SUMMARY
Hundreds of Chromatin Regulators (CRs) control chromatin structure and function by catalyzing and binding histone modifications, yet the rules governing these key processes remain obscure. Here, we present a systematic approach to infer CR function. We developed ChIP-string, a meso-scale assay that combines chromatin immunoprecipitation with a signature readout of 487 representative loci. We applied ChIP-string to screen 145 antibodies, thereby identifying effective reagents, which we used to map the genome-wide binding of 29 CRs in two cell types. We found that specific combinations of CRs co-localize in characteristic patterns at distinct chromatin environments, genes of coherent functions and distal regulatory elements. When comparing between cell types, CRs redistribute to different loci, but maintain their modular and combinatorial associations. Our work provides a multiplex method that substantially enhances the ability to monitor CR binding, presents a large resource of CR maps, and reveals common principles for combinatorial CR function.
INTRODUCTION
Gene regulation in eukaryotes relies on the functional packaging of DNA into chromatin, a higher-order structure composed of DNA, RNA, histones and associated proteins. Chromatin structure and function is regulated by post-translational modifications of the histones, including acetylation, methylation and ubiquitinylation (Kouzarides, 2007; Margueron and Reinberg, 2010; Ruthenburg et al., 2007).
Advances in genomic technologies – in particular Chromatin Immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) – have enabled researchers to characterize chromatin structure genome-wide in different mammalian cells (Barski et al., 2007; Birney et al., 2007; Heintzman et al., 2007; Mikkelsen et al., 2007; Zhang and Pugh, 2011; Zhou et al., 2011). The resulting maps have shown that distinct histone modifications often exist in well-defined combinations, corresponding to different genomic features (e.g., promoters, enhancers, gene bodies) or regulatory states (e.g., actively transcribed, silenced, poised). The number of chromatin types may in fact be relatively limited (Ernst and Kellis, 2010; Filion et al., 2010). For example, a study of chromatin landscapes across 9 different human cell types distinguished 15 dominant chromatin types or ‘states’ based on their combinatorial histone modifications (Ernst et al., 2011). The chromatin ‘state’ of each locus varies between cell types, reflecting lineage-specific gene expression, developmental programs or disease processes.
The human genome encodes hundreds of chromatin regulators (CRs) that add (‘write’), remove (‘erase’) or bind (‘read’) these modifications (Kouzarides, 2007; Ruthenburg et al., 2007). CRs are expressed in a tissue-specific manner, and play important roles in normal physiology and disease (Ho and Crabtree, 2010). For example, cancer genome projects have unveiled prevalent mutations in CR genes, suggestive of broad roles in tumor biology (Elsasser et al., 2011). It is compelling to hypothesize that combinatorial histone modification states are determined by different combinations of CRs.
Despite their importance, the target loci and specific functions of most mammalian CRs remain unknown. In contrast to histone modifications that are readily mapped by ChIP-seq, systematic localization of CRs has proven challenging. While recent studies in yeast (Venters et al., 2011) and fly (Filion et al., 2010) have profiled multiple CRs, few have been mapped in mammalian cells. Furthermore, the available profiles typically have lower signal-to-noise ratios than maps of histone modifications or transcription factors. This is likely due to the indirect associations between CRs and DNA, compounded by sub-optimal antibody reagents and ChIP procedures. This severely restricts our ability to identify the CRs that act at any given locus (Figure 1A), to determine how they impart distinct histone modifications, and to decipher how they influence genome function in cis.
Figure 1. A systematic approach for associating CRs to genomic loci and chromatin states.
(A) CR associations with chromatin modification states. Right: The chromatin regulator EZH2 (pink gear) is associated (arrow) with a locus marked by the histone modification H3K27me3 (small red ball). Left: For many other chromatin regulators (non-colored gears, CR) their target loci and associated modifications (different colored balls) remain unknown. (B) Our process consists of four steps (left to right): 1. ChIP assays were performed with 145 different antibodies targeting 92 distinct CRs and 15 modifications. 2. ChIP samples were screened for enrichment across 487 signature loci by ChIP-string to identify promising CR antibodies. 3. These antibodies were applied in ChIP-seq to generate genome-wide maps for 29 CRs. 4. We identified target loci and associated histone modifications for each CR. We found that the CRs partition into six modules with correlated binding patterns, and also exhibit instances of combinatorial binding.
Here, we developed a general methodology for identifying effective procedures to map CRs in mammalian cells (Figure 1B), and used the approach to study the localization of CRs in K562 cells and human embryonic stem (ES) cells. We first developed a meso-scale localization assay, ChIP-string, based on a signature readout of 487 loci representing diverse chromatin states. We used this approach to screen 145 antibodies, thereby identifying effective reagents, which we used to generate genome-wide binding maps for 29 CRs by ChIP-seq.
The resulting datasets provide a comprehensive view of the associations between CRs, and their relationships to histone modification states. We found that CRs bind in characteristic modular combinations, each associated with distinct modification patterns and genomic features, and often with different functional groups of genes. For example, HDAC1 and SAP30 co-bind sharply over transcription start sites (TSS) of cell cycle-related genes, while SIRT6 and CHD7 co-bind the proximal portions of highly active genes encoding ribosomal and chromatin architecture proteins. Other sets of CRs co-associate at distal elements or repressed loci. Remarkably, most modules combine CRs with opposing enzymatic activities that likely mediate homeostatic regulation of dynamic chromatin. When comparing different cell types, CRs often redistribute to different genomic regions, yet maintain their characteristic modular associations. Our work provides a new experimental approach to use ChIP in high-throughput screens, presents a valuable resource for studying CR location and function, and reveals new principles of chromatin organization.
