Abstract
The metazoan cell membrane is highly organized. Maintaining such organization and preserving membrane integrity under different conditions are accomplished through intracellular tethering to an extensive, flexible, protein network. Spectrin, the principal component of this network is attached to the membrane through the adaptor protein ankyrin, which directly bridges the interaction between β-spectrin and membrane proteins. Ankyrins have a modular structure which includes two tandem ZU5 domains. The first domain, ZU5A, is directly responsible for binding β-spectrin. Here, we present a structure of the tandem ZU5 repeats of human erythrocyte ankyrin. Structural and biophysical experiments show that the second ZU5 domain, ZU5B, does not participate in spectrin binding. ZU5B is structurally similar to the ZU5 domain found in the netrin receptor UNC5b supramodule, suggesting that it could interact with other domains in ankyrin. Comparison of several ZU5 domains demonstrates that the ZU5 domain represents a compact and versatile protein interaction module.
Keywords: Protein-protein interactions, protein structure, Ankyrin-R, UNC5b, crystallography
Introduction
Ankyrin proteins organize, transport, and anchor membrane protein complexes, such as the anion-exchange transporter in erythrocyte cell membranes, to the actin/spectrin cytoskeletal network1. The cellular roles and consequences of ankyrin dysfunctions have been linked to blood2; 3; 4, cardiac5; 6; 7, and neurological disorders8. In erythrocytes, the importance of ankyrin mediated attachment tothe spectrin cytoskeletal network is demonstrated by experiments where ankyrin dysfunction led to loss of red blood cell mechanical stability2.
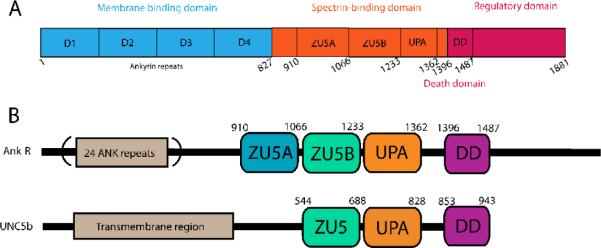
There are three major ankyrin isoforms in humans: ankyrin-R (Ank-R) encoded by the ANK1 gene found in erythrocytes9, ankyrin-B (Ank-B) encoded by the ANK2 gene localized primarily in the brain, and lastly, ankyrin-G (Ank-G) encoded by the ANK3 gene, which is ubiquitously expressed10; 11. Alternative splice variants of each isoform have also been identified, and are known to have distinct localization and functional properties12. Canonical ankyrins are modular proteins consisting of three regions: an N-terminal membrane domain comprised of ankyrin repeats responsible for attachment to membrane proteins, a central spectrin-binding domain, and a C-terminal regulatory domain (Figure 1A)10; 13. The ankyrin repeat is a well-studied protein interaction module that has been found not only in ankyrin, but also in many other proteins14. The structure of 12 tandem repeats of human ankyrin-R spanning the region immediately adjacent to the spectrin-binding domain is known and displays a crescent-shaped arrangement of the individual ankyrin repeats. In addition, the structures of many other ankyrin repeats from different sources have been elucidated14; all show a similar arrangement. There is scant structural information on the C-terminal domain, aside from the structure of the Death domain (DD) (PDB ID: 2YVI), which resembles other Death domains. Interestingly, DDs are found to interact with pro-apoptotic proteins. For example, the DD of the Ank-G190 isoform interacts with the pro-apoptotic Fas protein in kidney tubules15. Aside from the DD region, the regulatory domain of ankyrin is thought to be unstructured and flexible13. Intramolecular interactions by the regulatory domain have been suggested to provide functional diversity in different ankyrin isoforms13; 16. It has been shown, for instance, that the regulatory domain of ankyrin-B interacts with its own membrane binding domain. This interaction is crucial in driving ankyrin-B specific functions17; 18.
Figure 1.
