Abstract
AIM: To understand the interaction of human IQGAP1 and CDC42, especially the effects of phosphorylation and a cancer-associated mutation.
METHODS: Recombinant CDC42 and a novel C-terminal fragment of IQGAP1 were expressed in, and purified from, Escherichia coli. Site directed mutagenesis was used to create coding sequences for three phosphomimicking variants (S1441E, S1443D and S1441E/S1443D) and to recapitulate a cancer-associated mutation (M1231I). These variant proteins were also expressed and purified. Protein-protein crosslinking using 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide was used to investigate interactions between the C-terminal fragment and CDC42. These interactions were quantified using surface plasmon resonance measurements. Molecular modelling was employed to make predictions about changes to the structure and flexibility of the protein which occur in the cancer-associated variant.
RESULTS: The novel, C-terminal region of human IQGAP1 (residues 877-1558) is soluble following expression and purification. It is also capable of binding to CDC42, as judged by crosslinking experiments. Interaction appears to be strongest in the presence of added GTP. The three phosphomimicking mutants had different affinities for CDC42. S1441E had an approximately 200-fold reduction in affinity compared to wild type. This was caused largely by a dramatic reduction in the association rate constant. In contrast, both S1443D and the double variant S1441E/S1443D had similar affinities to the wild type. The cancer-associated variant, M1231I, also had a similar affinity to wild type. However, in the case of this variant, both the association and dissociation rate constants were reduced approximately 10-fold. Molecular modelling of the M1231I variant, based on the published crystal structure of part of the C-terminal region, revealed no gross structural changes compared to wild type (root mean square deviation of 0.564 Å over 5556 equivalent atoms). However, predictions of the flexibility of the polypeptide backbone suggested that some regions of the variant protein had greatly increased rigidity compared to wild type. One such region is a loop linking the proposed CDC42 binding site with the helix containing the altered residue. It is suggested that this increase in rigidity is responsible for the observed changes in association and dissociation rate constants.
CONCLUSION: The consequences of introducing negative charge at Ser-1441 or Ser-1443 in IQGAP1 are different. The cancer-associated variant M1231I exerts its effects partly by rigidifying the protein.
Keywords: CDC42, Cytoskeleton, Protein phosphorylation, Cancer-associated mutation, Protein-protein interaction
INTRODUCTION
The IQGAP family of proteins function at the interface between cellular signalling and the cytoskeleton[1-3]. They receive information from a variety of signalling molecules, including kinases, small GTPases, growth factor receptors and calcium sensors[4-21]. This information is relayed directly to the actin cytoskeleton through interaction with filamentous actin (F-actin) which promotes filament bundling and caps the “barbed ends” of the filaments[22-25]. There are also indirect influences on the actin cytoskeleton mediated through the Wiskott-Aldrich Syndrome Protein (WASP) family[24,26] and with microtubules mediated via cytoplasmic linker protein 170 (CLIP-170) and adenomatous polyposis coli (APC) protein[27,28]. The IQGAP proteins are named after two key regions within them-the calmodulin binding IQ-motifs and GTPase activating protein (GAP) related domain (GRD). Although the GRD does bind the small GTPases CDC42 and Rac1[4], it does not function as the GTPase activator. Indeed, the available evidence suggests that it inhibits the catalytic activity of small GTPases[4,7]. This is consistent with sequence data and structural predictions. GAPs function by inserting an “arginine finger” into the active site of small GTPases which acts as a proton donor in the enzymatic mechanism of GTP hydrolysis[29-31]. IQGAPs lack this arginine residue and are thus not expected to be able to enhance the rate of GTP hydrolysis.
Humans have three IQGAP isoforms, IQGAP1, IQGAP2 and IQGAP3 with the first of these being the best characterised[32]. Like family members from other species they share a common domain organisation in which the actin-binding calponin homology domain (CHD) is at the N-terminus of the protein, the IQ-motifs are approximately in the middle of the primary sequence and the GRD is towards the C-terminus (Figure 1). There is no complete, three-dimensional structure of an IQGAP available, but it is assumed that the various domains fold in such a way to enable communication between them. It is also anticipated that there is considerable capacity for conformational change in the molecule in order to receive, integrate, interpret and output signals. The structures of some isolated domains have been determined. The structure of the CHD from human IQGAP1 has been solved by NMR spectroscopy and an x-ray structure of part of the GRD is also available[33,34]. Molecular modelling has predicted largely α-helical structures for the IQ-motifs[16,35].
Figure 1.
