Abstract
The origin of new genes has long been considered a fundamental question in evolutionary biology. In eukaryotes, a major pathway for the ‘birth’ of new nuclear genes has been transfer of genes from the cytoplasmic organelles (mitochondria and plastids) to the nucleus. While the vast majority of gene transfer occurred shortly after endosymbiosis, the process continues today and is still driving the evolution of nuclear genomes. In tobacco (Nicotiana tabacum) a number of studies have indicated that DNA can transfer from the chloroplast to the nucleus at relatively high frequency. Less has been known, however, about how a newly transferred organelle gene can become activated in this new genetic environment. In a recent report we observed, in real-time, the activation of a plastid reporter gene newly transferred to the nucleus. A key observation from this study was that non-homologous repair is an important generator of novel sequence combinations which, in rare instances, can result in the nuclear activation of plastid genes. In addition, the activation of relocated genes can be aided by the fortuitous presence of plastid sequences able to promote nuclear expression.
Keywords: chloroplast, plastid, endosymbiosis, evolution, gene transfer, nupt
The cells of plant, algal and some protist lineages contain three genetic compartments. These are the nucleus, which houses the majority of the genes, and the two cytoplasmic organelles—mitochondria and plastids. The genomes of each of these compartments are derived from ertswhile free-living prokaryotes. In contrast to the nuclear genome, plastid (chloroplast) and mitochondrial genomes are vastly reduced in size when compared with those of their ancestors and extant relatives.1 This reduction in genome size is due both to the loss of redundant organelle genes and to mass relocation of organelle genes to the nucleus.1,2 The majority of this gene transfer probably occurred early in eukaryote evolution,1 but molecular and bioinformatic analyses (for some examples see refs. 3–10) have shown that transfer of functional genes and DNA fragments from the plastid to the nucleus continues today.
A look at almost any sequenced eukaryote nuclear genome reveals large tracts of DNA essentially identical to the extant plastid and/or mitochondrial genomes.4 These sequences are referred to as nupts (nuclear integrants of plastid DNA) and numts (nuclear integrants of mitochondrial DNA) respectively, or collectively as norgs (nuclear integrants of cytoplasmic organelle DNA). The norgs found in published genome sequences may only be the ‘tip of the iceberg’, as contig assembly tends to minimize clone length, meaning large duplications of organelle DNA can go undetected.11 This is particularly relevant, given that many de novo insertions of plastid DNA contain multiple copies of large regions of the plastome at a single locus.5,9 The common practice of discarding ‘contaminating’ organelle DNA when assembling nuclear genome sequences, must also contribute to the under-representation of norgs in published whole genome sequences.
Real-Time Experimental Reconstruction of Endosymbiotic Gene Transfer
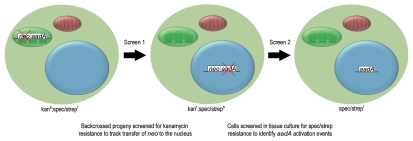
In tobacco (Nicotiana tabacum) it has been possible to observe experimentally, in real-time, the transfer of plastid DNA to the nucleus.5,9,10 These observations were made in plants containing, in their plastome, an aminoglycoside resistance gene (aadA) used to select the transplastomic lines, together with a closely linked kanamycin resistance gene (neo) designed for exclusive nuclear expression. As neo was not active in the chloroplast but would be active if relocated to the nucleus, transfer of this gene could be detected simply by screening seedling progeny, or cells in tissue culture, for kanamycin resistance (Fig. 1 and screen 1). As neo was already equipped for nuclear expression, these experiments did not measure the frequency of endosymbiotic gene transfer per se but merely the frequency at which plastid DNA transfers to the nucleus.
Figure 1.
Consecutive screens were used to identify endosymbiotic transfer of a chloroplast transgene. Screen 1, used transplastomic tobacco plants containing a plastid specific copy of aadA and a nucleus specific copy of neo in their plastid genome. These kanamycin sensitive (kans), spectinomycin and streptomycin resistant (spec/strepr) plants were backcrossed to female wild type and the resultant progeny tested for kanamycin resistance. Kanamycin resistant (kanr) progeny contained neo (and usually aadA) in their nuclear genome. These plants had wild type chloroplasts, inherited from their maternal parent and an inactive nuclear copy of aadA, making them sensitive to spectinomycin and streptomycin (spec/streps). In screen 2, cells from kanamycin resistant plants were screened for spectinomycin and streptomycin resistance to identify cells in which aadA had been activated in the nucleus.