A. Domain organization of canonical ankyrins. The ~89 kDa membrane binding domain consists of 24 tandem ankyrin repeats (blue), a 62 kDa spectrin-binding domain (orange) that includes the ZU5A, ZU5B, and UPA domains, and a 55 kDa C-terminal flexible regulatory domain (red) containing aDeath domain (DD). Numbering in the diagram corresponds to that of human ankyrin-R. B. Comparative domain organization of Ankyrin-R and UNC5b. UNC5b contains a large transmembrane region followed by ZU5 (light green), UPA (orange) and DD (purple) domains. The arrangement of these domains is similar to the one observed in ankyrins. In the latter, the first ZU5 domain (blue), ZU5A, mediates the interaction with β-spectrin repeats 14–15. UNC5b is known to form a supramodule involving ZU5-UPA-DD domains25. Based on the sequence similarity, it is expected that ankyrin forms a similar supramodule involving the ZU5B, UPA and DD domains, but not the ZU5A spectrin binding domain.
The region of the spectrin-binding domain responsible for binding spectrin has been mapped to a small sub-domain known as ZU519; 20. This domain interacts directly with repeats 14–15 of β-spectrin21; 22 and represents be the minimal binding fragment. The structure of the Ank-R ZU5 domain has been previously solved and shows a β-strand core with several surface loops23. Additionally, the manner in which this domain interacts with spectrin was elucidated from the structure of the complex of Ank-R ZU5 with repeats 13–15 of β-spectrin24. The Ank-R ZU5 domain comprises a small part of the spectrin-binding domain of ankyrin. Sequence analysis suggests that a second ZU5 domain and a UPA domain in the spectrin binding region, and the Death domain at the start of the regulatory region follow the spectrin-binding ZU5 domain. Thus, the spectrin-binding domain appears to be formed by two tandem ZU5 domains, hereafter termed ZU5A and ZU5B, followed by the UPA domain (Figure 1B). The functional role of the domains adjacent to the ZU5A domain, has not been established, although the DD has been reported to provide functional diversity and regulate intramolecular interactions13.
Recently, the structure of the C-terminal region of UNC5b, a protein showing sequence similarities to ankyrin, was solved25. Members of the UNC5 family of proteins are important in axonal guidance, angiogenesis, and apoptosis26; 27. The C-terminal region of UNC5b has similar domain architecture to the spectrin-binding and regulatory regions of canonical ankyrins. Both protein families include conserved ZU5B, UPA and DD domains, but only ankyrin contains the ZU5A domain involved in spectrin interaction (Figure 1B). Structural and biochemical studies of the UNC5b ZU5B-UPA-DD region show that it folds into a supramodule, where the Death domain interaction with the ZU5B domain forces the UNC5b supramodule into a closed conformation that inhibits UNC5b activity25. The UNC5b structure suggests that canonical ankyrin could also form a similar ZU5B-UPA-DD supramodule24; 25. Based on ankyrin supramodule models, it is neither clear whether the two tandem ZU5 repeats interact with each other nor is it evident that the ZU5B domain is involved in spectrin binding.
To investigate how the second ZU5 repeat in ankyrin may regulate function, we crystallized and solved an atomic structure of an ankyrin fragment spanning the two tandem ZU5 repeats, termed ZU5A-ZU5B. The structure reveals that the ZU5B domain resembles ZU5A domain and that the two domains interact minimally. Gel shift assays and Surface Plasmon Resonance (SPR) data indicate that the ZU5B domain does not affect binding to spectrin, in agreement with the structure of ZU5A-ZU5B, which shows the ZU5B domain extending away from the ZU5A spectrin-binding site. Finally, comparison of several ZU5 domain structures suggests that ZU5 domains are versatile interaction modules. The structural data serve as a starting point to understand the possible formation of an ankyrin supramodule involved in ankyrin functional regulation.