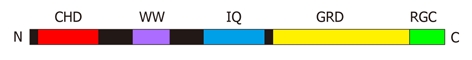
A schematic representation of human IQGAP1. The domains are shown on a linear representation of the protein sequence. The folded protein is unlikely to be arranged in a linear fashion and it is possible that some of these domains contact each other. CHD: Calponin homology domain (residues 44-159); WW: WW domain (679-712); IQ: IQ-motifs containing region (745-864); GRD: GAP-related domain (1004-1237); RGC: Ras-GAP C-terminal domain (1563-1657). The definitions of the domain boundaries are those of Briggs and Sacks[2].
In vitro biochemical studies on IQGAPs have tended to rely on fragments of the protein which are amenable to recombinant expression and purification in bacterial systems. CDC42 and Rac1 interaction with the GRD is promoted by the presence of GTP[4]. Phosphorylation of human IQGAP1 at Ser-1443, however, promotes interaction with CDC42 in the absence of nucleotide[36]. This phosphorylation, along with one at Ser-1441, promotes outgrowth of neurites[37].
Given the protein’s involvement in the transduction of information from signalling pathways to the cytoskeleton, it is not surprising that it has been implicated in various types of cancer[38,39]. However, only one cancer-associated mutation in the coding sequence of the Iqgap1 gene has been identified; this mutation results in the amino acid change M1231I[40]. It is not clear how this change affects the function of IQGAP1, although it does lie in the GRD prompting the hypothesis that it interferes with GTPase binding. However, this has not been tested experimentally.
Here, we identified a novel, biochemically amenable fragment from the C-terminal region of human IQGAP1 and confirmed that it is active, as judged by it ability to bind CDC42 in a crosslinking experiment. We then describe a detailed, quantitative investigation into the affinity of this interaction in the absence of added GTP. To probe the molecular consequences of phosphorylation in this region we used “phosphomicking” variants in which serine residues are replaced with negatively charged ones. We also recapitulated the cancer-associated variant M1231I in order to investigate its binding properties and carried out molecular modelling studies to provide further understanding the consequences of this alteration.
MATERIALS AND METHODS
Expression and purification of wild type and variant human IQGAP C-terminal region
The sequence encoding amino acids 877-1558 in human IQGAP1 was amplified by polymerase chain reaction (PCR) using the Kazusa cDNA clone KIAA0051[41] as a template. The sequence was inserted into the pET-46 Ek/LIC (Merck, Nottingham, United Kingdom) by ligation independent cloning according to the manufacturer’s instructions. Insertion into this vector introduces sequence encoding the amino acids MAHHHHHHVDDDDK at the 5′-end of the coding sequence. The complete coding sequence was verified (MWG Biotech, Ebersburg, Germany). The plasmid was transformed in to competent Escherichia coli (E. coli) HMS174(DE3). Colonies resulting from these transformations were picked and grown, shaking at 37 °C, overnight in 5 mL of Luria-Bertani medium supplemented with 100 μg/mL of ampicillin. This overnight culture was diluted in to 1 L of Luria- Bertani medium supplemented with 100 μg/mL of ampicillin and grown, shaking at 37 °C until the cell density, as estimated by the A600 nm reached 0.6 to 1.0 (typically 3-4 h). The culture was the induced by the addition of 1 mmol/L IPTG and grown for a further 3 h. Cells were harvested by centrifugation (4200 g for 15 min), resuspended in 20 mL of buffer R [50 mmol/L Hepes-OH, pH 7.5, 150 mmol/L sodium chloride, 10 %(v/v) glycerol] and stored, frozen at -80 °C until required.
These cell suspensions were thawed and then disrupted by sonication (three 30 s pulses of 100 W, with 30-60 s gaps in between to allow cooling of the cells). Cell debris was removed by centrifugation (20 000 g for 15 min) and the supernatant applied to a 1 mL nickel-agarose column (His-Select, Sigma, Poole, United Kingdom) which had been previously equilibrated in buffer A [50 mmol/L Hepes-OH, pH 7.5, 500 mmol/L sodium chloride, 10 %(v/v) glycerol]. The cell extract was allowed to pass through the column by gravity flow and the column was washed with 20 mL of buffer A. Protein was eluted with three 2 mL washes of buffer B (buffer A supplemented with 250 mmol/L imidazole). Protein containing fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and dialysed overnight at 4 °C against buffer D (buffer R supplemented with 2 mmol/L dithiothreitol). The protein concentration was determined by the method of Bradford[42], using bovine serum albumin as a standard. Aliquots (50-100 μL) of the protein were stored frozen at -80 °C.
