987; Hoffmann-Benning et al. 1994; Kutschera 2001, 2003; Edelmann & Kutschera 2002).
The effects of auxin on protein biosynthesis have been investigated in numerous studies using entire coleoptile (or stem) segments as experimental material (Dietz et al. 1990). However, as Fig. 1B, C show, this ‘simple’ model organ for auxin research is composed of four tissue systems (outer and inner epidermis; mesophyll, vascular bundles). Moreover, in the outer and inner epidermal cell layers, stomata of different size and shape are apparent, so that protein extracts obtained from excised segments (± IAA) represent mixtures of at least six different cell types with specific functions (i.e. sucrose transport in the phloem, gas exchange via the stomata, etc.). A recent report has shown that stomatal movements are, at least in part, caused by an elongation response of the guard cells (Meckel et al. 2007). These novel data document that organ growth and CO2/water exchange with the environment are interrelated processes. However, as pointed out by these authors, little is known about the interaction between stomatal movements and cell elongation.
In this study, we first confirmed for the coleoptile of rye that the ‘epidermal growth-control theory’ of primary auxin action, which was proposed two decades ago based on experiments with etiolated maize seedlings (Kutschera et al. 1987; Kutschera & Niklas 2007), accounts for the hormone-induced elongation response in Secale cereale (Fig. 3A, B). In a first set of proteomic experiments, we identified auxin-mediated up- and down-regulated proteins in intact coleoptile segments and thereafter searched for changes in these proteins detectable in the outer epidermis (on the flat side of the coleoptile, i.e. the cell layer without stomata). In a recent study we have shown that cessation of coleoptile elongation and loss of auxin sensitivity in etiolated rye seedlings is associated with degradation of the vacuolar H+ ATPases (V-ATPases) of the cells (Kutschera et al. 2010). These electrogenic proton pumps are integral components of the tonoplast and membranes of the Golgi-dependent secretory pathway. Here we document that, in entire coleoptiles, subunit A of a vacuolar proton ATPase is up-regulated by +53% under the influence of IAA (spot 3 in Fig. 4; Table 1). This finding indicates that the hormone may promote the biosynthesis of this membrane-associated protein. However, in the growth-controlling outer epidermis, this IAA effect was not detected. Hence, the significance of this proteomic change with respect to the regulation of organ expansion is unclear.
As Tables 1 and 2 indicate, at least two proteins of interest were identified in both the intact organ and the epidermal cells: an up- and a down-regulated protein, respectively (26S protease regulatory subunit S10B, +30% after 2.0 h, and a small Ras-related GTP-binding protein, −35% after 0.5 h of auxin treatment). The significance of these proteomic changes for the initiation of cell elongation may be as described below.
It is well known that the evolutionary conserved central eukaryotic protease, the 26S proteasome, degrades specific proteins via an ATP- and ubiquitin-dependent digestion mechanism (Voges et al. 1999; Smalle & Vierstra 2004). In plants, this multi-catalytic enzyme complex modulates organ growth through the selective removal of short-lived regulatory proteins (Huang et al. 2006; Kurepa et al. 2009; Zhang et al. 2011). In hormone-sensitive plant organs, the 26S proteasome is known to play a critical role in auxin signalling by the degradation of AUX/IAA transcription repressors, and hence affects IAA-modulated transcriptional regulation (Teale et al. 2006; Santner & Estelle 2010). Our data show that, in epidermal cells, one subunit of this proteolytic complex (S10B, an ATPase) is rapidly up-regulated by IAA and post-translationally modulated. We suggest that auxin regulates the stability of components of its signalling pathway, but more work is required to further support this interpretation.
