ABSTRACT
The strong restriction barrier present in Staphylococcus aureus and Staphylococcus epidermidis has limited functional genomic analysis to a small subset of strains that are amenable to genetic manipulation. Recently, a conserved type IV restriction system termed SauUSI (which specifically recognizes cytosine methylated DNA) was identified as the major barrier to transformation with foreign DNA. Here we have independently corroborated these findings in a widely used laboratory strain of S. aureus. Additionally, we have constructed a DNA cytosine methyltransferase mutant in the high-efficiency Escherichia coli cloning strain DH10B (called DC10B). Plasmids isolated from DC10B can be directly transformed into clinical isolates of S. aureus and S. epidermidis. We also show that the loss of restriction (both type I and IV) in an S. aureus USA300 strain does not have an impact on virulence. Circumventing the SauUSI restriction barrier, combined with an improved deletion and transformation protocol, has allowed the genetic manipulation of previously untransformable strains of these important opportunistic pathogens.
IMPORTANCE Staphylococcal infections place a huge burden on the health care sector due both to their severity and also to the economic impact of treating the infections because of prolonged hospitalization. To improve the understanding of Staphylococcus aureus and Staphylococcus epidermidis infections, we have developed a series of improved techniques that allow the genetic manipulation of strains that were previously refractory to transformation. These developments will speed up the process of mutant construction and increase our understanding of these species as a whole, rather than just a small subset of strains that could previously be manipulated.
IMPORTANCE
Staphylococcal infections place a huge burden on the health care sector due both to their severity and also to the economic impact of treating the infections because of prolonged hospitalization. To improve the understanding of Staphylococcus aureus and Staphylococcus epidermidis infections, we have developed a series of improved techniques that allow the genetic manipulation of strains that were previously refractory to transformation. These developments will speed up the process of mutant construction and increase our understanding of these species as a whole, rather than just a small subset of strains that could previously be manipulated.
Introduction
Functional genomic analyses of the Gram-positive opportunistic pathogens Staphylococcus aureus and Staphylococcus epidermidis have been limited by the inability to manipulate genetically the majority of clinical isolates. In general, wild-type strains of S. aureus and S. epidermidis have an impenetrable restriction barrier preventing the uptake of “foreign” DNA (1–3). A plethora of knowledge has been generated through the mutagenesis of “laboratory” S. aureus isolates such as Newman (4), 8325-4 (5), and more recently some strains of the community-acquired methicillin-resistant S. aureus (CA-MRSA) USA300 (6). For S. epidermidis, the transformable strains 1457 (7) and O-47 (8) have been the mainstay of genetic studies, but the genome sequences for these isolates are not currently available. The emergence of CA-MRSA and the high frequency of methicillin resistance found in hospital isolates of S. epidermidis have highlighted the need to understand better the evolution and biology of these medically important staphylococci at a genetic level (9, 10). The majority of MRSA isolates currently cluster into five clonal complexes (clonal complex 5 [CC5], CC8, CC22, CC30, and CC45) of which only CC5 and CC8 have been successfully manipulated genetically (9). Pathogenomic studies on CC5 and CC8 strains stem from the isolation of S. aureus RN4220 almost 30 years ago. This is a restriction-defective mutant that can still modify DNA, which was obtained by extensive chemical mutagenesis of strain 8325-4 (11). Strain RN4220 accepts DNA derived from wild-type Escherichia coli, unlike the parental strain 8325-4. Passage of DNA through RN4220 has allowed the subsequent transfer of DNA by electroporation or generalized phage transduction to a small subset of closely related strains, but not to more distantly related S. aureus or S. epidermidis isolates.
Experimental evidence presented by Waldron and Lindsay (3) suggested that the conserved type I restriction-modification (RM) system was solely responsible for the inability to transform S. aureus isolates with E. coli-derived plasmid DNA. They identified a premature stop codon in the type I restriction gene (hsdR) in S. aureus RN4220. Complementation with full-length hsdR expressed from a low-copy-number plasmid rendered strain RN4220 incapable of accepting plasmids electroporated from E. coli, reduced the rate of conjugation with Enterococcus faecalis, and prevented transduction with phage isolated from a distantly related strain of S. aureus. However, inactivation of hsdR (1, 2) in wild-type S. aureus isolates did not yield a transformable strain, suggesting at least one other pathway is involved. The gene responsible for preventing the transformation of S. aureus with E. coli-derived plasmid DNA was recently identified and characterized by Corvaglia et al. (1) in two clinical isolates of S. aureus (UAMS-1 and SA564). The biochemical properties of the conserved type IV modification-dependent restriction endonuclease (termed SauUSI) were subsequently characterized in detail (12). Cytosine methylated DNA was identified as the motif recognized by SauUSI, with plasmid isolated from a B strain of E. coli (which is naturally a dcm mutant) able to bypass the type IV barrier. Inactivation of both hsdR and sauUSI yielded a strain that was transformable at equivalent efficiency with plasmid DNA isolated either from E. coli or from the parental S. aureus strain. These data demonstrate that only these two pathways contribute to the restriction barrier in the strains analyzed (12).
Here we have characterized the RM systems of the widely used laboratory S. aureus strain Newman and corroborated the previous findings (1, 12). We have created a dcm mutation in the high-efficiency E. coli cloning strain DH10B (called DC10B). We show that direct transformation can be performed with plasmid DNA isolated from DC10B, but not the progenitor DH10B, into S. aureus strains from the majority of clonal complexes tested. Furthermore, we show that S. epidermidis contains an ortholog of SauUSI (termed McrR) which also recognizes cytosine methylation. McrR can also be bypassed with plasmid DNA isolated from DC10B. To take advantage of these findings, we have developed a robust method for creating mutations by allelic exchange in both laboratory strains and previously untransformable strains of S. aureus and S. epidermidis and have developed an improved electroporation protocol. The virulence in an intravenous mouse infection model of S. aureus USA300 strain NRS384 was not affected by the mutation of hsdR or sauUSI singly or in combination. These developments open up previously recalcitrant strains of S. aureus and S. epidermidis to genetic analysis, which will ultimately improve our understanding of these important pathogens.
RESULTS AND DISCUSSION
Bypassing the restriction barrier.
Transformation of S. aureus strain Newman by the protocol of Augustin and Götz (13) with 5 µg of shuttle plasmid pRMC2 DNA (14) isolated from the wild-type E. coli K-12 strain BW25113 (15) consistently failed to yield transformants. Similar results were observed with plasmid isolated from the high-efficiency cloning strain of E. coli K-12, DH10B. However, if the plasmid was isolated from the restriction-defective S. aureus strain RN4220 or from strain Newman itself, we would routinely obtain 104 CFU with the same concentration of pRMC2. These results show that a strong restriction barrier is present in the Newman strain, which impedes the uptake of E. coli K-12-derived plasmid DNA. When we applied the electroporation protocol for Staphylococcus carnosus developed by Löfblom et al. (16), we observed a 50-fold improvement in the transformation efficiency, and for the first time, a low number of transformants were obtained with plasmid DNA isolated from strain BW25113 (Fig. 1). A second advantage of the S. carnosus protocol is the reduced time required for production of competent cells (2 h instead of 4 h). The protocol is also applicable to S. epidermidis, albeit with a marked reduction in efficiency (a maximum of 103 CFU was obtained in S. epidermidis RP62a with pRMC2 DNA isolated from the same strain).
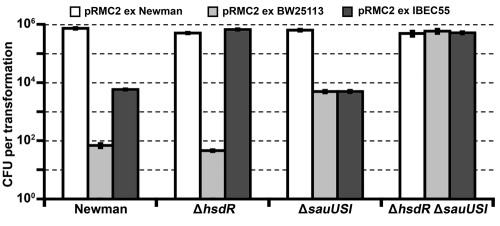
FIG 1 .