Results
ZU5A-ZU5Bbinding to spectrin
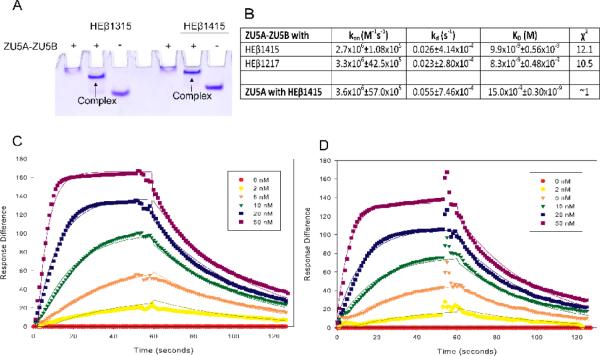
ZU5A is the minimal spectrin binding domain of ankyrin. In order to determine whether the presence of ZU5B affected binding, we measured the binding affinity of ZU5A-ZU5B to two different human erythrocyte β-spectrin fragments containing the minimal ankyrin binding domain21. Binding was characterized through native gel shift assays and SPR, as described previously21; 23; 24. The native gel shift assay was first conducted to confirm that spectrin and ZU5A-ZU5B interact. ZU5A-ZU5B protein and either human erythrocyte β-spectrin repeats 13–15 (HEβ1315) or 14–15 (HEβ1415) were mixed in a 1:1 molar ratio and run on a 10% native polyacrylamide gel. Due to its high pI (calculated pI: 8.44), the migration of the ZU5A-ZU5B is limited in this assay. The spectrin fragments have comparatively low pI values and therefore migrate quickly through the gel. Complex formation is demonstrated by the presence of a band with intermediate mobility. The results of this assay (Figure 2A) demonstrate that a complex of ZU5A-ZU5B fragments with β-spectrin fragments is formed as expected by previous studies21; 23.
Figure 2. The ZU5B domain does not affect spectrin/ankyrin binding.
A. Native gel shift assay of the ZU5A-ZU5Bfragment of ankyrin and spectrin. The ZU5A-ZU5B fragment was mixed with repeats 13–15 (HEβ1315) or 14–15 (HEβ1415) of human erythrocyte β spectrin to ascertain whether the fragments interact. The native gel shift shows a shift in spectrin mobility due to the interaction with ankyrin. The presence of absence of ZU5A-ZU5B is shown by + or − signs. As expected, the ankyrin fragment interacts with spectrin repeats containing the ankyrin binding region. B. Binding affinity and kinetic parameters data describing the ZU5A-ZU5B interaction with β-spectrin.Binding affinity and kinetic parameters measured by SPR between the ZU5A-ZU5B fragment and repeats 12–17 (HEβ1217) or repeats 14–15 of human erythrocyte β-spectrin. For comparison, parameters for the ZU5A only fragment are shown in the final row of the table21. The ZU5A-ZU5B interaction with human β-spectrin is comparable to the one observed between spectrin and ZU5A alone. C and D. SPR trace of ZU5A-ZU5B interaction with HEβ1217 and HEβ1415. Surface plasmon resonance data and fits for ZU5A-ZU5B binding to biotinylated HEβ1217 or HEβ1415 at various concentrations of ZU5A-ZU5B shown in the inset.
Whereas the gel shift assay demonstrated that the two protein fragments interact, it was not possible to ascertain whether the presence of the second ZU5 domain had a subtle effect on binding affinity. To characterize this interaction in a quantitative manner, the binding affinity and kinetics of the complexes formed between the ZU5A-ZU5B ankyrin fragment and different spectrin fragments were measured by SPR (Figure 2). Regardless of the fragment combination used, comparable binding affinities were observed to those previously reported using the minimal binding domains (ZU5A with HEβ1415) (Table I). The equilibrium dissociation constants (KD) of ZU5A and ZU5A-ZU5B binding to HEβ1415 were 15.221 and 9.9 nM, respectively. Binding by ankyrin to other spectrin fragments results in equilibrium dissociation constants in the same range (KD for ZU5A binding to HEβ1315 is 9.3 nM24 and for ZU5A-ZU5B binding to HEβ1217 is 8.3 nM). In addition, the association and dissociation rate constants (ka and kd) for the different ankyrin fragments are also comparable, demonstrating that the binding kinetics were minimally affected by the inclusion of the second ZU5 domain. These data indicate that either the presence of the ZU5B domain does not interfere with ZU5A binding to spectrin or that the contribution by the ZU5B domain is minimal. Moreover, these measurements are comparable to the 25 nM value measured for intact brain spectrin tetramers and brain ankyrin28.
Table I.