Mutations were introduced in to the coding sequence using the “QuikChange” protocol[43] and verified by DNA sequencing. Each variant protein was expressed in, and purified from, E. coli using essentially the same procedure as outlined above for the wild type.
Expression and purification of human CDC42
The complete coding sequence of human CDC42 was amplified by PCR using IMAGE clone 3626647[44] as a template and inserted into pET-46 Ek/LIC. The DNA sequence of the insert was verified. The expression and purification of the protein was carried out using the same protocol as for IQGAP1-CTD.
Crosslinking of CDC42 and IQGAP
GTP bound CDC42 was prepared by incubating a mixtures of 6 μmol/L CDC42, 0.9 mmol/L GTP and 0.9 magnesium chloride on ice for 30 min. Nucleotide-depleted (ND) CDC42 was prepared by incubating 6 μmol/L CDC42 with 5 mmol/L EDTA on ice for 30 min. Protein-protein crosslinking was carried out using 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). Methods were based on those previously used for the detection of interaction between the atrial myosin essential light chain and F-actin[45,46]. Untreated, GTP-loaded or ND CDC42 (3 μmol/L) was mixed with IQGAP(DR6) (3 μmol/L) and incubated for 30 min at 22 °C. EDC was then added to a final concentration of 0.6 mmol/L and the incubation continued for a further 60 min. Products were analysed by SDS-PAGE.
Surface plasmon resonance
Surface plasmon resonance was measured using a BIAcore 3000 instrument (BIAcore, Uppsala, Sweden). Prior to analysis all proteins were dialysed into HBS Buffer (BIAcore; 10 mmol/L Hepes, pH 7.4, 150 mmol/L NaCl). CDC42 was immobilised onto a CM5 sensor chip (BIAcore) using N-hydroxysuccinimide (NHS)/EDC chemistry. The surface was activated with a mixture of 100 mmol/L NHS and 400 mmol/L EDC for 30 min. CDC42 (25 μmol/L) was then flowed over the surface for two 7 min periods and the surface was then blocked and deactivated with 1 M ethanolamine for 30 min. Immobilisation of CDC42 resulted in a change in the response units (RU) of approximately 1400 RU.
Binding was measured by flowing 0.5 μmol/L to 2.5 μmol/L IQGAP(DR6) over the surface for 300 s (association phase) followed by buffer for 300 s (dissociation phase). In between binding measurements, the surface was regenerated by the injection of sodium hydroxide (5 mmol/L for 220 s). For each binding measurements controls were carried out in parallel in which the protein was flowed over a cell which had been activated with NHS/EDC and blocked with ethanolamine. To determine the response due to interaction between IQGAP(DR6) and CDC42, the readings for the controls were subtracted from the experimental ones. The association and dissociation rate constants (ka and kd, respectively) and the dissociation equilibrium constant (KD) were determined by non-linear curve fitting of the data using BIAevaluation software.
Molecular modelling
The structure of human IQGAP1, residues 962-1345 (PDB 3FAY)[34], was taken as a starting point for molecular modelling studies. This structure file describes one, unbroken polypeptide chain. The selenomethionine residues in this structure were altered to methionine using PyMol (www.pymol.org) and the resulting structure energy minimised using YASARA[47]. Residue 1231 in this minimised structure was altered to isoleucine, and the mutated structure re-minimised using YASARA. Polypeptide flexibility was estimated by generating 500 conformers in the momentum motion type mode of FIRST/FRODAN with an energy cut off of -1 kcal/mol[48,49].
RESULTS
Identification of a biochemically tractable C-terminal fragment of human IQGAP1
Previous reports demonstrated that a fragment beginning at residue 864 and continuing through to the C-terminus of the protein (residue 1657) can be expressed in, and purified from, E. coli, albeit at relatively low levels[4]. We noted that the structure of the Ras-GAP C-terminal domain (RGD), a 112 amino acid residue region at the extreme C-terminus of the protein has been deposited in the Protein Data Bank (PDB ID: 1X0H). From this we reasoned that there must be a domain boundary in the region of residue 1545. Therefore, a region beginning at residue 877 and finishing at 1558 was expressed. This fragment, which we named IQGAP1(DR6), can be purified with good yield, typically 1-2 mg per litre of E. coli culture (Figure 2A). Similar purities and yields were achieved with the various variant proteins also described in this work (data not shown). Full length, recombinant, human CDC42 could also be purified in good yield (Figure 2B).
Figure 2.