Transformation of S. aureus Newman and isogenic mutants defective in restriction. Concentrated pRMC2 DNA (5 µg) isolated from S. aureus Newman, E. coli BW25113 (dam+ dcm+), or E. coli IBEC55 (dam+) were electroporated into Newman or Newman restriction mutants. The transformation efficiency was expressed as the mean number of all transformants obtained in each experiment ± standard deviation (error bar) of three replicates. The graph shows data representative of the data from three independent experiments.
Construction of pIMAY.
The role of RM was assessed by Corvaglia et al. (1) in two clinical strains using the targetron system (17) for gene disruption rather than the more traditional allelic exchange approach for the creation of marked or unmarked mutations. The targetron system is useful for the rapid disruption of genes and for testing gene essentiality. However, allelic exchange allows precise control over the mutation allowing the deletion of entire genes, introduction of point mutations, insertion of hybrid genes, and gene restoration to eliminate the possibility of polar effects (18). The plasmid pE194ts replicon is commonly used for allelic exchange in staphylococci, as exemplified by pKOR1 (19). Chromosomal integration of the plasmid requires growth at the high temperature of 43°C (19). It was reported recently that mutations in the sae genes encoding a two-component system can be selected during growth at high temperature in the presence of the antibiotic erythromycin or chloramphenicol (20). To avoid these concerns, we have developed a new vector for allelic exchange in staphylococci called pIMAY (Fig. 2 and 3A). The vector utilizes the plasmid pWV01ts replicon (21), which is highly temperature sensitive in staphylococci and thus allows plasmid integrants to be selected at 37°C. PCR can easily be applied to (i) demonstrate that extrachromosomal plasmid is no longer present with primers external to the multiple cloning site (MCS) on pIMAY (IM151 and IM152 [IM151/IM152] [Fig. 3B]) and (ii) determine whether plasmid integration has occurred via the upstream or downstream region of homology cloned into pIMAY (Fig. 3C). Growth at a temperature permissive for pIMAY replication (below 30°C) and the induction of secY antisense RNA (derived from pKOR1) prevents growth of cells that retain the integrated plasmid and selects for cells that have lost the plasmid (19). Additionally, a high level of chloramphenicol (Cm) resistance is obtained by expression of cat from a strong promoter (22). This reduces the pressure for selection of variants with increased Cm resistance, which, combined with the low copy number of the plasmid, lessens the chance of tandem duplication occurring during chromosomal integration.
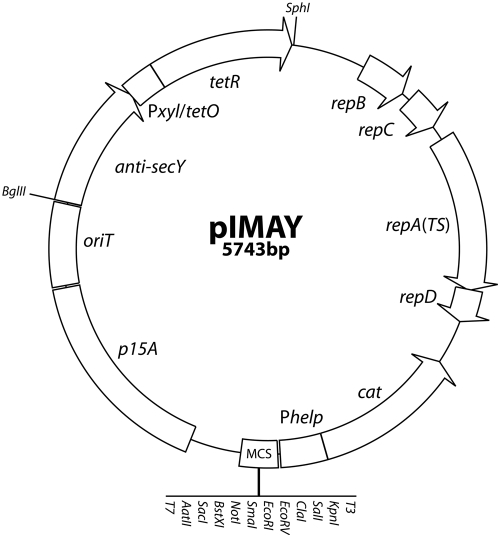
FIG 2 .
Genetic map of pIMAY. The E. coli/staphylococcal temperature-sensitive plasmid pIMAY comprises the low-copy-number E. coli origin of replication (p15A), an origin of transfer for conjugation (oriT), the pBluescript multiple cloning site (MCS), and the highly expressed cat gene (Phelp-cat) derived from pIMC (34). The temperature-sensitive replicon for Gram-positive bacteria (repBCAD) and the tetracycline-inducible antisense secY region (anti-secY) were amplified from pVE6007 (21) and pKOR1 (19), respectively. The restriction sites listed are unique. Primers (IM151/152) bind external to the MCS of pIMAY and are used to screen clones in E. coli (amplify 283 bp without a cloned insert) and to determine the presence of a replicating plasmid in staphylococci.
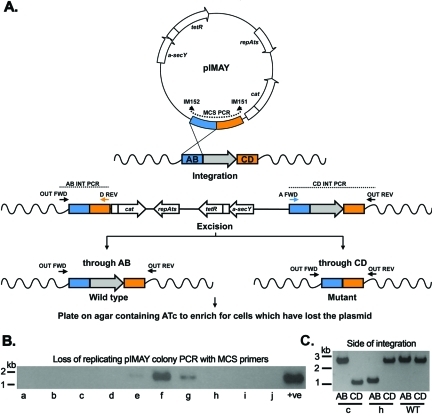
FIG 3 .
Schematic of allelic exchange with pIMAY. (A) A plasmid isolated from E. coli DC10B is transformed into staphylococci at 28°C, and single-crossover (SCO) integration was stimulated by growth at 37°C in the presence of chloramphenicol. The loss of replicating plasmid is assayed by colony PCR with MCS primers (IM151/IM152). Clones negative for replicating plasmid are then screened for the side of integration with a combination of chromosomal and cloning primers (e.g., OUT FWD/D REV [AB integration {AB INT}] or OUT REV/A FWD [CD integration {CD INT}]). The diagram details an integration event through the AB side (equivalent to clone h in panels B and C). A clone from either AB or CD integration event is grown at 28°C in broth without antibiotic selection to stimulate rolling circle replication and then plated on TSA with 1 µg/ml ATc. Expression of the secY antisense RNA (a-secY) inhibits growth of cells maintaining the plasmid. Plasmid excision through the AB side recreates the wild-type locus, while CD excision yields a mutated gene. (B) Colony PCR from 10 randomly chosen clones (clones a to j) after growth at 37°C for the presence of replicating plasmid. The absence of product indicates that the plasmid has integrated. Colony PCR from cells grown at 28°C is included as a positive control (+ve). (C) Two clones without replicating plasmid (clones c and h) were shown by colony PCR to have integrated either on the AB (upstream [clone h]) or CD (downstream [clone c]) side of the gene to be deleted. Wild-type (WT) genomic DNA was included as a control.
Deletion of hsdR and sauUSI in S. aureus Newman.
To assess the roles of hsdR (encodes the restriction component of a type I RM system) and sauUSI (encodes a type IV restriction gene) in S. aureus Newman, we created unmarked deletion mutations in each of the genes and a double mutant with pIMAY. No differences in delta toxin production, hemolysis, growth rate, or final growth yield were observed for the mutants compared to Newman (data not shown) indicating the integrity of Agr and Sae. We then transformed wild-type Newman and Newman ΔhsdR, Newman ΔsauUSI, and Newman ΔhsdR ΔsauUSI mutants with pRMC2 DNA isolated from either Newman, E. coli BW25113 (dam+ dcm+ hsd+) or E. coli IBEC55 (dam+ only) (15). In line with the experimental evidence presented for the clinical isolate UAMS-1 (1), the Newman ΔhsdR ΔsauUSI mutant exhibited the highest transformation efficiency, which was equivalent to wild-type Newman transformed with plasmid DNA isolated directly from Newman (Fig. 1). In contrast, a ca. 130-fold reduction in the transformation efficiency of strain Newman was observed with plasmid DNA isolated from E. coli IBEC55. Transformation of the ΔhsdR mutant with plasmid DNA isolated from strain IBEC55 yielded a high transformation efficiency equivalent to that of DNA isolated from Newman or transformation into Newman ΔhsdR ΔmcrR mutant. Our data have also shown that the role of the type I RM system in Newman is to prevent the uptake of DNA from other staphylococci as well as from foreign sources. Plasmid DNA isolated from either Dcm+ or Dcm− E. coli was transformed into the sauUSI mutant at a ca. 100-fold reduced level compared to plasmid DNA isolated from wild-type Newman (Fig. 1). The gene upstream of sauUSI is predicted to encode a nudix hydrolase which is potentially involved in the degradation of mutagenic nucleotide triphosphates (23). Deletion of this gene did not yield the same transformable phenotype observed in the ΔsauUSI strain (data not shown), which suggests that the putative nudix hydrolase is not required for SauUSI activity.