The transfer frequency was found to be remarkably high, particularly in the male germline, with one in 11,000 to 16,000 pollen grains containing a new nuclear copy of the gene.5,9 This frequency was at least 15-fold higher than that observed in the female germline9 and ∼300 fold higher than that observed in somatic tissue.10 The exceptionally high rate is thought to relate to the programmed degeneration and exclusion of plastids in developing male gametophytes,9 both a result and probable cause of uni-parental inheritance. During this process plastid DNA is presumably released into the cytoplasm from where it can transfect nuclear chromosomes. It follows that other factors that compromise organelle integrity, such as environmental stress,12 may also lead to an increase in the frequency with which plastid DNA enters the nucleus.
While the rate at which plastid DNA relocates is an important question, perhaps more interesting, from both evolutionary and biotechnological perspectives, is the rate at which a cytoplasmic organelle gene may become functionally active in the nucleus. This is not a trivial process as it requires not only transfer of DNA encoding a protein to the nucleus but also acquisition of a nuclear promoter and polyadenylation signal and, if the gene product is to be targeted back to the organelle, it must also acquire a sequence specifying a transit peptide or another mechanism of protein targeting.
We addressed the question of functional gene transfer using the kanamycin resistant lines generated in the genetic screens outlined above. In these lines the chloroplast fragments transferred to the nucleus were large and so, in almost all cases, aadA was co-transferred with neo to the nucleus. Although these plants contained a copy of aadA in the nucleus, they were sensitive to aminoglycosides, as the gene had a plastid promoter and terminator and was therefore inactive (these lines no longer contained aadA in the plastome as they were the progeny of backcrosses to female wild type; Fig. 1). By screening cells in tissue culture for aminoglycoside resistance we were able to regenerate plants in which aadA had undergone nuclear activation (Fig. 1 and screen 2), paralleling the pathway of genes functionally transferred during endosymbiotic evolution. This approach had been used once previously in a study by Stegemann and Bock.13 We extended their work, screening a much larger number of independent lines and using a significantly different arrangement of the experimental genes which enabled us to uncover more diverse and evolutionarily applicable activation events. Further, we reported the complete sequence of a de novo nupt and its flanking sequences, providing insight into how chloroplast DNA fragments are incorporated into nuclear chromosomes.
Plastid DNA Insertions Often Form Complex Loci
Insertions of plastid DNA are notoriously difficult to characterize.14 This is due to their large size and complex nature and also the presence of many copies of identical or highly similar sequences in the chloroplast and throughout the nuclear genome (due to historic transfer events). Despite these difficulties, we reported the first full sequence of a de novo nupt and showed that it was comprised of three fragments of plastid DNA from disparate regions of the plastome.15 Analysis of the plastid/plastid and plastid/nuclear sequence junctions indicated that the insertion was mediated by synthesis-dependent non-homologous end joining (NHEJ), probably at a site of DSB repair. Despite this tripartite structure, this particular locus is likely to represent a relatively simple chloroplast DNA insertion. Indeed, it was experimentally tractable because of its comparatively short length (∼17 kb) and the absence of any internal duplications. Several other lines had far more complex insertions containing approximately 10–15 copies of a >20 kb region of the chloroplast genome which together must exceed several hundred kilobases in length. Recent insertions of similar sizes have been observed in Arabidopsis11 and maize16,17 indicating that insertions of this size are not unique to tobacco. Clearly, the insertion of plastid DNA can generate remarkably large loci, containing complex arrangements of sequence from dispersed parts of the plastid genome. The probable generator of these loci is the cell's endogenous non-homologous DNA repair machinery. By ‘stitching’ together and integrating a selection of what must be a plethora of available DNA fragments, this nuclear repair pathway generates complex libraries of novel sequence combinations from which new nuclear genes can arise. Our results suggest cytoplasmic organelle DNA is a foremost contributor to the spectrum of fragments inserted during repair.