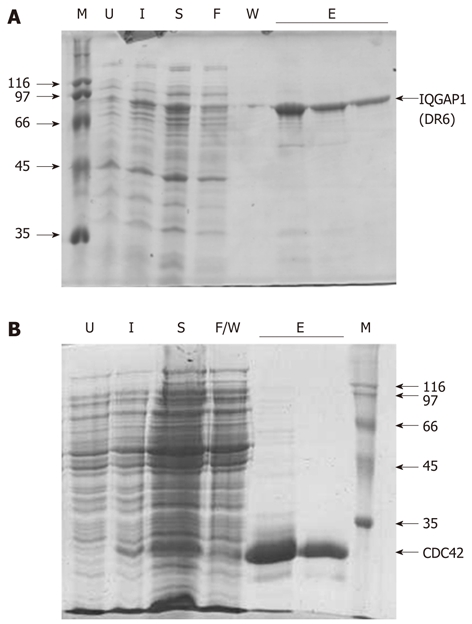
Proteins used in this study. The recombinant expression and purification of (A) IQGAP1(DR6) and (B) CDC42 monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In both (A) and (B), U, extract from cells prior to induction; I, extract from cells 2-3 h after induction; S, soluble material remaining after sonication; F, material which flowed through the column without binding; W, material removed in the washing steps; E, elutions; M, molecular mass markers (with their masses shown to the side of the gel in kDa). In the case of CDC42, F and W were combined into a single sample.
Interactions between CDC42 and IQGAP1(DR6)
Recombinant, human CDC42 was shown to interact with the C-terminal domain fragment. The two proteins could be cross-linked using the reagent EDC which is specific to carboxylate and amino groups (Figure 3). This demonstrates that the recombinant C-terminal fragment is likely to be folded and is functional. The amount of crosslinked product was greatest in the presence of GTP (Figure 3).
Figure 3.
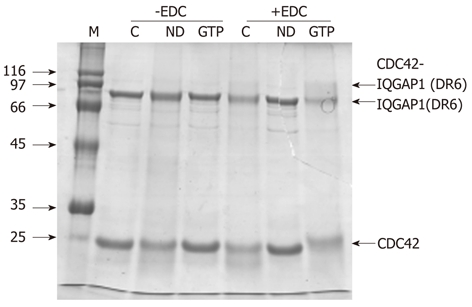
Interaction between IQGAP1 and CDC42. An interaction between IQGAP1(DR6) and CDC42 can be detected by EDC crosslinking in the presence of GTP. In the absence of EDC (lanes headed -EDC), a mixture of IQGAP(DR6) and CDC42 (lane C) behaves the same on SDS-PAGE as IQGAP1(DR6) and nucleotide-depleted CDC42 (lane ND) and as IQGAP1(DR6) and GTP-loaded CDC42 (lane GTP). In the presence of the crosslinker (lanes headed +EDC), crosslinking was not observed between IQGAP(DR6) and CDC42 (C) or IQGAP(DR6) and nucleotide-depleted CDC42 (ND). However an additional band, corresponding to approximately the combined molecular masses of IQGAP(DR6) and CDC42 is seen with GTP-loaded CDC42 (GTP).
Effects of phosphomimicking mutations
To investigate the effects of phosphorylation at serines 1441 and 1443, the phosphomimic variants S1441E, S1443D and S1441E/S1443D were constructed. These amino acid changes insert negative charges into the structure at the sites which can be phosphorylated. Similar mutants have been shown to recapitulate the effects of phosphorylation in an in vivo cell model[37]. Since it has been hypothesised that phosphorylation increases the affinity for CDC42 in the absence of GTP[36], this interaction was investigated by surface plasmon resonance. Interaction between the wild type and immobilised CDC42 could be detected by surface plasmon resonance in the absence of added GTP (Figure 4). Fitting of these data resulted in rate constants for the association and dissociation phases of the reactions (ka and kd) and, consequently, a value for the dissociation constant (KD) (Table 1). It was noted that these fits were not perfect with some non-random residuals (not shown). This may indicate that there is heterogeneity in the preparations and/or that the binding event is more complex. However, for the purposes of comparison, the simple bimolecular interaction model was used throughout.
Figure 4.