In S. aureus RN4220, premature stop codons are present in hsdR and sauUSI, producing a strain that can modify, but not restrict, foreign DNA. We restored the sauUSI mutation to the wild type, creating the RN4220 sauUSI+ strain, which was poorly transformable with E. coli-derived pRMC2 DNA with 102 transformants compared to 106 CFU for RN4220. This confirms a dominant role for sauUSI over hsdR as was also observed in Newman.
C-terminally His-tagged SauUSI is an endonuclease.
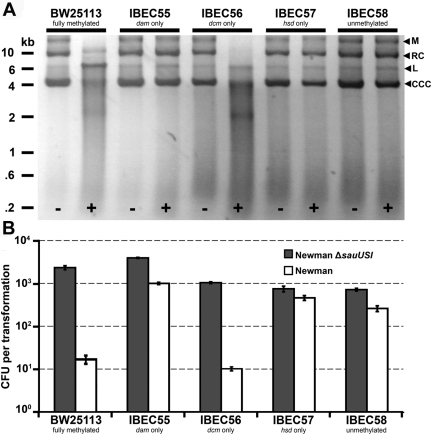
To characterize the barrier to transformation encoded by sauUSI, we assessed the function of the purified SauUSI protein. The full-length SauUSI protein of Newman was expressed in E. coli BL21(DE3) from pET21d+ (C-terminal hexahistidine tag vector) and purified by nickel affinity chromatography, yielding SauUSI-his. We then tested the effect of incubating SauUSI-his with pRMC2 DNA isolated from a panel of isogenic methylation-defective mutants of E. coli BW2511. The pRMC2 DNA profiles (Fig. 4A) demonstrate that SauUSI-his can digest plasmid DNA isolated from both E. coli BW25113 (dam+ dcm+ hsd+) and IBEC56 (dcm+ only), while pRMC2 isolated from E. coli IBEC55 (dam+ only), IBEC57 (hsd+ only), and IBEC58 (unmethylated) was unaffected. This indicates that cytosine methylation is the signal that is recognized by SauUSI. This occurs in all strains derived from E. coli K-12 (e.g., DH5α, TOP10, or XL1-Blue) but not in those derived from E. coli B (e.g., BL21). For this reason, we could express SauUSI-his in strain BL21 without toxicity. There was also a strict requirement for ATP, as DNA digestion was not observed when ATP was omitted from the buffer. The frequencies of transformation of S. aureus Newman and Newman ΔsauUSI with pRMC2 isolated from E. coli BW25113 and the methylation mutants were consistent with the digestion profiles, with wild-type Newman being transformed efficiently only if cytosine methylation was absent from the DNA (Fig. 4B).
FIG 4 .
Assays for the activity of SauUSI. (A) The shuttle plasmid pRMC2 was isolated from isogenic E. coli methylation mutants and incubated in NEB ligation buffer with (+) or without (−) purified SauUSI-his for 1 h at 37°C. The DNA was then purified and run on a 1% agarose gel. The positions of linearized (6.4 kb) (L), relaxed circular (RC), covalently closed circular (CCC), and multimers (M) of pRMC2 are indicated by the black arrowheads to the right of the gel. (B) Concentrated pRMC2 DNA (5 µg) isolated from isogenic E. coli methylation mutants was electroporated into either S. aureus Newman or Newman ∆sauUSI mutant. The transformation efficiency was expressed as the total number of transformants obtained in each experiment ± standard deviation (error bar) of three replicates. The graph shows data representative of the data from three independent experiments.
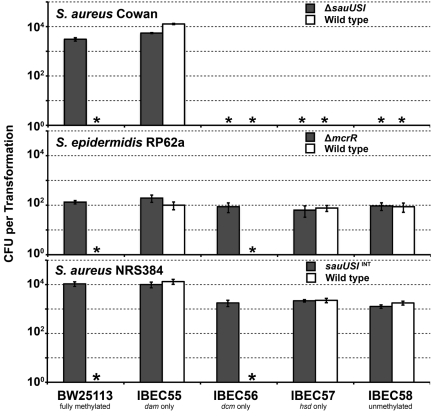
Isolating mutations in S. epidermidis RP62a and S. aureus Cowan.
We sought to apply these findings and to attempt allelic exchange in the genome-sequenced S. epidermidis strain RP62a (24) and S. aureus Cowan (25), two strains that we had not previously been able to manipulate genetically. We were able to transform both strains with pIMAY isolated from E. coli IBEC55 (at a very low efficiency for RP62a) and subsequently to construct mutations in the sauUSI-like genes in each. We called the gene of S. epidermidis RP62a mcrR for methylated cytosine recognition and restriction. Similar transformation profiles were obtained as observed for S. aureus Newman with regard to the involvement of cytosine methylation (Fig. 5). Additionally, the CA-MRSA strain NRS384 and the isogenic sauUSI targetron mutant also had the same transformation profile as Newman and Newman ΔsauUSI mutant (Fig. 5). However, Cowan exhibited a strict requirement for the presence of adenine methylation on the transforming plasmid DNA, as transformants could not be obtained in its absence in either the wild type or sauUSI mutant. Out of 15 S. aureus strains used in this study, only Cowan genomic DNA was sensitive to digestion with the restriction enzyme DpnI, which recognizes adenine methylation at a 5′ GATC 3′ motif. This suggests the presence of a novel adenine methylase expressed by Cowan and indicates the possibility of an additional restriction system in this strain.
FIG 5 .
Transformation of S. aureus Cowan, S. epidermidis RP62a, and S. aureus NRS384. pRMC2 DNA (5 µg) isolated from isogenic E. coli methylation mutants was electroporated into either the wild type or the corresponding ΔsauUSI or ΔmcrR mutant of the strain specified. The transformation efficiency is expressed as the total number of transformants obtained in each experiment with standard deviation of three replicates. An asterisk denotes that no transformants were detected.
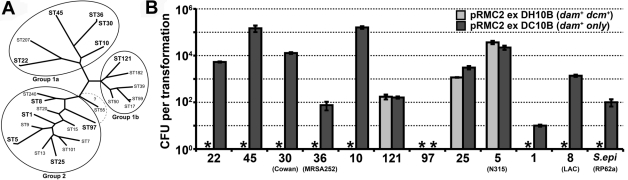
Creation of E. coli DC10B—a universal staphylococcal cloning host.
E. coli IBEC55 is not an ideal cloning host, as it does not contain mutations associated with the production of good-quality plasmid DNA (recA endA) and is not highly transformable. Through recombineering (recombination-mediated genetic engineering) (26) in a high-efficiency cloning strain of E. coli (DH10B), we created DC10B, a mutant with the dcm gene deleted. Plasmid DNA isolated from DC10B was capable of transforming Newman at the same efficiency as that of IBEC55 (data not shown). Eleven representative S. aureus strains from a diverse selection of multilocus sequence types (STs) (Fig. 6A) and S. epidermidis RP62a were tested as recipients for transformation with pRMC2 DNA isolated from either DH10B or DC10B (Fig. 6B). Strains from sequence type 1 (ST1), ST8, ST10, ST22, ST30, ST36, and ST45 could be transformed only with DNA from E. coli DC10B, while ST5, ST25, and ST121 yielded colonies irrespective of the plasmid source. This was expected for the ST5 strain N315, because a premature stop codon is present in the sauUSI gene. We have not investigated the restriction status of the transformable ST25 and ST121 clones. The isolate chosen from ST97 did not transform with plasmid DNA from either DH10B or DC10B. It is possible that this strain requires different growth conditions or treatments prior to electroporation to become competent (1) or an additional RM system may be present (27). When pRMC2 was reisolated from the different hosts, no evidence of deletion or rearrangement was observed (see Fig. S1 in the supplemental material). E. coli dam mutants have a higher frequency of spontaneous mutation compared to wild type or dcm mutants (41). Thus, the dcm mutant of DH10B is an ideal host for the construction of recombinant plasmids for subsequent direct transformation into S. aureus or S. epidermidis. We are currently investigating the possibility of bypassing the type I restriction barriers in S. aureus by E. coli to further improve the transformation efficiency. The knowledge gained here could be applied to other bacteria where DNA uptake is impeded by RM.