In addition, we discovered a rapid effect of IAA on the down-regulation of a small Ras-related GTP-binding protein in epidermal cells of the rye coleoptile. It has long been known that small GTP-binding proteins are involved in vesicle-mediated protein transport from the cytoplasm to the plasma membrane (Verma et al. 1994; Thiel & Battey 1998; Takai et al. 2001). Since auxin action on wall expansion in the grass coleoptile is related to the delivery of glycoproteins via the Golgi-dependent secretory pathway (Kutschera 2001, 2003; Edelmann & Kutschera 2002), we suggest that the rapid IAA-effect on the small Ras-related GTP-binding protein is a causal link in an unknown chain of events that leads from auxin perception to the rapid mechanical loosening and turgor-driven expansion of the growth-controlling outer epidermal wall (Xu et al. 2010). The finding that these evolutionary, highly conserved eukaryotic small GTP-binding proteins are required for Golgi-mediated vesicle traffic supports this suggestion (Verma et al. 1994; Thiel & Battey 1998; Takai et al. 2001). However, our quantitative proteomic analysis revealed no up-regulated glycoprotein in the outer epidermis, indicating that IAA does not rapidly promote the biosynthesis of novel cell wall components. As suggested previously (Kutschera 2003), IAA may stimulate the Golgi-mediated secretion of osmiophilic wall material that is synthesised at a rate independent of the auxin level within the cells.
In summary, the results of this study indicate that auxin signalling and action in the growth-controlling epidermis of the rye coleoptile are interconnected processes, but more work is required to further corroborate this general conclusion.
ACKNOWLEDGEMENTS
This project was supported by the United States Department of Energy (Grant DE-FG 02-04ER15525) and National Science Foundation (Grant NSF 0724688 to Z.-Y.W. and A.L.B), the National Institute of Health (Grant 2R01 G 11066258 to Z.-Y.W.) and the Alexander von Humboldt-Foundation, Bonn, Germany (AvH-Fellowships Stanford 2008–2010 to U. K.). Mass spectrometry analysis was performed by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director), supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources (NIH NCRR P41RR001614). Z. D. was supported, in part, by the Zhejiang Academy of Agricultural Sciences, China.
REFERENCES
- Chalkley RJ, Baker PR, Huang L, Hansen KC, Allen NP, Rexach M, Burlingame AL. Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer. Molecular and Cellular Proteomics. 2005;4:1194–1204. doi: 10.1074/mcp.D500002-MCP200. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Deng Z, Zhang X, Tang W, Oses-Prieto JA, Suzuki N, Gendron JM, Chen H, Guan S, Chalkley RJ, Peterman TK, Burlingame AL, Wang ZY. A proteomic study of brassinosteroid response in Arabidopsis. Molecular and Cellular Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz A, Kutschera U, Ray PM. Auxin enhancement of mRNAs in epidermis and internal tissues of the pea stem and its significance for control of elongation. Plant Physiology. 1990;93:432–438. doi: 10.1104/pp.93.2.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann HG, Schopfer P. Role of protein and RNA synthesis in the initiation of auxin-mediated growth in coleopotiles of Zea mays L. Planta. 1989;179:475–485. doi: 10.1007/BF00397587. [DOI] [PubMed] [Google Scholar]
- Edelmann HG, Bergfeld R, Schopfer P. Role of cell wall biogenesis in the initiation of auxin-mediated growth in coleoptiles of Zea mays L. Planta. 1989;179:486–494. doi: 10.1007/BF00397588. [DOI] [PubMed] [Google Scholar]
- Edelmann HG, Bergfeld R, Schopfer P. Effect of inhibition of protein glycosylation on auxin-induced growth and the occurrence of osmiophilic particles in maize (Zea mays L.) coleoptiles. Journal of Experimental Botany. 1995;46:1745–1752. [Google Scholar]
- Edelmann HG, Kutschera U. Long-term effect of auxin on cell elongation in rye coleoptiles: ultrastructural investigations. Journal of Applied Botany. 2002;76:159–162. [Google Scholar]
- Fröhlich M, Hodick D, Kutschera U. Thickness and structure of the cell walls in developing rye coleoptiles. Journal of Plant Physiology. 1994;144:714–719. [Google Scholar]
- Fröhlich M, Kutschera U. Chloroplast development in rye coleoptiles. Botanica Acta. 1994;107:12–17. [Google Scholar]
- Heyn ANJ. The physiology of cell elongation. Botanical Review. 1940;6:515–574. [Google Scholar]