After Insertion, Non-Homologous DNA Repair Generates Novel Sequence Arrangements
Activation of the dormant aadA gene in its new nuclear environment was revealed by screens for aminoglycoside resistance in somatic cells.13,15 The activation events were the result of local sequence rearrangements such as deletions, inversions, duplications and insertions that recruited nearby nuclear promoters and enhancers. Analyses of the resulting novel sequence junctions suggest that, similar to the insertion of plastid sequences, rearrangements causing “eukaryotization” of aadA are primarily the result of non-homologous end joining at DNA double strand breaks. Thus it is possible that integration may favor areas of the genome more susceptible to DSBs,18 a propensity which may also promote subsequent rearrangements at these loci. This is supported by the finding that large insertions of chloroplast DNA preferentially locate to pericentromeric regions6 which are known DSB hotspots.19 These regions also contain a high density of transposable elements20 which may further increase the frequency of sequence rearrangements. Accordingly, work in rice indicates that the nuclear genome continually integrates shuffles and eliminates chloroplast DNA sequences6 and in tobacco it has been shown that about 50% of nupts are deleted at high frequency within a generation of insertion.14
Notably, the majority of sequence rearrangements appear to be local in nature. This is indicated by the fact that in both studies13,15 all activations of aadA resulting from sequence rearrangements were due to the recruitment of the nearby 35S promoter, the promoter used to drive expression of the closely linked neo gene. In no case was activation due to the recruitment of native nuclear regulatory sequences. The activation frequencies determined in both screens are therefore dependent on their unique experimental arrangements and at most provide a rough upper limit of the activation frequency expected in the absence of a closely linked strong nuclear promoter. Given the large size of our screen, it is now obvious that recruitment of a native nuclear promoter is very rare, probably too rare to investigate through experimental simulation.
Chloroplast DNA Contains Elements Able to Promote Nuclear Expression
A surprise finding of this work was the observation that chloroplast DNA can contain fortuitous sequence elements able to promote nuclear expression. All cells from two of the lines screened were found to be uniformly resistant to aminoglycosides, indicating that in these plants aadA was expressed in the absence of any secondary sequence changes. It appears that, in these two lines, aadA was transferred to the nucleus in sufficiently high copy number that weak nuclear activity of the psbA promoter21 upstream of aadA resulted in significant nuclear expression. Interestingly, we also found that polyadenylation can occur, albeit inefficiently, at two sites within the psbA 3′ UTR of aadA transcripts. Both sites fortuitously match the loose AU-rich plant polyadenylation consensus sequence.22 Polyadenylation of nuclear transcripts from a consensus sequence within the psbA terminator was also observed in the study by Stegemann and Bock.13 They suggest that the AT-rich nature of chloroplast non-coding regions may provide many fortuitous polyadenylation sites, greatly aiding the process of gene activation after DNA transfer. This may also be true of other AT-rich regulatory elements such as TATA and CAAT boxes, both of which are found within the psbA promoter.21 Whether this weak nuclear expression is a unique characteristic of the psbA promoter, or is true more widely of chloroplast promoters, remains to be seen.
Implications and Overview
The use of chloroplast biotechnology will undoubtedly play an important role in securing food supply for an increasing world population.23 An often-advocated advantage of chloroplast biotechnology is that transgene containment is provided through maternal inheritance of chloroplasts.24 However, to fully understand the level of transgene containment provided, the rate of functional gene transfer to the nucleus must be considered. In our estimates, while it is clear that functional transfer of a chloroplast transgene to the nucleus is possible, provided the chloroplast promoter has no latent nuclear activity, paternal chloroplast leakage remains the major threat to escape.25,26 As little is currently known about the nuclear activity of chloroplast promoters, it may be prudent to assess chloroplast promoters for nuclear activity prior to use in chloroplast biotechnology. In addition, we observed that nuclear activation of a chloroplast transgene primarily occurs via the recruitment of a closely linked nuclear promoter. This point should be considered if simultaneously transforming nuclear and chloroplast genomes.27 This process has the potential to co-integrate nuclear and chloroplast transgenes at a single nuclear locus, inevitably increasing the chance of generating an active nuclear copy of the chloroplast transgene.
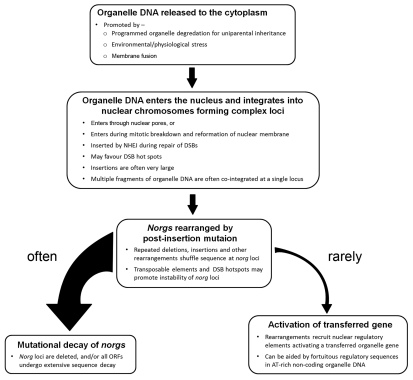
Over the past decade a very dynamic picture of organelle DNA in the nuclear genome has arisen, leading to the current model of functional gene transfer (Fig. 2). Organelle DNA is incorporated into the nuclear genome at a strikingly high rate and the resulting loci are often very large, containing multiple copies of DNA fragments from disparate regions of the organelle genomes. Once in the nucleus, these sequences evolve rapidly, generating many new sequence arrangements. In some rare instances these new sequence combinations give rise to new nuclear genes. The process of gene activation may be aided by the presence of latent gene regulatory elements in the AT-rich chloroplast non-coding DNA. While these new nuclear genes may be inefficiently expressed initially, a low level of expression may be sufficient to selectively maintain the gene while more affective transcription and polyadenylation is established through further mutation. The rarity of these events may deter experimental investigation by laboratory scientists but this should not lead to complacency among gene regulation authorities. Broad acre crops surely offer the cell numbers required to uncover endosymbiotic evolutionary gene transfer events.