RESULTS
ChIP-string: meso-scale location analysis for chromatin proteins
We developed a new method to determine, in multiplex, the enrichment of many CRs or histone modifications at hundreds of representative loci. We reasoned that such a signature binding profile would be highly informative. First, querying several hundred regions is less biased than sampling a handful of loci as typically done by ChIP-PCR. Second, it yields a ‘signature’ pattern that could help determine if a CR is consistently associated with loci sharing a chromatin state. Third, a signature can be measured faster and at a much lower cost that a genome-wide profile and is thus appropriate for screening antibodies and ChIP conditions for difficult targets or for perturbation screens using RNA interference or small molecules.
As a signature readout, we assembled a panel of 487 genomic loci representing different types of chromatin environments. To choose the regions, we used genome-wide chromatin state annotations for human ES and K562 cells, derived from multiple histone modification maps (Ernst et al., 2011). We selected representative loci for each of the major states in the two cell types; e.g., active or repressed promoters, transcripts, distal elements, etc. (Experimental Procedures; Table S1). We reasoned that individual CRs would localize to subsets of these representative loci, and thus enable us to distinguish an effective CR ChIP assay.
To measure enriched binding at the signature loci, we developed the ChIP-string method. ChIP-string leverages the nCounter Analysis System platform (NanoString Technologies), originally developed for multiplex quantification of RNA molecules. We designed a probe-set complementary to the signature loci, and adapted the nCounter operating procedures for ChIP DNA (Experimental Procedures). We validated ChIP-string by analyzing histone modification ChIPs and comparing the measurements to ChIP-seq data (Figures 2A and S1A).
Figure 2. Screening CR-antibodies by ChIP-string.
(A) Comparison of H3K4me3 ChIP-string to ChIP-seq data. Scatter plots compare ChIP-seq read density (X-axis) against ChIP-string counts (Y-axis). The adjacent panels reflect ChIP-string experiments performed with 5 ng of DNA (left), 0.5 ng of DNA (middle), or after whole genome amplication of 0.5 ng of DNA (right). Spearman correlations (R) are indicated at lower right. (B) ChIP-string assays that scored positively associate with distinct histone modification ‘states’. Columns represent ChIP-string data for 21 CR antibodies (bold) and 12 histone modification antibodies (non-bold). Relative enrichments are indicated for the 200 most informative loci (rows); white indicates no enrichment, black indicates high enrichment. The probes were clustered, and then sorted by the ‘chromatin states’ of the corresponding locus (initiation – green; elongation or enhancer – purple; Polycomb-repressed – red; heterochromatin – light blue). The experiments (columns) were ordered by hierarchical clustering and then fine-tuned by visual inspection (Experimental Procedures). Supporting data for ChIP-string and antibody specificity are presented in Figure S1.
We evaluated the sensitivity of ChIP-string by conducting the assay with successively smaller quantities of ChIP DNA. We found that a minimum of ~5 ng of DNA is needed to maintain quantitative accuracy. While histone modification ChIPs typically yield more than 5 ng of DNA, CR ChIPs yield much smaller quantities (<1 ng), even when millions of cells are used as starting material. We therefore implemented a rapid genomic amplification step to ChIP DNA prior to nCounter detection, and confirmed that it faithfully maintains enrichment for a majority of the signature loci (Figure 2A).
ChIP-string screen identifies effective reagents for mapping CRs
We applied ChIP-string to 126 CR antibodies, 17 histone modification antibodies and 2 IgG control antibodies (Table S2). We also analyzed 16 other control samples of un-enriched chromatin input. We used chromatin from K562 cells, with the exception that ES cell-specific CRs were profiled using chromatin from ES cells. In 21 cases, more than one antibody was tested for the same target protein, allowing us to evaluate different epitopes. Overall, we screened ~150 samples. We normalized the data by sample and then by probe, using an approach analogous to methods applied to microarray data. We then standardized the measurements in each sample, creating a scale of relative probe enrichment that is comparable across samples (Experimental Procedures).
Next, we distinguished effective CR antibodies from those yielding non-specific enrichment. We calculated correlation coefficients between each pair of ChIP-string experiments, and hierarchically clustered the data. A substantial majority of the CR binding signatures (~80) either clustered with IgG control antibodies, or formed separate clusters with overall weak signals before standardization (Figure S1B). Furthermore, none of these experiments correlated well with any specific histone modification or chromatin state. Although we cannot rule out that they enrich regions not captured by our probe-set, we designated these CR antibodies as ‘failed’ in our screen.
The remaining CR ChIP-string experiments were clearly distinct from the IgG and input control experiments (Figure S1B), exhibiting both a larger number of enriched probes as well as higher enrichment values. In many cases, these CR experiments enriched subsets of loci in patterns reminiscent of individual histone modifications or chromatin states (Figure 2B). Regardless, we designated all of these remaining CR antibodies as ‘passed’. Notably, an alternative analysis procedure, which used different statistical methods in pre-processing and antibody assessment, led to highly similar results, supporting the robustness of the screen. This alternative procedure can be used even when only few antibodies are tested (Experimental Procedures).
We carried out ChIP-seq for 39 CR antibodies that passed the screen, and a sample of 9 ‘failed’ CR antibodies. Of the 39 ‘passed’ antibodies, 34 (~90%) yielded high-quality genome-wide profiles as reflected by robust enrichment of specific genomic loci, whereas none of the failed antibodies yielded high-quality data. These results indicate that ChIP-string provides an independent and objective means to identify effective reagents for CR mapping.