FIG 6 .
Transformation of strains from a diverse selection of S. aureus sequence types and an S. epidermidis isolate. (A) Phylogenetic relatedness of the S. aureus strains selected for transformation, adapted from reference 35 with permission of the publisher. Representative sequence types used for transformation are highlighted in bold type. (B) Concentrated pRMC2 DNA (5 µg) isolated from E. coli DH10B (dam+ dcm+) or DC10B (dam+) was electroporated into strains from different S. aureus STs (denoted by the number on the x axis) including strains Cowan, MRSA252, N315, LAC, and S. epidermidis RP62a. The transformation efficiency was expressed as the total number of transformants obtained in each experiment ± standard deviation of three replicates. An asterisk denotes that no transformants were detected. The graph shows data from one experiment.
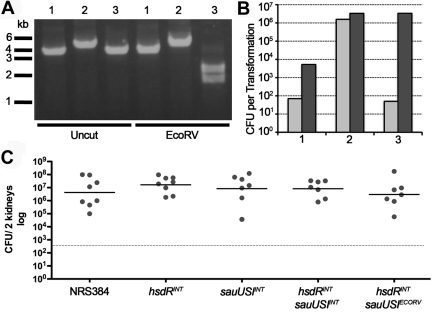
Impact on virulence of restriction mutants of S. aureus USA300 strain NRS384.
We sought to assess the role that RM systems might play in influencing the ability of bacteria to grow in vivo. Instead of using the laboratory strain Newman, we constructed restriction-deficient mutations in the CA-MRSA strain NRS384 (NARSA USA300-014 clone) using the targetron system. The double mutant (hsdRINT sauUSIINT) was reverted by allelic exchange to restore wild-type sauUSI (Fig. 7A). The strains were phenotypically identical with no morphological or growth rate differences. The transformation profiles were similar to those observed for Newman. Restoration of sauUSI in the double mutant dramatically reduced the transformation frequency (Fig. 7B). In the AJ mouse intravenous infection model, no significant differences in the bacterial numbers recovered from the kidneys at day 7 were observed (Fig. 7C). Under the conditions tested, the conserved type I RM and type IV restriction system do not affect the virulence of S. aureus.
FIG 7 .
Transformation and virulence of restriction mutants of S. aureus USA300 strain NRS384. (A) PCR profiles (primers IM110/IM111) of the sauUSI region amplified from NRS384 (lanes 1), NRS384 hsdRINT sauUSIINT (lanes 2), and NRS384 hsdRINT sauUSIECORV (lanes 3) without or with EcoRV digestion. (B) Concentrated pRMC2 DNA (5 µg) isolated from E. coli DH10B (dam+ dcm+) (light grey bars) or DC10B (dam+) (dark grey bars) was electroporated into the strains described above, and transformants were enumerated. (C) Intravenous injection of 2 × 106 CFU into 6- to 7-week-old female A/J mice. On day 7 of infection, the mice were euthanized, both kidneys were aseptically removed, and the bacterial CFU were enumerated as described in Materials and Methods. Each symbol represents the value for an individual mouse, and the short black line represents the mean for the group of mice. The broken line denotes the limit of detection at 333 CFU for the two kidneys.
Cytosine methylation—a barrier for DNA transfer?
Previously, it was shown that plasmid DNA isolated from Enterococcus faecalis (a major potential reservoir of vancomycin resistance) could not be electroporated into wild-type S. aureus unless a deletion in sauUSI was first constructed (1). This suggests that a barrier to plasmid transfer between E. faecalis and S. aureus might be cytosine methylation. From the genome sequences of E. faecalis strains submitted to public databases, we could identify a putative cytosine methyltransferase in all strains examined. However, when genomic DNA was isolated from E. faecalis strains OG1RF, V583, and JH2-2 (grown in brain heart infusion broth [BHI] at 37°C), only DNA from OG1RF could be digested with purified SauUSI (data not shown), suggesting that some E. faecalis strains but not others could potentially act as reservoirs for the transfer to DNA to S. aureus. Naturally occurring vancomycin-resistant S. aureus clones are all in ST5 (CC5) (28). Both genome-sequenced isolates from CC5 (29) contain a truncated version of the sauUSI gene, leading to the possibility that acquisition of vanA mediated by conjugation from enterococci could occur in strains that lack functional sauUSI. Further investigation into the sauUSI status of vancomycin-resistant strains of S. aureus is required.
Summary.
The availability of the E. coli dcm mutant (DC10B) along with an improved transformation protocol and the construction of the temperature-sensitive plasmid pIMAY has dramatically accelerated our ability to generate mutations in staphylococci. We have been successful in creating mutations in S. aureus strains Newman, NRS384 (USA300), and Cowan and S. epidermidis RP62a as described above but also in the hospital-acquired MRSA (HA-MRSA) strain MRSA252 (CC30) and two additional CA-MRSA strains (TCH1516 and LAC, both CC8). From the start of cloning to the confirmation of the mutation takes 2 weeks, under conditions that are less stressful to the bacterium to be altered than previously published protocols. Breaking the restriction barrier opens up clinical isolates such as MRSA lineages CC22, CC30, and CC45 to genetic manipulation (e.g., allelic exchange and transposon mutagenesis) and will ultimately improve our understanding of staphylococcal genetics, potentially leading to novel methods to combat these deadly opportunistic pathogens.
MATERIALS AND METHODS
Media and reagents.