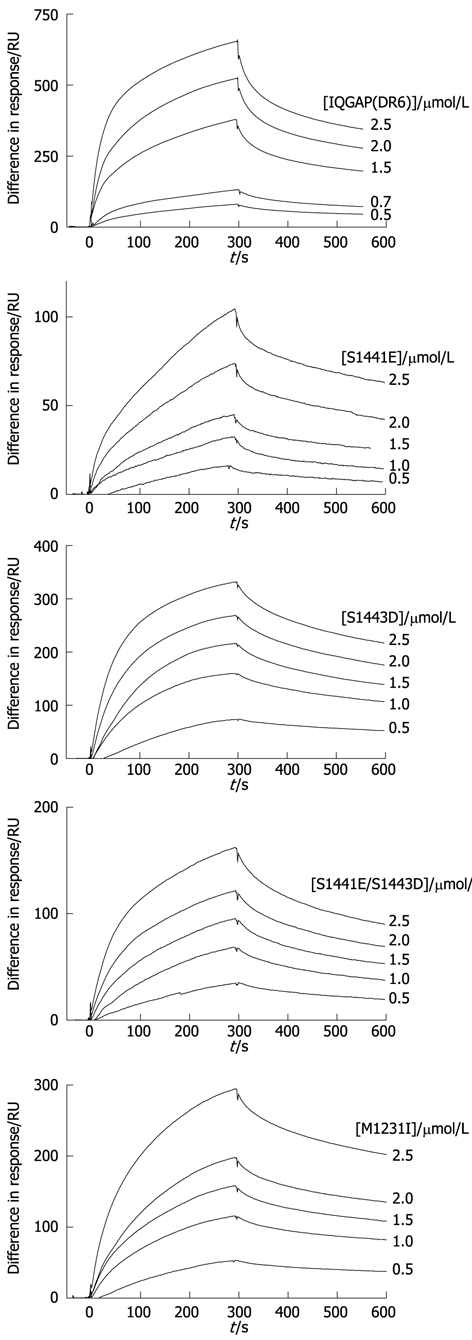
Interaction of IQGAP-CTD and CDC42 can be detected by surface plasmon resonance. Typical sensorgrams resulting from flowing IQGAP1(DR6) wild type and variants over immobilised CDC42 (for conditions, see Materials and Methods). From top to bottom, wild type, S1441E, S1443D, S1441E/S1443D, M1231I. Protein concentrations are shown to the right of the sensorgrams.
Table 1.
Binding parameters for the interaction of IQGAP1(DR6) and CDC42
| IQGAP1(DR6) variant | ka/l.mol-1.s-1 | kd/s-1 | KD/μmol/L |
| Wild type | 4900 ± 100 | (6.5 ± 0.5) × 10-3 | 1.3 ± 0.13 |
| S1441E | 12 ± 1 | (2.7 ± 0.03) × 10-3 | 220 ± 21 |
| S1443D | 5800 ± 100 | (4.7 ± 0.2) × 10-3 | 0.81 ± 0.048 |
| S1441E/S1443D | 3600 ± 100 | (4.0 ± 0.2) × 10-3 | 1.1 ± 0.086 |
| M1231I | 1800 ± 100 | (1.7 ± 0.2) × 10-3 | 0.90 ± 0.16 |
These were determined by surface plasmon resonance. The values are reported ± their standard errors as determined by the BIAevaluation fitting programme (see Materials and Methods).
All three phosphomimic variants also bound to CDC42 in the absence of additional nucleotide. However, in the case of S1441E, the affinity was reduced by two orders of magnitude. This arises mainly because of a reduction in the association rate constant. It should be noted that this reduced value (12 l.mol-1.s-1) is very low and, therefore, may be subject to greater error than the other values. Interestingly, the double mutant (S1441E/S1443D) binds with a similar affinity to the wild type (Table 1).
Effects of the cancer-associated mutation, M1231I
The ability of the disease-associated variant to interact with CDC42 was tested by surface plasmon resonance. These experiments suggest that it is able to do so with similar affinity to the wild type protein. However, both the association and dissociation rate constants are reduced compared to wild type (Table 1).
To help understand the biochemistry of the M1231I variant protein, a molecular model was constructed based on the crystal structure of the GRD. This suggested that the overall fold is not greatly changed by the substitution of this methionine for isoleucine (rmsd between the wild type and variant protein 0.564 Å over 5556 equivalent atoms). The residue lies towards the surface of the protein, away from the predicted GTPase binding site. In addition to gross structural changes, the functions of proteins can be affected by the flexibility of the molecule[50]. Computational estimation of the backbone flexibility of the molecule suggested that the M1231I variation results in changes in flexibility at a number of sites within the protein (Figure 5A). The site with the greatest loss of flexibility is a loop (Ser-1212 to Leu-1217) which links the α-helix containing residue 1231 with residues predicted to play a key role in the CDC42 binding site (Tyr-1192 to Arg-1194; Figure 5B). This loss of flexibility may affect the dynamics of small GTPase interaction.
Figure 5.