A compendium of genome-wide CR maps
We used 29 of the CR antibodies that ‘passed’ our screen to generate 42 ChIP-seq datasets of the genome-wide distributions of 27 CRs in K562 cells and 15 CRs in ES cells (Figures 3A and S2; Experimental Procedures). We confirmed the specificity of each of these antibodies by Western blots (Figure S1C). We used two independent peak-calling procedures to collate enriched sites for each CR in each cell type (Table S3). The number of sites ranged from 1,680 for HP1γ to 30,993 for RBBP5 to 39,180 for RNA polymerase II phosphorylated at serine 5 (RNAPIIS5P), with a median of 9,194 sites per CR. The vast majority of enriched regions were between 1 and 2 kb in size.
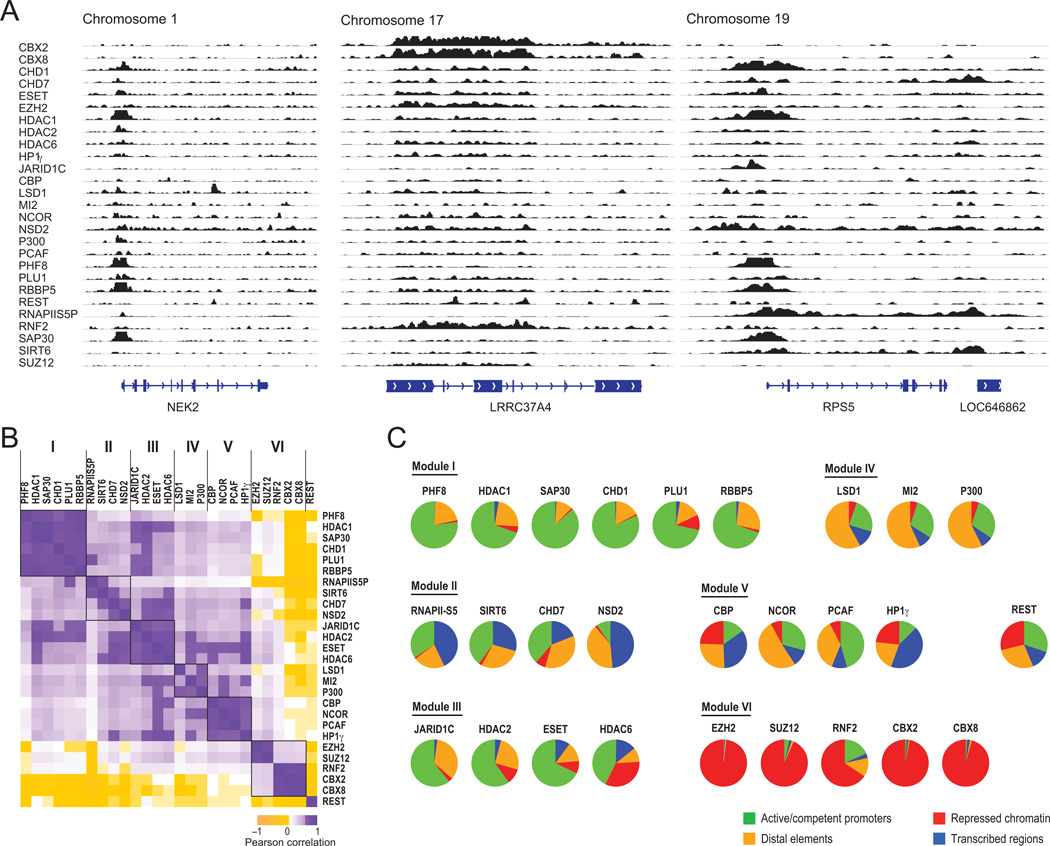
Figure 3. CR binding maps reveal modular organization and coherent associations with chromatin states.
(A) Binding of CRs at representative genomic loci in K562 cells. ChIP-seq profiles for 27 CRs are shown across three loci (Chromosome 1: 211,833,852-211,852,044; Chromosome 17: 43,580,509-43,600,984; Chromosome 19: 58,895,062-58,910,985). Examples of tracks derived from human ES cells (H1) are shown in Figure S2. (B) CRs partition into modules with correlated binding profiles. Correlation matrix reflects pair-wise correlations of binding peaks between CR datasets. Purple: positive correlation between CRs; yellow: negative correlation. White: no correlation. Correspondence is evident among CRs within each of the six CR Modules (demarcated by black squares). (C) For each CR, a pie chart indicates the proportion of binding sites that reside in regions with a given chromatin state annotation (green – active/competent promoter; gold – distal regulatory element/candidate enhancer; red – repressed chromatin; blue – transcribed region). CRs within a common module have similar distributions of binding.
CR binding patterns reveal a modular organization
Comparing the enrichment profiles of the CRs, we found that CRs bind in characteristic combinations. Specifically, we calculated correlations between each pair of CR binding profiles over all regions showing a significant peak in at least one dataset (Experimental Procedures). This allowed us to not only compare the different bound locations, but also to consider the shapes of the binding peaks in cases where the locations overlap. We then hierarchically clustered the CRs based on all pair-wise correlations. The resulting dendrogram and correlation matrix reveal striking associations between groups of CRs. These are reflected in six major modules (Figure 3B), each containing between three and six CRs with similar binding profiles. The six modules encompass all of the CR profiles, except REST (RE1-binding protein), whose profile is dissimilar to all others. Although REST is extensively implicated in CR recruitment, it is the only sequence-specific DNA binding protein in our compendium, which may explain its failure to conform to the modular organization seen for the other 28 CRs.