Summary of crystallographic data
| Crystal | ZU5A-ZU5B Native 1 | ZU5A-ZU5B SeMet | ZU5A-ZU5B Native 2 |
|---|---|---|---|
| Space group | C2 | C2 | C2 |
| Unit cell | a= 277.8Å b= 40.5 Å c= 94.7 Å α=γ= 90° β= 92.3Å |
a= 279.5 Å b= 40.9 Å c= 95.3 Å α=γ= 90Å β= 92.0Å |
a= 160.1 Å b= 40.5 Å c= 178.1 Å α=γ= 90Å β= 112.7° |
| Data Collection | |||
| Detector type/source | MarCCD/APS | MarCCD/APS | MarCCD/APS |
| Wavelength (Å) | 0.97872 | 0.97626 | 0.97872 |
| Resolution* | 36.94–2.60 (2.72–2.60) | 38.70–2.20 (2.30–2.20) | 29.60–2.00 (2.09–2.00) |
| Measured reflections | 91617(11302) | 226787 (27584) | 360576 (38385) |
| Unique reflections | 32774 (4022) | 55529 (6721) | 71819 (8411) |
| Completeness (%) | 99.0 (99.8) | 99.9 (100.0) | 99.3 (96.0) |
| Anomalous completeness (%) | - | 97.8 (99.2) | - |
| Multiplicity | 2.8 (2.8) | 4.1 (4.1) | 5.0 (4.6) |
| Anomalous multiplicity | - | 2.1 (2.1) | - |
| Rmeas+ | 0.057 (0.389) | 0.104 (0.591) | |
| Mean ((I/σ(I))§ | 18.3 (4.2) | 12.8 (3.0) | |
| Phasing † | |||
| Resolution | 37.06–2.20 | ||
| Phasing powerisomorphous (acentric/centric) | 0.835/0.733 | ||
| Phasing poweranomalous | 0.563 | ||
| Rcullis-isomorphous | 0.255/0.248 | ||
| Rcullis-anomalous | 0.931 | ||
| FOM (acentric/centric) | 0.256/0.108 | ||
| Refinement | |||
| Resolution | 24.90–2.21 (2.26–2.21) | 29.68–2.00 (2.05–2.00) | |
| No. of reflections working/test | 51531/3282 | 71813/4994 | |
| Rfactor (%) | 22.26 (27.1) | 17.31 (19.06) | |
| Rfree (%) | 25.88 (31.2) | 20.57 (20.02) | |
| Protein atoms | 7736 | 7825 | |
| Water molecules | 189 | 686 | |
| Ethanol molecules | 7 | 18 | |
| Bond lengths | 0.011 | 0.010 | |
| Bond angles | 1.311 | 1.05 |
Structure determination and overall structure of ZU5A-ZU5B
A structure of ZU5A-ZU5B fragment of human erythrocyte ankyrin was solved by X-ray crystallography. Selection and design of the ankyrin subdomains were based on sequence alignment and previous observations indicating the presence of a secondary ZU5 domain after the ZU5A minimal spectrin-binding domain (F. Bazan, personal communication). The structure was solved in two closely-related crystal forms by a combination of molecular replacement29 and SAD phasing30. In each crystal form, there are three monomers in the asymmetric unit with almost identical conformations. The root mean square deviation (RMSD) for all common Cα between the three monomers in the two forms is 0.586 Å and there are no marked differences between them aside from packing interactions. For these reasons, only the higher resolution structure is described herein.
In the structure, the three molecules in the asymmetric unit (A, B, and C) have the same overall fold. Chains B and C superpose very well with an RMSD value of 1.1 Å for all common Cα atoms. Chain A shows some variations when compared to the two other chains with RMSD values of 1.4 Å and 1.9 Å when the common Cα are superposed to chains B and C, respectively. Chains A and C are related by an almost perfect 2-fold axis (175°) and this pseudo-dimer corresponds to the crystallographic dimer formed by the B subunit. Thus, it appears that the preferred mode of interaction of the protein in the crystal is by dimerization (A and C or B and its crystallographic partner) (Supplemental Figure S1A). The two domains have a small interface surface area (~520 Å2)31. No other possible ZU5A/ZU5B pair in the crystal has a larger interface surface area, ruling out the possibility of a domain swapped molecule (Supplemental Figure S1A). It is likely that the dimers observed are crystallographic artifacts and do not represent the oligomerization state in solution.