Bacterial strains, plasmids, and oligonucleotides used in this study are described in Table 1. E. coli, S. aureus, and S. epidermidis were routinely cultured at 37°C in L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl), Trypticase soy broth (TSB) (Difco) or brain heart infusion broth (BHI) (Difco). For growth on agar, L broth or brain heart infusion broth was solidified with 1.5% agar, yielding LBA and BHIA, respectively. The following antibiotics and concentrations were used: chloramphenicol (Cm), 10 µg/ml; kanamycin, 50 µg/ml; erythromycin (Em), 25 µg/ml; and carbenicillin, 100 µg/ml (Sigma).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Bacterial strain, plasmid, or oligonucleotidea | Description (relevant genotype or phenotype) or sequence (5′ to 3′)b | Source, reference, or RE sitec |
|---|---|---|
| Escherichia coli strains | ||
| DH10B (K-12 strain) | dam + dcm + ΔhsdRMS endA1 recA1 | Invitrogen |
| BW25113 (K-12 strain) | dam + dcm + hsdMS+ hsdR514 | 15 |
| IBEC55 | Δdcm ΔhsdMS in the BW25113 background; Dam methylation only | 15 |
| IBEC56 | Δdam ΔhsdMS in the BW25113 background; Dcm methylation only | 15 |
| IBEC57 | Δdam Δdcm in the BW25113 background; Hsd methylation only | 15 |
| IBEC58 | Δdam Δdcm ΔhsdMS in the BW25113 background; no methylation | 15 |
| DC10B | Δdcm in the DH10B background; Dam methylation only | This study |
| BL21(DE3) (B strain) | F– ompT hsdSB (rB– mB–) gal dcm (DE3); IPTG-inducible T7 RNA polymerase | Novagen |
| Staphylococcus strains | ||
| Newman | ST8; CC8 isolated in 1952 human clinical MSSA; genome sequenced | 4 |
| Newman ΔhsdR | Newman with a deletion of Sae0139 | This study |
| Newman ΔsauUSI | Newman with a deletion of Sae2386 | This study |
| Newman ΔhsdR ΔsauUSI | Newman with a deletion of both Sae0139 and Sae2386 | This study |
| Newman ΔhsdR sauUSIECORV | Restoration of Sae2386 in the ΔhsdR ΔsauUSI background | This study |
| Newman Δnudix | Newman with a deletion of Sae2385 | This study |
| NRS384 | USA300-14 clone obtained from NARSA | NARSA collection |
| NRS384 hsdRINT | Targetron insertion at nucleotide 735 of hsdR | This study |
| NRS384 sauUSIINT | Targetron insertion at nucleotide 739 of sauUSI | This study |
| NRS384 sauUSIINT hsdRINT | Targetron insertion in hsdR made in the NRS384sauUSIINT background | This study |
| NRS384 sauUSIECORV hsdRINT | Restoration of the sauUSI mutation with a silent EcoRV site in the double insertion mutant background | This study |
| RN4220 | ST8; CC8; chemically mutagenized derivative of 8325-4, transformable with E. coli DNA; premature stop codon in both hsdR and sauUSI | 11 |
| RN4220 sauUSI+ | Nonsense mutation in sauUSI corrected to wild type with 8325-4 sequence | This study |
| Cowan | ST30; CC30 MSSA;high-level protein A producer; ATCC 12598 | 25 |
| Cowan ΔsauUSI | Deletion of sauUSI | This study |
| N315 | ST5 CC5 MSSA; genome sequenced | 29 |
| LAC | ST8 CC8 CA-MRSA; USA300 | 6 |
| MRSA252 | ST36 CC30 MRSA; genome sequenced | 36 |
| Oxford 13 | ST22 CC22 | 37 |
| Oxford 19 | ST10 CC16 | 37 |
| Oxford 71 | ST1 CC1 | 37 |
| Oxford 159 | ST25 CC25 | 37 |
| Oxford 207 | ST15 CC15 | 37 |
| Oxford 233 | ST45 CC45 | 37 |
| Oxford 560 | ST121 CC51 | 37 |
| Oxford 3177 | ST97 CC16 | 37 |
| RP62a | Methicillin-resistant, biofilm-forming Staphylococcus epidermidis isolate; genome sequenced | 24 |
| RP62a ∆mcrR | Deletion of Serp2052; able to accept DNA at a low frequency from wild-type E. coli | This study |
| Enterococcus faecalis strains | ||
| OG1RF | Rifampin- and fusic acid-resistant E. faecalis clone derived for OG1 | 38 |
| JH2-2 | Rifampin- and fusic acid-resistant E. faecalis clone derived for JH2 | 39 |
| V583 | Vancomycin-resistant clinical isolate of E. faecalis | 40 |
| Plasmids | ||
| pNL9164 | Temperature-sensitive targetron plasmid for S. aureus pT181 replicon; Ampr Eryr | Sigma |
| pNL9164(hsdR) | pNL9164 retargeted for hsdR of NRS384 | This study |
| pNL9164(sauUSI) | pNL9164 retargeted for sauUSI of NRS384 | This study |
| pKD4 | Plasmid for amplification of frt-kan-frt for E. coli gene deletion; Ampr Kanr | 26 |
| pKD46 | E. coli temperature-sensitive plasmid containing λ red recombinase genes under the control of an arabinose-inducible promoter; Ampr | 26 |
| pCP20 | E. coli temperature-sensitive plasmid containing flp required for antibiotic marker excision; Ampr Cmr | 10 |
| pIMC | Site-specific integrating vector; p15A low-copy-number origin of replication; RP4 conjugative origin of transfer and Phelp-driven chloramphenicol resistance marker; pBluescript MCS; Cmr | 34 |
| pKOR1 | Temperature-sensitive shuttle vector for allelic exchange in S. aureus; Ampr Cmr | 19 |
| pVE6007 | pWV01ts-derived plasmid that cannot replicate in E. coli; Cmr | 21 |
| pIMC5 | Temperature-sensitive Gram-positive replicon from pVE6007 with an E. coli replicon; MCS and antibiotic resistance from pIMC; Cmr (IM46/IM47/IM48/IM49) | This study |
| pIMAY | pIMC5 with tetracycline; inducible secY antisense from pKOR1; Cmr (IM72/IM73) | This study |
| pIMAYΔhsdR | A deletion encompassing the entire hsdR gene (between the ATG and TAA codons); amplified from Newman (IM93/IM3/IM4/IM94) | This study |
| pIMAYΔsauUSI(CC8) | A deletion encompassing the entire sauUSI gene (between the ATG and TAA codons); amplified from Newman (IM89/IM90/IM91/IM92) | This study |
| pIMAYΔsauUSI(CC30) | A deletion encompassing the entire sauUSI gene (between the ATG and TAA codons); amplified from Cowan (IM89/IM90/IM91/IM150) | This study |
| pIMAYΔmcrR(S.epi) | A deletion encompassing the entire mcrR gene (between the ATG and TAA codons); amplified from RP62a (IM216/IM217/IM218/IM219) | This study |
| pIMAY sauUSIEcoRV | A silent EcoRV site was introduced into the middle of the sauUSI gene (with DNA flanking for gene restoration in the ΔsauUSI mutant) (IM89/IM350/IM351/IM92) | This study |
| pIMAY(RN4220sauUSI+) | A 1-kb fragment amplified from Newman surrounding the premature stop codon in RN4220 sauUSI (IM108/IM109) | This study |
| pIMAYΔnudix | A deletion encompassing the entire putative nudix gene (between the ATG and TAA codons); amplified from Newman (IM222/IM223/IM224/IM225) | This study |
| pET21d+ | C-terminal hexahistadine tagging vector; Ampr | Novagen |
| pET21d+sauUSI | The entire sauUSI gene amplified from Newman and fused to a C-terminal His tag (IM196/IM197) | This study |