CR-Modules associate with distinct genomic features and chromatin environments
We next studied the relationship of the CRs and CR-Modules to genomic features, including promoters, transcribed regions, and distal regulatory elements. CRs within each module exhibit remarkably similar patterns of association to genomic features and chromatin states (Figure 3C), which are distinct between modules. Each localization pattern is consistent with known biology, while also providing insight into CR functions. We discuss each module below.
Module I (PHF8, RBBP5, PLU1, CHD1, HDAC1 and SAP30; promoters) is characterized by preferential binding at promoters (Figure 3C), with 65% to 80% of binding sites overlapping transcriptional start sites (TSSs). The targets carry H3K4me3 and other modifications related to competent (i.e., non-repressed) promoters, but exhibit a wide range of transcriptional activity based on RNA-seq. The results are consistent with known biology: RBBP5 is a core component of MLL complexes that catalyze H3K4me3 (Smith and Shilatifard, 2010), while PHF8 and CHD1 both bind this modification (Flanagan et al., 2005; Kleine-Kohlbrecher et al., 2010; Sims et al., 2005). In addition, the module contains PLU1 (JARID1B), an H3K4me3 demethylase.
Module I also includes HDAC1 and SAP30, core members of the SIN3 histone deacetylase complex with exquisitely similar binding profiles (R = 0.92). Although deacetylases have generally been linked to repression, the robust occupancy of these factors at non-repressed TSSs is consistent with a prior study that localized deacetylases to many active genes (Wang et al., 2009b). Importantly, such co-association of CRs with ‘activating’ and ‘repressive’ characteristics in one module is also seen in other modules and nearly all classes of target loci (see below). The co-binding patterns likely reflect widespread roles for opposing CRs in fine-tuning chromatin structure at regulatory loci. Such co-association of CRs with ‘activating’ and ‘repressive’ characteristics in one module is also seen in other modules and nearly all classes of target loci (e.g. promoters, candidate enhancers, etc.). This ‘bi-functional’ co-binding patterns likely reflect widespread roles for opposing CRs in fine-tuning chromatin structure at regulatory loci.
Module II (RNAPIIS5P, SIRT6, NSD2, CHD7; transcribed regions) is characterized by binding to active promoters as well as proximal and distal transcripts (Figure 3C). In particular, the cobinding patterns suggest interplay between initiating RNAPII (Smith and Shilatifard, 2010) and SIRT6 (R = 0.70): 78% of SIRT6-enriched windows reside over the TSS or within the first 5 KB of an active gene (compare to 75% for RNAPIIS5P). Another member of the module, NSD2, also localizes to active transcripts but with greater preference for distal, elongating regions (49% of enriched intervals are within actively transcribed regions). This may reflect interplay between NSD2, a histone methyltransferase, and the elongation mark H3K36 methylation (Nimura et al., 2009). Finally, CHD7 binds promoters, transcribed regions, and some distal elements.
Module III (JARID1C, HDAC2, HDAC6, ESET; promoters) comprises four CRs with catalytic activities typically associated with repression. These factors co-localize with active and competent promoters, similar to Module I, but also bind repressed targets. JARID1C (SMCX) is an H3K4 demethylase closely related to PLU1 (Module I). HDAC2 and HDAC6 complement HDAC1 (Module I) at active promoters, but also associate with Polycomb-repressed targets. Finally, the H3K9 methyltransferase ESET expands the spectrum of known heterochromatic CRs at promoters. These binding patterns suggest prevalent roles for ‘repressive’ CRs at sites of dynamic chromatin activity.
Module IV (P300, MI2, LSD1; candidate enhancers) includes three CRs that preferentially bind distal regulatory elements, including sites with enhancer-like chromatin (Figure 3C). Consistent with prior reports (Heintzman et al., 2007), over 70% of P300 sites are distal from TSSs and ~50% of those distal regions are enriched for modifications that correlate with enhancer activity, such as H3K4me1 and H3K27Ac (Birney et al., 2007; Ernst et al., 2011; Heintzman et al., 2007). Moreover, ~55% of distal P300 sites coincide with highly conserved sequences (Lindblad-Toh et al., 2011). Module IV also contains two members of the NuRD repressor complex – MI-2 and LSD1 (Wang et al., 2009a). Both CRs bind distal elements, with ~30% overlap to P300 peaks. LSD1 is a demethylase specific for mono- and di-methylated H3K4 (Shi et al., 2004), two characteristic methylation states of enhancer chromatin (Birney et al., 2007; Heintzman et al., 2007). These novel associations support a model in which chromatin at distal regulatory elements is tightly regulated by opposing enzymatic activities, as observed for promoters above.
Module V (NCOR, PCAF, CBP, HP1γ; candidate enhancers, other distal features) contains CRs that bind a more diverse set of elements. NCOR, PCAF and CBP each bind thousands of distal elements, many with enhancer-like characteristics, such as P300 binding. Considering all distal P300 sites, 48% are co-bound by CBP, 45% by NCOR and 35% by PCAF (p < 10−15 in all cases). Nevertheless, these three CRs also bind many other loci, accounting for their overall lower correlation with P300 and separate module. CBP is closely related to P300 and has also been shown to bind enhancers (Kim et al., 2010). PCAF and NCOR are antagonistic regulators associated with nuclear hormone receptor activity and repression, respectively (Perissi et al., 2010). Although they are typically studied at promoters, their co-localization patterns suggest that they also act at enhancers. The partitioning of distal element CRs into separate modules suggests a high degree of specificity among enhancers and their regulators.
HP1γ, a heterochromatin protein that physically interacts with H3K9me3, occupies diverse chromatin environments. Its association to this module reflects frequent co-binding of distal elements with CBP. However, HP1γ also correlates with CRs in other modules, and binds repetitive elements, Polycomb-repressed regions and ZNF gene clusters (O'Geen et al., 2007).