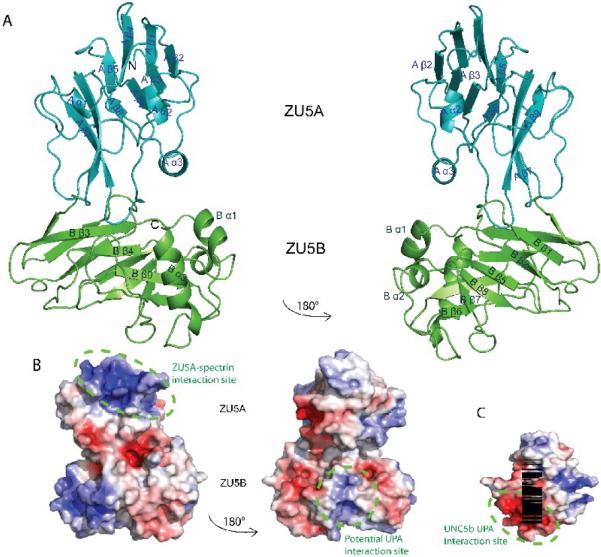
The ZU5A-ZU5B fragment of Ank-R shows two distinct regions corresponding to each of the ZU5 domains. Both ZU5 domains are formed by a compact β-sheet rich core with different numbers of outer loops and helices (Figure 3A). The linker between the two domains is formed by a loop joining the last β strand in ZU5A with the first β strand in ZU5B. Overall, the ZU5B domain extends away from the ZU5A domain making few interactions. The ZU5A domain adopts the same fold as described previously23 with two anti-parallel β sheets of four strands and five strands each. Similarly, the ZU5B domain consists of two anti-parallel β sheets, except that each sheet is made of five β strands. In addition, whereas ZU5A has two short helices, ZU5B has three helices, two short and one long helix near the C-terminus. In general, the β core of both domains is very similar and all loops and helices are solvent-exposed. In the structure of the ZU5A domain23, it was noted that the N- and C-termini are located on opposite ends of the β-strand core. However, in the ZU5B domain the N- and C-termini are located on the same side of the β-core and relatively close to each other.
Figure 3. Structure of the ZU5A-ZU5B fragment of human erythrocyte ankyrin-R.
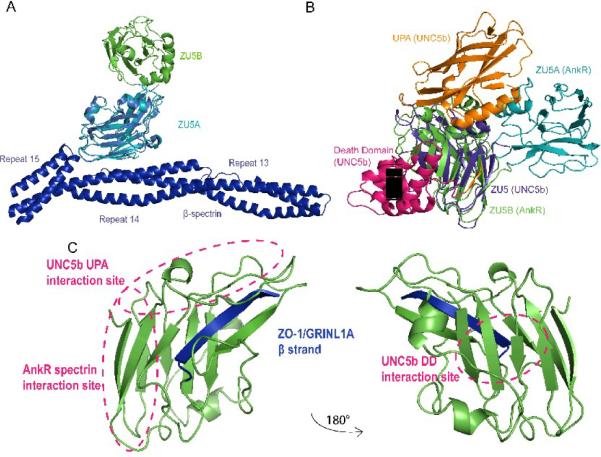
A. Ribbon diagram of the ZU5A-ZU5B fragment. N and C-termini are as labeled on figure. The structure shows two independent, compact, and similar well-folded β-sheet rich cores, each with different numbers of loops and helices. The ZU5B domain (green) extends away from the ZU5A (turquoise) and interacts minimally with it. The ZU5A and ZU5B domains do not interact extensively.The interface between the two domains is small and involves few contacts. The contact surface area is 514 Å231, emphasizing the minimal interaction between the domains. B. Electrostatic surface charge of ZU5A-ZU5B. The ZU5A-ZU5B fragment shows a positive patch on the surface on the ZU5A domain that corresponds to the spectrin binding site23; 24 (circled in purple). In contrast, ZU5B does not exhibit a marked charged character and does not have the equivalent positively charged patch associated with spectrin binding. C. Electrostatic surface charge of the ZU5 domain of UNC5b (PDB ID: 3G5B). The electrostatic map of the ZU5 domain of UNC5b displays a negative patch corresponding to the region of interaction with the UPA domain (circled in green). A similarly charged patch is observed on the surface of the ankyrin ZU5B domain, suggesting that the ankyrin UPA domain could make a similar interaction with ZU5B. The UNC5b domain is shown in the same orientation as the right panel of the ZU5A-ZU5B diagram for ease of comparison. The molecular surface of the molecule is shown with the equipotential electrostatic surface mapped onto it at ± 5 kBT/ec (blue and red, respectively)33.