Figure 2.
A model for functional transfer of organelle genes to the nucleus.
Acknowledgments
This research was supported under the Australian Research Council's Discovery Projects funding scheme (project number DP0986973).
Abbreviations
- DSB
double strand break
- NHEJ
non-homologous end joining
- numt
nuclear integrant of mitochondrial DNA
- norg
nuclear integrant of cytoplasmic organelle DNA
- nupt
nuclear integrant of plastid DNA
References
- 1.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 2.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 3.Palmer JD, Adams KL, Daley DO, Qiu YL, Whelan J. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 4.Hazkani-Covo E, Zeller RM, Martin W. Molecular Poltergeists: Mitochondrial DNA Copies (numts) in Sequenced Nuclear Genomes. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- 6.Matsuo M, Ito Y, Yamauchi R, Obokata J. The rice nuclear genome continuously integrates, shuffles and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell. 2005;17:665–675. doi: 10.1105/tpc.104.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D. Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet. 2007;23:597–601. doi: 10.1016/j.tig.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Sheppard AE, Ayliffe MA, Blatch L, Day A, Delaney SK, Khairul-Fahmy N, et al. Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol. 2008;148:328–336. doi: 10.1104/pp.108.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stegemann S, Hartmann S, Ruf S, Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc Natl Acad Sci USA. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupar RM, Lilly JW, Town CD, Cheng Z, Kaul S, Buell CR, et al. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci USA. 2001;98:5099–5103. doi: 10.1073/pnas.091110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegemann S, Bock R. Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell. 2006;18:2869–2878. doi: 10.1105/tpc.106.046466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheppard AE, Timmis JN. Instability of Plastid DNA in the Nuclear Genome. PLoS Genet. 2009:5. doi: 10.1371/journal.pgen.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd AH, Timmis JN. The origin and characterization of new nuclear genes originating from a cytoplasmic organellar genome. Mol Biol Evol. 2011;28:2019–2028. doi: 10.1093/molbev/msr021. [DOI] [PubMed] [Google Scholar]
- 16.Lough AN, Roark LM, Kato A, Ream TS, Lamb JC, Birchler JA, et al. Mitochondrial DNA transfer to the nucleus generates extensive insertion site variation in maize. Genetics. 2008;178:47–55. doi: 10.1534/genetics.107.079624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roark LM, Hui Y, Donnelly L, Birchler JA, Newton KJ. Recent and Frequent Insertions of Chloroplast DNA into Maize Nuclear Chromosomes. Cytogenet Genome Res. 2010;129:17–23. doi: 10.1159/000312724. [DOI] [PubMed] [Google Scholar]
- 18.Lenglez S, Hermand D, Decottignies A. Genome-wide mapping of nuclear mitochondrial DNA sequences links DNA replication origins to chromosomal double-strand break formation in Schizosaccharomyces pombe. Genome Res. 2010;20:1250–1261. doi: 10.1101/gr.104513.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-strand-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 20.Hall AE, Kettler GC, Preuss D. Dynamic evolution at pericentromeres. Genome Res. 2006;16:355–364. doi: 10.1101/gr.4399206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelissen M, Vandewiele M. Nuclear transcriptional activity of the plastid psbA promoter. Nucleic Acids Res. 1989;17:19–29. doi: 10.1093/nar/17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li QS, Hunt AG. The polyadenylation of RNA in plants. Plant Physiol. 1997;115:321–325. doi: 10.1104/pp.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327:818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- 24.Daniell H. Transgene containment by maternal inheritance: Effective or elusive? Proc Natl Acad Sci USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elghabi Z, Ruf S, Bock R. Biolistic co-transformation of the nuclear and plastid genomes. Plant J. 2011;67:941–948. doi: 10.1111/j.1365-313X.2011.04631.x. [DOI] [PubMed] [Google Scholar]