| Oligonucleotides | ||
| IM46 (pVE6007 F) | ATATGCATGCGTT TTAGCGTTTATTTC GTTTAGTTATCGG | SphI |
| IM47 (pVE6007 R) | GTATTGCTATTAATC GCAACATCAAACC | |
| IM48 (pIMC F) | GATGTTGCGATTAATAGC AATACATTCTATAATAGA AGGTATGGAGGATG | |
| IM49 (pIMC R) | AGATCTCCTCTCGC CTGTCCCCTCAGTTC AGTAATTTCC | BglII |
| IM72 (anti secY F) | ATATAGATCTTGATC TAATGATTCAAACCCTTGTG | BglII |
| IM73 (anti secY R) | ATATGCATGCTGAAG TTACCATCACGGAAAAAGG | SphI |
| IM93 (ΔhsdR-AFwd) | ATATGGTACCGTGGC CACACATTACAGTATTCCC | KpnI |
| IM2 (ΔhsdR-B) | CATTCATATCCCCTT CCATACACTTTCTATTGC | |
| IM3 (ΔhsdR-C) | TATGGAAGGGGATATGA ATGTAATGATTCAGCCCC CTCGCTAGATTAGTG | |
| IM94 (ΔhsdR-DRev) | ATATGAGCTCATTCAT CTTTGTATTCTTTCATGTTTCC | SacI |
| IM5 (hsdR-outF) | AGTCATAGTGAATTGCA GTCAATTGC | |
| IM6 (hsdR-outR) | ATATAACAAGAACTTA ATTTCAGCCG | |
| IM89 (ΔsauUSI-AFwd) | ATATGGTACCGTGTAT GAAAATGCATGGAGTAGAGC | KpnI |
| IM90 (ΔsauUSI-B) | CATATTATCCCTCAGT CATAATTTTATTAACG | |
| IM91 (ΔsauUSI-C) |
CGTTAATAAAATTATG
ACTGAGGGATAATATGT
AATGTAAACCGAAAAATG
AATGTTAGTAAAG |
|
| IM92 (ΔsauUSI-DRev) | ATATGAGCTCCCAA TCCTCTGGATTCCAT ATTCTTTCC | SacI |
| IM150 (CC30 sauUSI-DRev) | ATATGAGCTCAAA CTCTTCGTCACGAAATCCTTCC | SacI |
| IM110 (sauUSI-OUT F) | ACAGCCCCAAGACA ATACTTTTCAC | |
| IM111 (sauUSI-OUTR) | ATACAGGACCAATCC TCTGGATTCC | |
| IM108 (RNsauUSIcomp)F | ATATGGTACCGTGCATT AGATGTTAGAGAAGTAAACC | KpnI |
| IM109 (RNsauUSIcomp)R | ATATGAGCTCATTTAATG ATACTGCATCCAATGAATTG | SacI |
| IM350 (384sauUSIcomp)B | GATATCACTTTCTAATG CTGCTTGTAACC | EcoRV |
| IM351 (384sauUSIcomp)C | ACAAGCAGCATTAGAAA GTGATATCTTATGTCCATT TCATTATTTTGGTGTG | EcoRV |
| IM196 (His-SauUSI F) | ATATCCATGGGTAGATTA CTAAATGATTTCAATC | NcoI |
| IM197 (His-SauUSI R) | ATATCTCGAGATTTGTTA GATAACGATATATATCATCTC | XhoI |
| IM216 (Se ΔmcrR-AFwd) | ATATGTCGACTCTAATAT ATTAAGTATGTAAACCACG | SalI |
| IM217 (Se ΔmcrR-B) | CAATCTAATTCTCCTCTATTATACG | |
| IM218 (Se ΔmcrR-C) | GTATAATAGAGGAGAATTA GATTGTAATTACTTATACTA AATTATTATTTATTG | |
| IM219 (Se ΔmcrR-DRev) | ATATGAATTCTGAATCACA GATCAAAAATGAAGACC | EcoRI |
| IM220 (Se mcrR-OUTF) | GAATTGAAAATTTTAGG TATTCAGATGG | |
| IM221 (Se mcrR-OUTR) | AAACCTTTAATAATTA TCAAGACAGC | |
| IM222 (Δnudix-AFwd) | ATATGGTACCACCTTCACC AAGACCGAATTTTCC | KpnI |
| IM223 (Δnudix-B) | CATAAGACTCACCCTTCA ATTTAAAATC | |
| IM224 (Δnudix-C) | TTAAATTGAAGGGTGAGTC TTATGTAATATGAGTAGATT ACTAAATGATTTC | |
| IM225 (Δnudix-DRev) | ATATGAGCTCATAGTA GACAGTAAAACATTATGC | SacI |
| IM226 (nudix OUT F) | TTTAAATAACGCGCTAAACCTAATGC | |
| IM227 (nudix OUT R) | CACTATCAACTAAATCGCCATTTTTC | |
| IM261 (Ec Δdcm F) |
TGTAATTATGTTAACCTG
TCGGCCATCTCAGATGGC
CGGTGAAATCTATGGTG
TAGGCTGGAGCTGCTTC |
|
| IM262 (Ec Δdcm R) |
TTGTGCCTCTTGCTGACG
CAACGCCACCGCCTGTTTG
ATTTTTGGCTCAAGGTCCAT
ATGAATATCCTCCTTAG |
|
| IM251 (Ec Δdcm OUTF) | AGAAGAGACGCGTCGCCTGCTCC | |
| IM252 (Ec Δdcm OUTR) | TACTGGTCACGTTGGGAAAATATCTC | |
| IMS80 (hsdR IBS) | AAAAAAGCTTATAATTA TCCTTACTTCTCCCGCAT GTGCGCCCAGATAGGGTG | HindIII |
| IMS81 (hsdR EBS1d) |
CAGATTGTACAAATGTGGTG
ATAACAGATAAGTCCCGCAT
ACTAACTTACCTTTCTTTGT |
BsrGI |
| IMS82 (hsdR EBS2) | TGAACGCAAGTTTCTAATT TCGGTTAGAAGTCGATAG AGGAAAGTGTCT | |
| IMS83 (hsdR OUT F) | AGTATACGACTTACCTCAA | |
| IMS84 (hsdR OUT R) | TCAGTTGTTTCTGCCACG | |
| IMS85 (sauUSI IBS) | AAAAAAGCTTATAATTATC CTTAAAAGACAAGGCGGT GCGCCCAGATAGGGTG | HindIII |
| IMS86 (sauUSI EBS1d) |
CAGATTGTACAAATGTGGTG
ATAACAGATAAGTCAAGGCG
TTTAACTTACCTTTCTTTGT |
BsrGI |
| IMS87 (sauUSI EBS2) | TGAACGCAAGTTTCTAATTTC GGGTTTCTTTCCGATAGAGG AAAGTGTCT | |
| IMS90 (sauUSI OUT F) | ATGAGTAGATTACTAAATG | |
| IMS91 (sauUSI OUT R) | CGTTACTACGTTTGAACC | |
| IMSuni | CGAAATTAGAAACTTGCGTTCAGTAAAC | |
| IM151 (pIMAY MCS F) | TACATGTCAAGAATAAACTGCCAAAGC | |
| IM152 (pIMAY MCS R) | AATACCTGTGACGGAAGATCACTTCG | |
S. aureus and S. epidermidis gene designations are taken from http://kegg.jp. For oligonucleotides, anti secY stands for antisense secY RNA.
The description (relevant genotype, phenotype, or other characteristic) is shown for bacterial strains and plasmids. MSSA, methicillin-sensitive S. aureus.
The primers used in the construction of recombinant plasmids are shown in parentheses at the end of the entry. The sequences for primers are shown. Restriction sites are indicated by underlining. Regions of homology for SOE PCR with the B primer are shown in italic type, and regions of homology for recombineering in E. coli are shown in bold type.
The source or reference is shown for bacterial strains and plasmids. The restriction enzyme (RE) site is shown for oligonucleotides.
Oligonucleotides and DNA sequencing were purchased from IDT. Restriction enzymes and LigaFAST T4 DNA ligase were purchased from NEB and Promega, respectively. High-fidelity PCR was performed with KOD Hotstart DNA polymerase (Novagen) or Phusion DNA polymerase (Finnzymes) on genomic DNA isolated with the Genelute bacterial genomic DNA kit (Sigma). Plasmids and PCR products were purified using WizardPlus kits (Promega). To isolate plasmid DNA from S. aureus, a 10-ml overnight culture was treated with 100 µg lysostaphin (Ambi Products, New York) in P1 buffer for 30 min at room temperature and then processed as recommended by the manufacturer (GeneJET plamsid miniprep kit; Fermentas).
For colony PCR, a small amount of colonial growth was touched to the side of a PCR tube and microwaved for 5 min at 800 W. The tube was placed on ice, Phire Hotstart II master mix (Finnzymes) was added to the PCR tube, and thermocycling conditions were conducted as recommended by the manufacturer.
Construction of temperature-sensitive plasmids pIMC5 and pIMAY.
A new temperature-sensitive vector was constructed for allelic replacement mutagenesis in S. aureus. The vector is based on the replicon of the lactococcal plasmid pWV01ts (from pVE6007 [21]) rather than the commonly used staphylococcal pE194ts replicon (30). A hybrid vector (pIMC5) was created by spliced overlap extension (SOE) PCR. It comprises (i) the repBCAD(Ts) genes from pVE6007 and (ii) the E. coli backbone p15A rep and pBluescript KS multiple cloning site and the highly expressed chloramphenicol acyltransferase marker from pIMC (31). The counterselection marker encoding tetracycline-inducible antisense secY RNA was amplified from pKOR1 and introduced between the novel BglII and SphI sites to form pIMAY.
Electroporation.