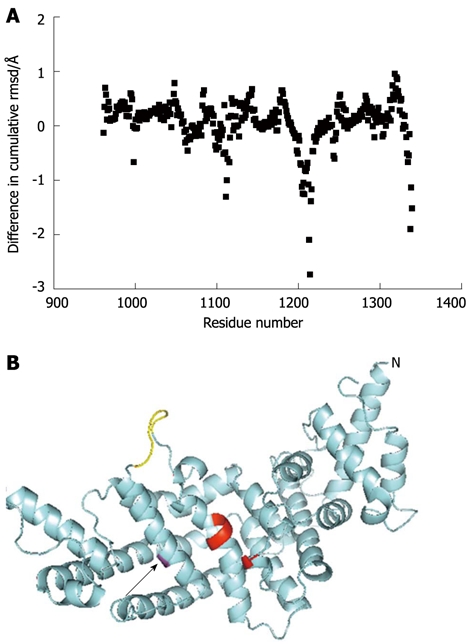
Predicted structural consequences of the M1231I, cancer associated, mutation. A: The predicted effects on backbone flexibility as determined by FIRST/FRODAN (see Materials and Methods) are plotted as the difference in cumulative flexibility between the M1231I variant and wild type. Thus, a negative value represents in loss in flexibility of the variant compared to wild type. The greatest predicted rigidification occurs around residue 1214 and is indicated with an arrow; B: A model of the GAP-related domain of the M1231I variant is shown in cyan, with Ile-1231 in magenta and indicated with an arrow. The N-terminus of the fragment is marked (N) and the C-terminus can be seen close to this in space. Key residues predicted to be involved in CDC42 interaction (Thr-1046 and Tyr-1192 to Arg-1194) are shown in red. A loop (Ser-1212 to Leu-1217) predicted to be more rigid in the variant compared to the wild type is shown in yellow. It links the helix containing residue 1231 and part of the CDC42 binding site.
DISCUSSION
These experiments establish a new fragment from the C-terminal region of human IQGAP1 which is amenable to biochemical analysis. The fragment interacts with CDC42, and the strength of interaction is increased in the presence of GTP. Interestingly, previous work has demonstrated, using isothermal titration calorimetry, an interaction between a C-terminal fragment of IQGAP (residues 962-1345) and GTP-loaded CDC42, but not with CDC42 purified in the absence of added nucleotides (assumed to be GDP-loaded)[34]. This may indicate that the additional residues present in the IQGAP1(DR6) fragment are important in CDC42 interaction in the absence of GTP. The phosphomimicking variants suggest that phosphorylation of the two serine residues has different effects. While the S1443D variant has slightly increased affinity for CDC42, the affinity of S1441E is decreased and introduction of a negative charge at both sites restores the affinity to essentially wild type levels. This suggests that there may be crosstalk between the two serines within the C-terminal domain. The results from the cancer-associated variant emphasise the importance of considering changes in protein flexibility, as well as changes to overall structure.
ACKNOWLEDGMENTS
We thank Elaine Hamilton and Kai Chi Chan for their assistance in the early stages of this project.
COMMENTS
Background
IQGAP family proteins act as “molecular telephone exchanges” collecting information from a variety of cellular signalling pathways, integrating this information and passing it on to the cytoskeleton. They are multi-domain proteins and biochemical studies have tended to concentrate on elucidating the roles of the individual domains. Given their role in organising the cytoskeleton in response to cellular signalling, it is not surprising that IQGAPs are implicated in a number of diseases, including bacterial infections and cancers.
Research frontiers
Human IQGAP1’s activity can be regulated by phosphorylation at serines 1441 and 1443. There is also a cancer-associated mutation, M1231I. All these residues lie in the C-terminal, GTPase-binding region of the protein. The molecular consequences of these changes on the affinity of IQGAP1 for CDC42 have not been investigated in detail previously.
Innovations and breakthroughs
We demonstrate that phosphomimicking alterations at residues 1441 and 1443 have different effects on the affinity for CDC42. The M1231I variant has similar affinity for wild type, but the association and dissociation rate constants are both reduced. Molecular modelling suggests that this variant does not cause any significant structural changes to the protein, but that it does reduce mobility in a key loop which links the residue 1231 to the putative CDC42 binding site.
Applications
The biochemically amenable, functional fragment of human IQGAP1 which we identify will have application in further biochemical studies on this protein. If further cancer-associated mutations are discovered in Iqgap1 it will be interesting to compare results and see if these also alter the flexibility of the protein.