Module VI (EZH2, SUZ12, CBX2, CBX8, RNF2; Polycomb-repressed) comprises core components of Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). Binding occurs almost exclusively in regions enriched for H3K27me3, which typically correspond to transcriptionally-inactive, GC-rich promoters (Ku et al., 2008; Lee et al., 2006; Simon and Kingston, 2009). However, RNF2 (RING1B), an E3 ubiquitin ligase also present in other protein complexes (Vidal, 2009), shows a limited extent of binding outside of H3K27me3-marked regions.
Together, these findings portray diverse regulatory functions for CRs, and identify combinations of regulators that co-bind, and likely co-regulate, common genomic targets. In specific examples, coordination involves multiple CRs in the same protein complex. However, in most cases, CRs in a module show only partial, albeit significant, overlap, consistent with both shared and unique regulatory functions.
Fine-scale analysis of CR binding patterns across promoters
To evaluate the extent and significance of the modular CR organization and whether it is also guided by combinatorial principles, we next systematically examined CR binding patterns at individual loci. We inspected all promoters bound by more than one regulator. We focused on promoters because roughly half of CR binding events occur within 3 kb of a TSS, and nearly all CRs show some binding across such regions. We clustered the 1,081 promoters that are highly enriched for at least two CRs (Experimental Procedures) by the combinatorial binding of the 18 CRs with substantial promoter occupancy. We also grouped the CRs based on their localization patterns across these loci. This promoter-focused grouping (CR groups) is largely consistent with the CR Modules deduced from genome-wide correlations. However, this fine-scale analysis highlights differential associations of individual CRs with TSSs and flanking regions, as well as differential relations to gene activity.
A first group of CRs – PLU1, CHD1, SIRT6, and CHD7 – exhibits binding profiles characteristic of RNAPII initiation (Figure 4A). Although enriched across all transcriptionally-competent promoters, these CRs are most strongly bound at highly active promoters undergoing productive initiation and elongation, as indicated by the high expression levels of the corresponding genes. Their broad binding distributions over TSSs emulate RNAPIIS5P (Figure 4A, B). Fine binding patterns thus identify additional CRs with close connections – and possible direct physical interactions – with initiating RNAPII (Smith and Shilatifard, 2010).
Figure 4. Fine-scale CR binding profiles distinguish coherent gene sets.
(A) Combinatorial binding patterns of CRs at individual promoters are associated with distinct expression and function. Fine-scale binding profiles are shown for CRs (vertical sections) across 1,081 target promoters (rows) after hierarchical clustering and re-ordering of major promoter clusters based on expression (original ordering shown in Figure S3). The profiles depict enrichment Z-scores (red – high, white – low) for 300 bp windows within 3 kb of TSSs. Promoter clusters derived based on common CR binding profiles (indicated by thick white horizontal lines) share transcriptional status, and the corresponding genes often share coherent functions (labels on right, curated from enriched functional gene sets, listed in Table S4B). Left bar shows RNA expression levels (log2(FPKM)) derived from RNA-seq data: orange – high; blue – low; white – median. CR labels colored according to the dendrogram shown above. (B–D) Composite profiles for CRs with similar binding patterns at shared promoters. Profiles reflect average binding of the indicated CRs (Y axis) over co-bound promoters, centered on TSSs (X axis). (B) Peaks that surround TSSs, but dip at the TSS itself for CHD1 (purple) and SIRT6 (blue). (C) Sharp peaks over TSSs for HDAC2 (purple), PHF8 (green) and SAP30 (blue). (D) bimodal peaks of Polycomb CRs – diffuse peaks over TSSs for PRC2 members EZH2 (purple) and SUZ12 (blue) along with TSS-excluded peaks for PRC1 members CBX2 (green) and CBX8 (brown). (E) Composite profiles for CRs with distinct binding patterns at shared promoters. PLU1 (purple) has a sharp peak over TSSs, while CHD1 (blue) has a broader peak that extends downstream. Note that the promoters in this composite differ from panel B. (F–G) Composite profiles show distinct patterns for the same CR at promoter sets with different activity levels. (F) HDAC2 at PRC2-repressed (blue) and transcriptionally–competent promoters (purple). (G) HDAC1 at transcriptionally-competent (cluster 5, purple) and active (cluster 1, blue) promoters.
A second, larger CR group – ESET, HDAC6, JARID1C, HDAC2, HDAC1, SAP30 and RBBP5 – bind active and competent promoters (Figure 4A) in sharp peaks that precisely coincide with TSSs (Figure 4C). In addition to facilitating RNAPII engagement, these CRs may help maintain chromatin integrity around the nucleosome-free TSSs (Jiang and Pugh, 2009), by fine-tuning modifications of the flanking −1 and +1 nucleosomes.
A third group of CRs – EZH2, SUZ12, RNF2, CBX2 and CBX8 – includes core components of PRC2, which catalyzes H3K27me3, and PRC1, which binds H3K27me3 and mediates chromatin compaction (Margueron and Reinberg, 2010; Simon and Kingston, 2009) (Figure 4A). These CRs bind inactive promoters, many of which correspond to genes involved in development or signaling. Remarkably, PRC2 and PRC1 subunits exhibit distinct fine-scale binding profiles over the promoters (Figure 4D). PRC2 components (EZH2, SUZ12) peak over TSSs, potentially reflecting interactions with DNA sequences in these nucleosome-depleted regions. In contrast, PRC1 components (CBX2, CBX8) bind broadly across the same regions, likely promoted by physical interactions with flanking H3K27me3-marked nucleosomes (Figure 4D). Notably, Polycomb-repressed promoters are the only set of genomic elements in our study that are not subject to opposing chromatin regulatory activities, as they are bound exclusively by repressive CRs in K562 cells.