Structural comparison of the ZU5 domains
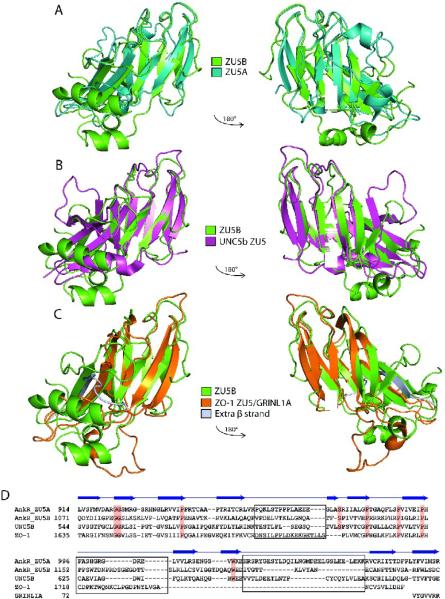
Structural superposition of ZU5A onto ZU5B reveals the overall similarities and significant differences between these two domains (Figure 4A). In general, the structures of the two domains are closely related; the RMSD difference between all main chain atoms in the common regions of the two domains is 1.4 Å. The largest difference between the two domains is the extra helix at the C-terminal end of ZU5B(B α3, Figure 3A), spanning residues 1222–1233, which travels on the side of the domain and occupies roughly the same region as the loop/helix/loop region (1113–1128) after the fourth strand in ZU5A. In ZU5A the corresponding residues (956–972) form aloop joining the fourth and fifth β-strands and are found next to the main body of the protein. Thus, the last helix in ZU5B is in the same overall position as the 956–967 loop in ZU5A. The position of the ZU5B helix sends the last few residues in the opposite direction compared to that observed in ZU5A. In addition, a loop joining A β7 and A α3 and spanning residues 1022 to 1045 in ZU5A (Figure 3A) is much shorter in ZU5B. This loop covers a side of the β sheet in ZU5A, but leaves it exposed in ZU5B. Interestingly, all of the large structural differences are on the side of the ZU5A domain that is not in contact with spectrin. The spectrin binding region in ZU5A bears a remarkably similar fold to the corresponding region in ZU5B.
Figure 4. ZU5 domains have a similar structure containing a β-sheet core.