Terminology
Cancer-associated mutation: A mutation in the gene sequence encoding a protein which is associated with a higher risk of cancer. Care should be taken not to assume that the mutation causes cancer. CDC42: A small GTPase involved in regulating the cytoskeleton in eukaryotic cells; Crosslinking: A method for detecting interactions between proteins. It is especially useful for capturing short-lived, transient interactions (which are often found in signalling complexes); IQGAP: A family of cytoskeletal scaffolding proteins. Humans have three-IQGAP1, IQGAP2 and IQGAP3.
Peer review
This manuscript is worthy of publishing because the contents do accord with the Journal scopes and the biochemical characterization provides insight to understand IQGAP/CDC43 interaction. The technique is sound; but the discussion is not thorough.
Footnotes
Peer reviewers: Guillermo Montoya, Dr., Department of Structural Biology and Biocomputing, Spanish National Cancer Research Centre (CNIO), Spanish National Cancer Research Centre (CNIO) Melchor Fdez. Almagro 3, 28029 Madrid, Spain; Yi Wang, Associate Professor, Department of Biochemistry, Baylor College of Medicine, One Baylor Plaza, BCM125, Houston, TX 77030, United States; Chin-Chuan Wei, Professor, Department of Chemistry, Southern Illinois University Edwardsville, PO Box 1652, Edwardsville, IL 62026, United States; Yu Jiang, Associate Professor, Pharmacology and Chemical Biology, University of Pittsburgh, Pittsburgh, PA 15261, United States
Supported by The Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom, No. BB/D000394/1; Action Cancer, Northern Ireland, United Kingdom, No. PG2 2005
S- Editor Cheng JX L- Editor A E- Editor Zhang DN
References
- 1.Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16:242–249. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs MW, Sacks DB. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 2003;542:7–11. doi: 10.1016/s0014-5793(03)00333-8. [DOI] [PubMed] [Google Scholar]
- 4.Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- 5.Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]
- 6.Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders AJ. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen D, Campbell LJ, Littlefield K, Evetts KA, Li Z, Sacks DB, Lowe PN, Mott HR. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: interfaces differ between the complexes. J Biol Chem. 2008;283:1692–1704. doi: 10.1074/jbc.M707257200. [DOI] [PubMed] [Google Scholar]
- 8.Jeong HW, Li Z, Brown MD, Sacks DB. IQGAP1 binds Rap1 and modulates its activity. J Biol Chem. 2007;282:20752–20762. doi: 10.1074/jbc.M700487200. [DOI] [PubMed] [Google Scholar]
- 9.Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci USA. 2007;104:10465–10469. doi: 10.1073/pnas.0611308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25:7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Sacks DB. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J Biol Chem. 2003;278:4347–4352. doi: 10.1074/jbc.M208579200. [DOI] [PubMed] [Google Scholar]
- 12.Mataraza JM, Briggs MW, Li Z, Frank R, Sacks DB. Identification and characterization of the Cdc42-binding site of IQGAP1. Biochem Biophys Res Commun. 2003;305:315–321. doi: 10.1016/s0006-291x(03)00759-9. [DOI] [PubMed] [Google Scholar]
- 13.Swart-Mataraza JM, Li Z, Sacks DB. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J Biol Chem. 2002;277:24753–24763. doi: 10.1074/jbc.M111165200. [DOI] [PubMed] [Google Scholar]
- 14.Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 15.Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, Sacks DB. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem. 1997;272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 16.Atcheson E, Hamilton E, Pathmanathan S, Greer B, Harriott P, Timson DJ. IQ-motif selectivity in human IQGAP2 and IQGAP3: binding of calmodulin and myosin essential light chain. Biosci Rep. 2011;31:371–379. doi: 10.1042/BSR20100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbele GO, Deloulme JC, Gentil BJ, Delphin C, Ferro M, Garin J, Takahashi M, Baudier J. The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. J Biol Chem. 2002;277:49998–50007. doi: 10.1074/jbc.M205363200. [DOI] [PubMed] [Google Scholar]
- 18.Pathmanathan S, Barnard E, Timson DJ. Interactions between the budding yeast IQGAP homologue Iqg1p and its targets revealed by a split-EGFP bimolecular fluorescence complementation assay. Cell Biol Int. 2008;32:1318–1322. doi: 10.1016/j.cellbi.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Pathmanathan S, Elliott SF, McSwiggen S, Greer B, Harriott P, Irvine GB, Timson DJ. IQ motif selectivity in human IQGAP1: binding of myosin essential light chain and S100B. Mol Cell Biochem. 2008;318:43–51. doi: 10.1007/s11010-008-9855-9. [DOI] [PubMed] [Google Scholar]