Fine-scale promoter analysis reveals combinatorial complexity of CR associations
Although the promoter clustering largely corresponds to the modular organization discerned from genome-wide correlations, it also reveals several exceptions that may reflect combinatorial CR binding. In some cases, different CRs bind the same promoters but with distinct binding structures. For example, despite largely overlapping targets, CHD1 and PLU1 exhibit markedly different binding patterns. CHD1 peaks sharply over TSSs, while PLU1 extends well into transcribed regions (Figure 4E).
In other cases, a CR is associated with different CR groups under different promoter contexts (Figure 4A, F–G). Particularly striking examples of such combinatorial partitioning involve deacetylase complexes (Yang and Seto, 2008). SIN3 complex members HDAC1, HDAC2 and SAP30 bind promoters of genes that oscillate during the cell cycle with an intensity that distinguishes them from all other targets (Figures 4A and G, cluster 5). In addition, HDAC1, HDAC2 and JARID1C (members of the CoREST complex (Tahiliani et al., 2007)) co-bind along with HDAC6 to repressed PRC2 targets (Figure 4A, clusters 6–8). This association may reflect physical interactions between CoREST and Polycomb complexes (Ren and Kerppola, 2011; Tsai et al., 2010) and/or direct interactions between HDAC2 and PRC2 (van der Vlag and Otte, 1999). The fine binding patterns of these CRs vary dramatically based on the context of the target gene’s activity or the co-binding CRs. For example, HDAC2 binds sharply over transcriptionally-competent TSSs, but distributes broadly over Polycomb-repressed promoters (Figure 4A clusters 5 and 13, and Figure 4F). This is consistent with a model in which histone deacetylases act as fine-tuners of accessible chromatin at competent TSSs, but as enforcers of hypo-acetylated chromatin domains at Polycomb-repressed loci.
Combinatorial CR binding patterns are associated with refined functional distinctions
We next explored whether individual CRs or CR combinations might be associated with specific cellular processes. The promoter-based analysis revealed 15 ‘combinatorial binding’ gene clusters, each of which shares binding by a combination of CRs, as well as a fine CR location structure around their TSSs (Figure 4A and S3, horizontal blocks). The genes in many of these clusters are characterized by shared functional attributes (Figure 4A, labels on right, Table S4). In particular, genes with similar expression levels but distinct biological functions are often bound by distinct combinations of CRs. For example, the ‘Protein Metabolism’ cluster (Figure 4A, cluster 2; 110 genes) is comprised of highly expressed genes whose promoters are co-bound by SIRT6, CHD1, PLU1 and RNAPIIS5P. A distinct cluster consists of 84 genes with similarly high expression, but whose promoters are co-bound by these CRs along with CHD7 and HDAC6, is enriched for genes involved in chromatin architecture (cluster 1). A separate binding cluster (cluster 5; 92 genes), enriched for cell cycle gene promoters, is unremarkable in terms of its intermediate expression levels, but prominently co-bound by HDAC1, HDAC2 and SAP30. The physical association of these core SIN3 components with these promoters offers a mechanistic explanation for documented roles for this repressor complex and histone deacetylase activity in cell cycle progression (David et al., 2008; Minucci and Pelicci, 2006). Interestingly, the promoters in the ‘stress response’ cluster are co-bound by most of the ‘activating’ and ‘repressive’ CRs in our panel, which may play important roles in the notable capacity of these genes to rapidly change their activity in response to stimuli.
CRs occupy different loci in ES cells, but maintain their modular associations
We next explored whether the co-localization patterns and associations observed in K562 cells can be generalized to other cell types. We considered several layers of CR organization. First, we asked whether CRs distribute to different genomic locations consistent with changed gene expression programs. Second, we asked whether the associations between individual CRs and chromatin modification states change between cell types. Third, we asked whether the modular relationships between CRs are maintained in different cell types.
To examine each of these possibilities, we generated ChIP-seq data for 15 CRs in human ES cells and analyzed their localization patterns (Figure 5A and B). We used the same computational methods as in K562 cells to identify regions of enrichment, which yielded similar overall statistics (Table S3).
Figure 5. Comparisons of CR binding and modular associations in K562 and ES cells.
(A) CRs distribute to distinct loci in ES and K562 cells. For each CR (left) or histone modification (right), bar graph indicates proportions of binding intervals that are ES cell-specific (‘H1 only’, light blue), K562-specific (‘K562 only’, gray) or overlapping between cell types (‘H1 and K562’, navy). The bars are vertically centered according to the overlap regions. (B) CR-CR associations and CR-histone modification associations are largely preserved between K562 and ES cells. Scatter plot presents the correlations in localization profiles between every pair of CRs (black dots) and every CR-histone modification pair (red dots) in either K562 cells (X axis) or ES cells (Y axis). Linear regression lines and correlation coefficients are indicated for each type of combination. (C) CRs associate with similar chromatin states in ES and K562 cells with some distinctions. Pie charts indicate the proportion of CR binding sites that correspond to a given chromatin state annotation; green – active/competent promoter, gold – distal regulatory element/candidate enhancer, red – repressed chromatin (including bivalent state), blue – transcribed region. CRs are grouped according to the modules in Figure 3. (D) Combinatorial binding profiles are shown for CRs in ES cells. Fine-scale binding profiles are shown for each CR across 1189 target promoters (rows) in ES cells, after hierarchical clustering and re-ordering of major promoter clusters as in Figure 4A (original ordering shown in Figure S4). Functional gene set annotations (right) curated from enriched sets (Table S5B). CR labels colored as in Figure 4A, with ES cell-specific CRs in black.