- Hoffmann-Benning S, Komparens KL, Kende H. Characterization of growth-related osmiophilic particles in corn coleoptiles and deepwater rice internodes. Annals of Botany. 1994;74:563–572. [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26 S proteasome is required for specifying leaf adaxial identity. The Plant Cell. 2006;18:2479–2492. doi: 10.1105/tpc.106.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Wang S, Li Y, Zaitlin D, Pierce AJ, Smalle JA. Loss of 26 S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiology. 2009;150:178–189. doi: 10.1104/pp.109.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U. Stem elongation and cell wall proteins in flowering plants. Plant Biology. 2001;3:466–480. [Google Scholar]
- Kutschera U. Auxin-induced cell elongation in grass coleoptiles: a phytohormone in action. Current Topics in Plant Biology. 2003;4:27–46. [Google Scholar]
- Kutschera U. Acid growth and plant development. Science. 2006;311:952–953. doi: 10.1126/science.311.5763.952b. [DOI] [PubMed] [Google Scholar]
- Kutschera U. The outer epidermal wall: Design and physiological role of a composite structure. Annals of Botany. 2008;101:615–621. doi: 10.1093/aob/mcn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U, Bergfeld R, Schopfer P. Cooperation of epidermis and inner tissues in auxin-mediated growth of maize coleoptiles. Planta. 1987;170:168–180. doi: 10.1007/BF00397885. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Niklas KJ. The epidermal-growth-control theory of stem elongation: an old and a new perspective. Journal of Plant Physiology. 2007;164:1395–1409. doi: 10.1016/j.jplph.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Deng Z, Oses-Prieto JA, Burlingame AL, Wang Z-Y. Cessation of coleoptile elongation and loss of auxin sensitivity in developing rye seedlings: A quantitative proteomic analysis. Plant Signaling and Behavior. 2010;5:509–517. doi: 10.4161/psb.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xu S-L, Oses-Prieto JA, Putil S, Xu P, Wang R-J, Li KH, Maltby DA, An L-H, Burlingame AL, Deng Z-P, Wang Z-Y. Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis. Molecular Plant. 2011;4:361–374. doi: 10.1093/mp/ssq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckel T, Gall L, Semrau S, Hormann U, Thiel G. Guard cells elongate: Relationship of volume and surface area during stomata movement. Biophysical Journal. 2007;92:1072–1080. doi: 10.1529/biophysj.106.092734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. The Plant Journal. 2010;61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H. Rapid auxin-induced cell expansion and gene expression: A four-decade-old question revisited. Plant Physiology. 2010;152:1183–1185. doi: 10.1104/pp.109.149591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P. Biomechanics of plant growth. American Journal of Botany. 2006;93:1415–1425. doi: 10.3732/ajb.93.10.1415. [DOI] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A, Bechthold M, Frahry G, Wagner A. Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta. 2002;214:821–828. doi: 10.1007/s00425-001-0699-8. [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26 S proteasome proteolytic pathway. Annual Review of Plant Biology. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiological Reviews. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang Z-Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008a;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Oses-Prieto JA, Suzuki N, Zhu S, Zhang X, Burlingame AL, Wang Z-Y. Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Molecular and Cellular Proteomics. 2008b;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. Auxin action: Signalling, transport and the control of plant growth and development. Nature Reviews Molecular Cell Biology. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Thiel G, Battey N. Exocytosis in plants. Plant Molecular Biology. 1998;38:111–125. [PubMed] [Google Scholar]
- Verma DPS, Cheon C-I, Hong Z. Small GTP-binding proteins and membrane biogenesis in plants. Plant Physiology. 1994;106:1–6. doi: 10.1104/pp.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickel P, Baumeister W. The 26 S proteasome: a molecular machine designed for controlled proteolysis. Annual Review of Biochemistry. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Went FW. Auxin, the plant-growth hormone. Botanical Review. 1935;1:163–182. [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu M-J, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143:99–110. doi: 10.1016/j.cell.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Luo D, Zeng M, Huang H, Cui X. Convergence of the 26 S proteasome and the REVOLUTA pathways in regulating inflorescence and floral meristem functions in Arabidopsis. Journal of Experimental Botany. 2011;62:359–369. doi: 10.1093/jxb/erq277. [DOI] [PMC free article] [PubMed] [Google Scholar]