- 20.Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA, Wang N, et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species--dependent endothelial migration and proliferation. Circ Res. 2004;95:276–283. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
- 21.Heil A, Nazmi AR, Koltzscher M, Poeter M, Austermann J, Assard N, Baudier J, Kaibuchi K, Gerke V. S100P is a novel interaction partner and regulator of IQGAP1. J Biol Chem. 2011;286:7227–7238. doi: 10.1074/jbc.M110.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin MJ, Cromer DA, King ME, Bloom GS. The mechanism for regulation of the F-actin binding activity of IQGAP1 by calcium/calmodulin. J Biol Chem. 2002;277:12324–12333. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- 23.Osman MA, Cerione RA. Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates cdc42p effects on the actin cytoskeleton. J Cell Biol. 1998;142:443–455. doi: 10.1083/jcb.142.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelikan-Conchaudron A, Le Clainche C, Didry D, Carlier MF. The IQGAP1 protein is a calmodulin-regulated barbed end capper of actin filaments: possible implications in its function in cell migration. J Biol Chem. 2011;286:35119–35128. doi: 10.1074/jbc.M111.258772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateer SC, Morris LE, Cromer DA, Benseñor LB, Bloom GS. Actin filament binding by a monomeric IQGAP1 fragment with a single calponin homology domain. Cell Motil Cytoskeleton. 2004;58:231–241. doi: 10.1002/cm.20013. [DOI] [PubMed] [Google Scholar]
- 26.Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282:426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- 27.Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 30.Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 31.Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 32.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 33.Umemoto R, Nishida N, Ogino S, Shimada I. NMR structure of the calponin homology domain of human IQGAP1 and its implications for the actin recognition mode. J Biomol NMR. 2010;48:59–64. doi: 10.1007/s10858-010-9434-8. [DOI] [PubMed] [Google Scholar]
- 34.Kurella VB, Richard JM, Parke CL, Lecour LF, Bellamy HD, Worthylake DK. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J Biol Chem. 2009;284:14857–14865. doi: 10.1074/jbc.M808974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathmanathan S, Hamilton E, Atcheson E, Timson DJ. The interaction of IQGAPs with calmodulin-like proteins. Biochem Soc Trans. 2011;39:694–699. doi: 10.1042/BST0390694. [DOI] [PubMed] [Google Scholar]
- 36.Grohmanova K, Schlaepfer D, Hess D, Gutierrez P, Beck M, Kroschewski R. Phosphorylation of IQGAP1 modulates its binding to Cdc42, revealing a new type of rho-GTPase regulator. J Biol Chem. 2004;279:48495–48504. doi: 10.1074/jbc.M408113200. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, McNulty DE, Marler KJ, Lim L, Hall C, Annan RS, Sacks DB. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J Biol Chem. 2005;280:13871–13878. doi: 10.1074/jbc.M413482200. [DOI] [PubMed] [Google Scholar]
- 38.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cell Signal. 2009;21:1471–1478. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 40.Morris LE, Bloom GS, Frierson HF, Powell SM. Nucleotide variants within the IQGAP1 gene in diffuse-type gastric cancers. Genes Chromosomes Cancer. 2005;42:280–286. doi: 10.1002/gcc.20150. [DOI] [PubMed] [Google Scholar]
- 41.Suyama M, Nagase T, Ohara O. HUGE: a database for human large proteins identified by Kazusa cDNA sequencing project. Nucleic Acids Res. 1999;27:338–339. doi: 10.1093/nar/27.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 44.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 45.Timson DJ, Trayer IP. The rôle of the proline-rich region in A1-type myosin essential light chains: implications for information transmission in the actomyosin complex. FEBS Lett. 1997;400:31–36. doi: 10.1016/s0014-5793(96)01314-2. [DOI] [PubMed] [Google Scholar]
- 46.Timson DJ, Trayer HR, Trayer IP. The N-terminus of A1-type myosin essential light chains binds actin and modulates myosin motor function. Eur J Biochem. 1998;255:654–662. doi: 10.1046/j.1432-1327.1998.2550654.x. [DOI] [PubMed] [Google Scholar]
- 47.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77 Suppl 9:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farrell DW, Speranskiy K, Thorpe MF. Generating stereochemically acceptable protein pathways. Proteins. 2010;78:2908–2921. doi: 10.1002/prot.22810. [DOI] [PubMed] [Google Scholar]
- 49.Mamonova T, Hespenheide B, Straub R, Thorpe MF, Kurnikova M. Protein flexibility using constraints from molecular dynamics simulations. Phys Biol. 2005;2:S137–S147. doi: 10.1088/1478-3975/2/4/S08. [DOI] [PubMed] [Google Scholar]
- 50.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]