The CRs differ substantially in their genomic location between the cell types, though the degree of overlap varies between CRs (Figure 5A, left). The patterns of re-localization are reminiscent of histone modifications (Figure 5A, right), which dynamically change between these cell types (Ernst et al., 2011) consistent with differential transcriptional programs.
Despite substantial differences in CR localization, the underlying CR organization is maintained between the cell types (Figure 5B). First, the degree of co-binding between pairs of CRs is conserved between the two cell types (R=0.64). Similarly, the degree of correspondence between a given CR and a given histone modification is also well correlated (R=0.79). Thus, CR-CR associations as well as CR-histone modification associations are globally preserved between cell types.
Furthermore, the relationships between individual CRs and genomic annotations remain largely unchanged. For most CRs, the distribution of binding between promoter, transcribed, distal and repressed regions is highly concordant between K562 and ES cells (Figure 5C). Conservation of binding patterns is also evident when comparing the fine-scale promoter profiles of CRs in ES cells (Figure 5D) to those in K562 cells (Figure 4A). Consistent patterns of binding are evident for CRs associated with competent TSSs (e.g., PHF8, RBBP5, SAP30), productive initiation (e.g. CHD1, SIRT6) and Polycomb repression (e.g., EZH2, SUZ12). Gene sets distinguished based on combinatorial binding profiles are also similar between the cell types (Figures 5D and S4; Table S5).
Notably, when there are changes in CR localization, they tend to be shared by members of the same module and to relate to a fundamental difference in chromatin structure between cells (Figure 5C). For example, although Module I CRs (e.g., PHF8, CHD1, RBBP5) are restricted to active and competent promoters in K562 cells, they also associate with Polycomb-repressed promoters in ES cells (Figure 5C and D). The presence of multiple ‘activating’ CRs at these inactive targets is consistent with the enrichment of the underlying chromatin for opposing (‘bivalent’) histone modifications. These CRs likely contribute to the poised character of the corresponding genes, many of which are induced during ES cell differentiation (Bernstein et al., 2006). In addition, P300 binds substantially fewer sites in ES cells than in K562 cells (Figure 5A and C), possibly reflecting a lower prevalence of enhancer-like chromatin in ES cells (Ernst et al., 2011).
Overall, our analysis suggests that the modular and combinatorial structures of CRs, and their association with histone modification states, are constitutive features of the chromatin regulatory network. Thus, changes in CR binding tend to be coordinated at the level of modules, and to correspond to changes in the underlying chromatin landscape.
DISCUSSION
Modular and combinatorial organization of the CR network
Despite their large number and the importance of chromatin organization to gene regulation, the localization and function of individual CRs remains poorly understood. Studies of histone modification patterns have revealed a relatively limited number of chromatin configurations or ‘states’ that distinguish different types of genome regulatory elements. It has been compelling to hypothesize that specific CRs contribute to the establishment and maintenance of these states in different cell types, and that they work in a combinatorial fashion, akin to transcription factors, which are encoded in a comparable number in the genome. However, it has been difficult to develop detailed models of CR function given the limited availability of comprehensive measurements and the paucity of effective capture reagents.
Here, we presented a first systematic view of CR localization across the human genome in two cell types, and a general methodology for studying the targeting and functions of such regulators. We reveal several major principles for the organization of the CR network in mammalian cells (Figure 6). (1) Coherent modules of CRs co-bind to common target loci that share specific chromatin states; the modules often consist of modifying enzymes that catalyze ‘activating’ and ‘repressive’ modifications, offering a means for precise tuning of chromatin and gene regulation. (2) In addition to these global associations, the same CR may associate with different modules at different target loci, suggesting complex functional relationships, indicative of combinatorial regulation. (3) Specific combinations of CRs bind sets of genes with related functions, suggesting functional specificity. (4) When comparing different cell types, CRs distribute to different loci, often in conjunction with changes in chromatin states; however, (5) they largely retain their modular associations.
Figure 6. Principles of CR organization.
Model of the association of CRs (gears) with histone modifications (little colored balls) across (A) actively transcribed genes, (B) transcriptionally competent TSSs, and (C) Polycomb-repressed regions. CRs colored as in Figures 4A and 5D.
In many respects, this view is reminiscent of the organization of sequence-specific transcription factors networks. In particular, the association of CRs within modules – each related to different chromatin modification states, functional gene groups, and expression patterns – is consistent with the modular organization of transcription factor networks in organisms from yeast to human (Yosef and Regev, 2011). Nevertheless, we cannot rule out the possibility that other CRs, not tested in our study, might adopt different, possibly non-modular organizations, analogous to that of REST, which do not conform to any CR modules defined here.
Due to this organization, changes in the expression of an individual CR may affect the function of one or more modules in which it participates, with potentially widespread consequences for gene expression and cellular phenotype. Such network properties could help explain how dynamic changes in CR expression guide differentiation processes, and how genetic inactivation of CRs promotes tumor progression. However, the binding modules derived here do not predict how the removal of specific components will affect other participants or downstream targets. Further study is therefore needed to derive more detailed functional models of the direct physical interactions between CRs and the associated binding hierarchies.
The organizational principles also suggest how CR binding can tune expression programs. Each of the CR-Modules targeting transcriptionally-active or competent (non-repressed) promoters contain proteins with opposing activities – those that catalyze the addition of modifications associated with active/accessible chromatin and those that catalyze their removal. Such opposing activities in bi-functional modules may underlie homeostasis at active chromatin loci, and allow precise tuning of gene expression. Since distal enhancers are also bound by activating and repressive CRs, they too may be subject to similar fine-tuning. Genomic loci targeted by Polycomb proteins in K562 cells are an outlier in this regard as they appear to be exclusively subject to repressive histone modifiers.