Electroporation was conducted essentially as described by Löfblom et al. (16). Overnight cultures of S. aureus or S. epidermidis were grown in 10 ml of either TSB or BHI (in 50-ml tubes) and then diluted to an optical density at 578 nm (OD578) of 0.5 in fresh prewarmed media. The cultures were reincubated for 30 min and then chilled in an ice slurry for 10 min, with all subsequent steps performed at 4°C on ice. The cells were harvested at 4,000 × g for 10 min, and the pellets were resuspended in an equal volume of autoclaved ice-cold water. The centrifugation and resuspension steps were repeated. The cells were then repeatedly centrifuged and resuspended first in 1/10, then in 1/25, and finally in 1/200 the volume of autoclaved ice-cold 10% (wt/vol) glycerol. Aliquots of 50 µl were frozen at −70°C. For electroporation, cells were thawed on ice for 5 min and then left at room temperature for 5 min before being centrifuged (5,000 × g for 1 min) and resuspended in 50 µl of 10% glycerol and 500 mM sucrose (filter sterilized). Plasmid DNA (5 µg) was precipitated with pellet paint (Novagen) and added to the cells, transferred to a 1-mm electroporation cuvette (Bio-Rad) at room temperature, and pulsed at 21 kV/cm, 100 Ω, and 25 µF. The cells were incubated in 1 ml of TSB supplemented with 500 mM sucrose (filter sterilized), incubated at 28°C or 37°C for 1 h before plating on BHIA plus 10 µg of Cm per ml (Cm10), and incubated at 28°C or 37°C.
Allelic exchange with pIMAY.
Deletion constructs for the hsdR, sauUSI, sauUSIECORV, and mcrR genes and a putative nudix hydrolase-encoding gene (Table 1) were PCR amplified as follows. A sequence upstream of the gene to be deleted was amplified with oligonucleotides A and B (A/B) (up to the start codon) and the downstream sequence with oligonucleotides C/D (down from the stop codon) separately. The upstream and downstream PCR products were diluted 1:20, and 1 µl of each was used as the template in a second SOE PCR with the A/D primers. Deletion constructs were cleaved at endonuclease sites introduced into A and D primers during PCR and ligated into pIMAY cut with the same enzymes and then transformed into E. coli DC10B. The plasmid DNA was sequenced. The DNA was then electroporated into the target strain and plated onto BHIA plus Cm10 at 28°C.
To correct the premature stop codon in the sauUSI gene of S. aureus RN4220, the wild-type sequence from S. aureus 8325-4 was amplified as a 1-kb fragment centered on the RN4220 premature stop codon and then processed as described above.
To complement the sauUSI mutation in S. aureus NRS384 hsdRINT sauUSIINT, the sauUSI deletion mutant was reverted to the wild type by allelic exchange. To differentiate NRS384 hsdRINT from the NRS384 hsdRINT sauUSIECORV complemented strain, a new EcoRV restriction site (http://emboss.bioinformatics.nl/cgi-bin/emboss/silent) was introduced into the complementation construct without altering the coding sequence. Phenotypically, no differences were observed between NRS384 hsdRINT and NRS384 hsdRINT sauUSIECORV mutants.
To integrate pIMAY into the chromosome, a single colony from the transformation plate was homogenized in 200 µl of TSB. The suspension was diluted 10-fold to 10−3, and 100 µl of each dilution was spread on BHIA plus Cm10 and incubated overnight at 37°C. For S. epidermidis, a colony from the transformation plate was inoculated into BHI plus Cm10 and grown overnight at 37°C, and diluted suspensions of the bacterial growth were plated for single colonies. For both S. aureus and S. epidermidis, large colonies were streaked on BHIA plus Cm10 and incubated overnight at 37°C, and colony PCR analysis was performed to determine (i) the absence of extrachromosomal plasmid DNA (with MCS oligonucleotides IM151/152) (Fig. 3B) and (ii) whether plasmid integration had occurred upstream or downstream of the gene (OUT F/D Rev oligonucleotides or OUT R/A Fwd oligonucleotides, e.g., hsdR IM5/94 or IM6/93) (Fig. 3C). Overnight cultures of both the upstream or downstream crossover that were free of replicating plasmid were grown at 28°C without chloramphenicol and then plated onto BHIA containing 1 µg/ml anhydrotetracycline (Vetranal; Sigma) (BHIA plus ATc). The plates were incubated at 28°C for 2 days. Large colonies were patched on BHIA plus ATc and BHIA plus Cm10 and grown at 37°C overnight. Chloramphenicol-sensitive colonies were screened by colony PCR with oligonucleotides to identify clones containing the desired mutation (OUT F/OUT R [e.g., ∆hsdR-IM5/6]). Putative mutants were validated by PCR amplification of genomic DNA flanking the deletion and DNA sequencing.
Purification of SauUSI.
The entire sauUSI gene was amplified from S. aureus Newman genomic DNA with primers IM196 and IM197, digested with NcoI and XhoI, and cloned into the C-terminal hexahistidine tag vector pET21d+. Plasmid DNA was transferred from E. coli DH10B to BL21(DE3) for protein expression. An overnight culture was diluted 1:100 in fresh L broth (500 ml) and grown at 37°C to an OD600 of 1.0. The temperature of the culture was reduced to 28°C, and expression of SauUSI was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 3 h, the induced cells were harvested by centrifugation (4,000 × g for 20 min at 4°C). The cell pellet was suspended in 10 ml of native E. coli lysis buffer (50 mM HEPES [pH 8], 500 mM NaCl, 10 mM imidazole, 5% glycerol, 1 complete protease inhibitor tablet [Roche] containing 125 U Benzonase [Novagen] and 10 µg of lysozyme). The cells were frozen at −70°C and then freeze-thawed 3 times before centrifugation (20,000 × g for 30 min at 4°C). The supernatant was filter sterilized (0.45-µm filter) before it was passed twice through Histrap FF column (GE Healthcare) equilibrated with native lysis buffer at 1 ml/min. The column was then washed with 25 ml of wash buffer (native E. coli lysis buffer containing 30 mM imidazole). The protein was eluted from the column in ten 500-µl aliquots of native lysis buffer containing 250 mM imidazole. The SauUSI-his eluates were visualized on a 10% SDS-polyacrylamide gel, and the protein-containing aliquots were combined. The imidazole in the buffer was diluted (112,500 times) by centrifugation three times through a 50-kDa-molecular-size-cutoff filter (Millipore) with the volume made up to 15 ml with 50 mM HEPES [pH 8], 500 mM NaCl, 5 mM MgCl2, and 5% glycerol (32). Aliquots were stored at 4°C.
Activity of SauUSI.
pRMC2 plasmid DNA (1 µg) isolated from isogenic E. coli strains derived from strain BW25113 containing different methylation enzymes were mixed with SauUSI-his (ca. 425 ng) in ligation buffer (50 mM Tris-Cl [pH 7.5], 10 mM MgCl2, 10 mM dithiothreitol [DTT], 1 mM ATP [final concentration]) and incubated at 37°C for 30 min. To separate the DNA from SauUSI, an equal volume of membrane binding solution (Promega) was added, and the DNA was ethanol precipitated with pellet paint. The samples were then run on a 1% agarose gel and compared to an untreated plasmid DNA control.
Recombineering in E. coli DH10B.