Specific hypotheses for CR function
The network organization suggests many specific hypotheses regarding the functions or molecular mechanisms of individual CRs or CR complexes. For example, in both K562 and ES cells, components of the SIN3 repressor complex are strikingly enriched at cell cycle gene promoters, providing a potential mechanistic explanation for known roles for deacetylases in cell cycle progression. In another example, we find that several repressive CRs bind both to competent TSSs and to Polycomb-repressed targets. These repressors, which include histone deacetylases and an H3K4 demethylase, likely enforce the hypo-acetylated and H3K4 unmethylated state characteristic of these repressed loci.
A meso-scale assay for CR-DNA interactions
Our ChIP-string assay, essential for this large-scale mapping, opens the way to further functional studies of these and other CRs in many cell systems. It allowed us to screen hundreds of antibody-condition combinations for CR localization, through which we identified a new set of effective reagents for mapping CRs. We expect this screening approach will help overcome the current paucity of ChIP-seq grade antibodies for studying the several hundred CRs that control chromatin structure and function. The modest success rate of antibodies tested here suggests that this will also require substantial efforts to develop antibodies against different CR epitopes as well as new types of affinity reagents.
The multiplexed assay for CR binding and histone modifications also has the potential to greatly enhance functional studies of chromatin. Its rapid turnaround and low cost will enable systematic studies of perturbations induced by small molecules or RNA interference, which have traditionally been restricted to downstream phenotypic readouts such as protein or RNA expression. In particular, this will help assess the functional impact of the components and organization of the CR network.
A systematic resource of genome-wide CR binding profiles
Our dataset provides an important resource for studying CRs, at an unprecedented scope. Prior studies of CR binding typically considered very few factors, and used varied procedures and cell types, all of which precluded systematic comparisons. In contrast, our resource allows direct comparison of many CRs in the same cell and between cell types. It also provides a reference to which users may compare their CR or transcription factor profiles, with the potential to predict binding partners and cellular functions. It should therefore enable the large community of chromatin biologists to develop and test mechanistic hypotheses, ultimately leading to a more comprehensive understanding of chromatin organization and gene regulation.
EXPERIMENTAL PROCEDURES
Data access
All raw data, mapped reads and integrated profiles are available at http://www.broadinstitute.org/software/crome/.
Chromatin regulator antibodies
We collated a list of 515 proteins with annotated functions related to histone modification, histone binding or chromatin remodeling. We obtained a total of 128 antibodies to these proteins, which we tested in the ChIP-string assay. A list of all antibodies annotated by their performance in ChIP-string and ChIP-seq is provided in Table S2. The specificity of all antibodies used in ChIP-seq was confirmed by Western blots (Figure S1C). Roughly 20 million K562 cells or H1 ES cells were used for each ChIP assay. Detailed procedures are in Supplemental Information.
Representative genomic loci and nCounter probe design
We chose a set of genomic loci designed to be representative of diverse chromatin environments. We used a hidden Markov model (Ernst and Kellis, 2010) and ChIP-seq maps for 10 chromatin marks in K562 and ES cells (Ernst et al., 2011) to identify 10 major chromatin states and annotate the genome accordingly. For each state in each cell type, we randomly selected 20 loci and used the corresponding sequences for probe design (Table S1).
We modified the nCounter Analysis System platform (NanoString Technologies) to measure enriched genomic DNA from ChIP experiments (ChIP-string). Detailed descriptions of probe-set design and ChIP-string procedures are in Supplemental Information.
ChIP-string data analysis and processing
We devised two alternative analysis methods for the ChIP-string screen. The first (‘original’) approach, optimal for large-scale screens, was used to score the screen and select ChIP experiments for sequencing. The second (‘alternative’) approach is suitable for both large- and small-scale studies, even for those testing just a few antibodies. The results of the two approaches on our screen data agree very closely. Detailed descriptions are in Supplemental Information.
ChIP-seq analysis
ChIP-seq was performed as described (Mikkelsen et al., 2007), followed by identification of enriched intervals, which were correlated to genomic elements and chromatin states. Pearson correlations were calculated between every pair of CRs, based on signal distributions across enriched intervals, and used to produce pair-wise cluster maps. CR binding profiles were used to hierarchically cluster promoters. Expression values were derived from RNA-seq data. Full details are provided in Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank B. Knoechel, C. Ye, J. Jaffe, and R. Mostoslavsky for helpful discussions, G. Geiss and R. Boykin for help with assay development, R. Raychowdhury and the Broad Sequencing Platform for technical assistance, X. Li and Y. Shi for Plu1 antibody, D. Jang and T. Liefeld for building the CRome portal, and L. Gaffney for figure preparation. OR and AG were supported by an EMBO fellowship. The work was supported by an ENCODE grant from the National Human Genome Research Institute (U54 HG004570 to B.E.B.), by an NHGRI CEGS grant (A.R. and B.E.B), Howard Hughes Medical Institute (A.R. and B.E.B.), an NIH PIONEER award (A.R.), and the BWF (A.R. and B.E.B.). A.R. is a researcher of the Merkin Foundation for Stem Cell Research at the Broad Institute.
REFERENCES
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331:1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin M, Parker B, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. Evolutionary constraint in the human genome based on 29 eutherian mammals. Nature. 2011 In Press. [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature reviews Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS genetics. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Molecular cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Molecular cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–370. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009a;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009b;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.