Strain DH10B is a high-efficiency cloning strain of E. coli K-12 (Invitrogen). Even though it has the recA1 mutation, in our hands, it is still amenable to genetic manipulation by recombineering. The plasmid pKD46 (26) was transformed into strain DH10B using the electroporation protocol of Sheng et al. (33) and selected on 100 µg/ml carbenicillin at 30°C. Strain DH10B carrying pKD46 was made electrocompetent as described above, except that the cells were grown at 30°C (instead of 37°C), and once the culture reached an OD600 of 0.4, filter-sterilized arabinose (Sigma) was added to a final concentration of 0.2%. The culture was incubated for 1 h to induce exo, beta, and gam expression, and the cells were washed and stored at −70°C. The plasmid pKD4 (26) was used as a template to PCR amplify the kanamycin resistance marker flanked by two flippase recognition target (FRT) sites. The forward and reverse primers (IM261/IM262) were tailed with 50 nucleotides complementary up to the dcm gene start codon and 23 codons down from the stop codon to generate an in-frame deletion within the dcm gene. The linear amplimer was electroporated into the electrocompetent E. coli DH10B(pKD46), and transformants were selected on LBA containing 50 µg/ml of kanamycin at 37°C. One kanamycin-resistant clone was selected and plated on LBA at 43°C to eliminate pKD46. An ampicillin-sensitive derivative was made competent and transformed at 30°C with pCP20 (26) (selected on LBA with 10 µg/ml Cm) to excise the kanamycin resistance marker. Finally, pCP20 was eliminated by plating at 43°C to yield E. coli DH10BΔdcm (called DC10B). The loss of cytosine methylation was confirmed phenotypically through the inability of SauUSI to digest DC10B plasmid or genomic DNA and transformation of wild-type S. aureus and S. epidermidis strains with a shuttle plasmid isolated from DC10B.
Creation of targetron insertion mutants in S. aureus NRS384.
Targetron insertion mutants were created in S. aureus NRS384 following the protocol of the manufacturer (Sigma) (17). Primers were designed to retarget the intron for hsdR (IMS80/IMS81/IMS82) or sauUSI (IMS85/IMS86/IMS87). To amplify the 350-bp retargeted amplicon for hsdR or sauUSI, the above primers were combined with IMSuni. The amplicon was digested with HindIII/BrsGI and cloned into similarly cut pNL9164. pNL9164hsdR or pNL9164sauUSI was then passaged through S. aureus RN4220 at 30°C before being electroporated into NRS384 at 30°C. A single colony was streaked on Trypticase soy agar (TSA) plus 25 µg of Em per ml (Em25) containing 10 µM CdCl2 and incubated overnight to induce intron mobilization. Single colonies were screened for insertion of the intron into hsdR (IMS83/IMS84) or sauUSI (IMS90/IMS91) by colony PCR. The plasmid was cured by overnight growth in broth at 43°C followed by plating onto TSA at 30°C. Colonies were patched onto TSA and TSA plus Em to identify plasmid-free isolates. The double mutant was created in the sauUSIINT background.
Intravenous infection of A/J mice.
Overnight cultures of S. aureus NRS384, hsdRINT, sauUSIINT, hsdRINT sauUSIINT, and hsdRINT sauUSIECORV strains were diluted 1:100 in TSB and grown to an OD600 of 0.5 to 0.6. The cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS) and resuspended to an OD600 of 1 (5 × 108 CFU/ml). The inoculum was diluted to 2 × 107 CFU/ml, and 100 µl was injected into the tail vein of a 6- to 7-week-old female A/J mouse. Eight mice were used for each strain. At day 7 postinfection, the mice were euthanized, and the total bacterial loads in both kidneys were determined by serial dilution and plating.
Nucleotide sequence accession number.
The nucleotide sequence of pIMAY was deposited in GenBank under accession number JQ621981.
SUPPLEMENTAL MATERIAL
HindIII cleavage of pRMC2 DNA isolated from the different S. aureus STs or S. epidermidis RP62a. Plasmid (pRMC2 derived from E. coli DC10B) was isolated from strains transformed in Fig. 5B and digested with HindIII. Strains are named according to the ST number or RP for RP62a. Arrowheads highlight the pRMC2 fragments of 2.9 kb, 2.65 kb, and 850 bp. Download Figure S1, TIF file, 0.4 MB.
ACKNOWLEDGMENTS
We thank Luciano Marraffini for providing strain RP62a, Alice Prince for strain LAC, Barbara Murray for strain OG1RF, Jodi Lindsay for strain JH2-2, Dag Anders Brede for providing V583, Luis Servin-Gonzalez for the E. coli methylation mutants in the BW25113 background, and Carsten Kroger for recombineering plasmids and advice.
We acknowledge the Science Foundation Ireland (grant number 08/IN.1/B1845) for financial support.
Footnotes
Citation Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3(2):e00277-11. doi:10.1128/mBio.00277-11.
REFERENCES
- 1. Corvaglia AR, et al. 2010. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. U. S. A. 107:11954–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Veiga H, Pinho MG. 2009. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl. Environ. Microbiol. 75:3034–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waldron DE, Lindsay JA. 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Neill AJ. 2010. Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 51:358–361 [DOI] [PubMed] [Google Scholar]
- 6. Voyich JM, et al. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907–3919 [DOI] [PubMed] [Google Scholar]
- 7. Mack D, Siemssen N, Laufs R. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heilmann C, Gerke C, Perdreau-Remington F, Götz F. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Otto M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat. Rev. Microbiol. 7:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreiswirth BN, et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 12. Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. 2011. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39:5597–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Augustin J, Götz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203–207 [DOI] [PubMed] [Google Scholar]
- 14. Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129 [DOI] [PubMed] [Google Scholar]
- 15. González-Cerón G, Miranda-Olivares OJ, Servín-González L. 2009. Characterization of the methyl-specific restriction system of Streptomyces coelicolor A3(2) and of the role played by laterally acquired nucleases. FEMS Microbiol. Lett. 301:35–43 [DOI] [PubMed] [Google Scholar]
- 16. Löfblom J, Kronqvist N, Uhlén M, Ståhl S, Wernérus H. 2007. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 102:736–747 [DOI] [PubMed] [Google Scholar]
- 17. Yao J, et al. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foster TJ. 1998. Molecular genetic analysis of staphylococcal virulence. Methods Microbiol. 27:433–454 [Google Scholar]
- 19. Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 20. Sun F, et al. 2010. Aureusimines in Staphylococcus aureus are not involved in virulence. PLoS One 5:e15703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Riedel CU, et al. 2007. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl. Environ. Microbiol. 73:3091–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galperin MY, Moroz OV, Wilson KS, Murzin AG. 2006. House cleaning, a part of good housekeeping. Mol. Microbiol. 59:5–19 [DOI] [PubMed] [Google Scholar]
- 24. Gill SR, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cowan ST, Shaw C, Willams RE. 1954. Type strain for Staphylococcus aureus Rosenbach. J. Gen. Microbiol. 10:174–176 [DOI] [PubMed] [Google Scholar]
- 26. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stobberingh EE, Schiphof R, Sussenbach JS. 1977. Occurrence of a class II restriction endonuclease in Staphylococcus aureus. J. Bacteriol. 131:645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu W, et al. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 30. Villafane R, Bechhofer DH, Narayanan CS, Dubnau D. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74:3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graslund S, et al. 2008. Protein production and purification. Nat. Methods 5:135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheng Y, Mancino V, Birren B. 1995. Transformation of Escherichia coli with large DNA molecules by electroporation. Nucleic Acids Res. 23:1990–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monk IR, Casey PG, Cronin M, Gahan CG, Hill C. 2008. Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC: a new site-specific integrative vector. BMC Microbiol. 8:96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cooper JE, Feil EJ. 2006. The phylogeny of Staphylococcus aureus—which genes make the best intra-species markers? Microbiology 152:1297–1305 [DOI] [PubMed] [Google Scholar]
- 36. Holden MT, et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bourgogne A, et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9(7):R110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 41. Palmer BR, Marinus MG. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HindIII cleavage of pRMC2 DNA isolated from the different S. aureus STs or S. epidermidis RP62a. Plasmid (pRMC2 derived from E. coli DC10B) was isolated from strains transformed in Fig. 5B and digested with HindIII. Strains are named according to the ST number or RP for RP62a. Arrowheads highlight the pRMC2 fragments of 2.9 kb, 2.65 kb, and 850 bp. Download Figure S1, TIF file, 0.4 MB.