A. Superposed structures of the ankyrin ZU5A domain on the ZU5B domain. Ribbon diagram of ZU5A (turquoise) superposed on ZU5B (green). The C-terminus of the protein is labeled. The overall β-sheet rich core is similar in both structures, but ZU5B shows differences in the positioning of helices and loops. In addition, the ZU5B domain has an additional helix. B. Superposed structures of ankyrin ZU5B on the ZU5 domain of UNC5b (PDB ID: 3G5B). Ribbon diagram of ankyrin-R ZU5B (green) superposed on the ZU5 domain of UNC5b (pink). The N-terminus of ZU5B and C-terminus of ZU5 of UNC5b are labeled. The comparison shows a clear similarity in the β-sheet rich core, but ZU5B has a larger structure due to the presence of longer loops and two extra two helices. C. Superposed structures of ankyrin-R ZU5B on the ZU5 domain of ZO-1 in complex with GRINL1A peptide (PDB ID: 2KXS). Ribbon diagram of ZU5B of ankyrin-R (green) superposed on the ZU5 domain of ZO-1 in the ZO-1/GRINL1A complex (orange). The N-terminus of ZU5B and the N-terminus of ZU5/GRINL1A of ZO-1 are labeled. The ZO-1 ZU5 domain forms an incomplete ZU5 fold that is completed by a region at the N-terminus of the protein and the GRINL1A peptide at the C-terminus32. The comparison shows region that the ZO-1 ZU5 domain is structurally very similar to the ZU5 domain of ZU5A of ankyrin-R in the β core region. Extra residues (1–14) included in the ZO-1/GRINL1A structure, which are not part of the ZO-1 or GRINL1A sequence, are shown forming an extra β strand (silver). D.Structure-based sequence alignment of four known ZU5 domains: ZU5A and ZU5B of Ankyrin-R, ZU5 of UNC5b, and ZU5/GRINL1A of ZO-1. The sequence alignment is strictly based on structural similarities. Some regions of the proteins are structurally dissimilar (shown in black boxes) precluding sequence alignment. Highly-conserved residues across the ZU5 domains are shaded in red. The β-strands observed in the ZU5B domain of ankyrin-R are indicated above the sequences. The alignment of the GRINL1A peptide is also shown.
Comparisons of the ZU5A domain in the ZU5A-ZU5B fragment with the ZU5A structure alone23 and in complex with spectrin24 show that the ZU5A domains superpose very well, with an RMSD of 1.2 Å for the main chain atoms of the entire domain in both cases. In the ZU5AZU5B fragment, theA β6-A β7 loop (999–1001) that is disordered in both the free ZU5A and ZU5A in complex with spectrin structures is present. Another surface loop (928–933), A β2-A β3, adopts a slightly different conformation in all structures. The latter is in direct contact with spectrin in the complex structure so it is not surprising that it is mobile. The other major difference is the position of the first four N-terminal residues, which in the ZU5A-ZU5B fragment and the free ZU5A structures are found in a position that would interfere with spectrin binding. This movement is probably due to the few contacts presents between the first amino acids and the rest of the protein.
Structural similarity was also investigated between the ZU5 of UNC5b and ZU5A and ZU5B of Ank-R (Figure 4B) and the ZU5 domain of ZO-1. Again, in all cases the overall ZU5 fold is similar and dominated by the β-sheet core. The RMSD between Ank-R ZU5A and ZU5B domains and the ZU5 domain of UNC5b is 1.2 and 1.4 Å, respectively, for all the main chain atoms in the common regions. Overall, the three ZU5 domains are similar in the common core regions with the major differences in the loops and helices. As mentioned above, Ank-R ZU5A has a long loop linking A β7 and A α3 (1023–1046) that is not present in either ZU5B or UNC5b ZU5. This loop is in an equivalent position to the Death domain binding site of UNC5b ZU5. Thus, the ZU5A domain does not present a Death domain binding surface. In addition, the equivalent of the 940–948 (A β3-A β4) loopin ZU5A is longer in UNC5b. This loop in ankyrin contacts spectrin directly and would interfere with binding in UNC5b. An additional helixin ZU5B,B α1 (residues 1115 to 1124), has no equivalent in either ZU5A or UNC5b ZU5. The helix is located in a region where UNC5b ZU5 interacts with the UPA domain and would likely interfere with binding in the observed conformation. Overall, important elements for spectrin binding by ankyrin ZU5A and DD binding by ZU5 in UNC5b are different in the two domains. In contrast, ZU5B does not have the additional loop present in ZU5A and is more similar to UNC5b ZU5, although it has an additional helix at the putative UPA interface region. Nevertheless, the overall similarity supports that the ZU5B domain is likely to be the functionally equivalent of the UNC5b ZU5.
To study the interaction of ZU5B with other domains further, the binding of ZU5A-ZU5B to purified Ankyrin-R DD domain was investigated both by native gel shift assays (data not shown) and gel filtration analysis (Supplemental Figure S2). Both approaches showed no interaction between ZU5A-ZU5B and DD. Whereas this approach showed the interaction between spectrin and ankyrin, in this case no shifts were detected. The gel filtration elution profiles of mixtures of ZU5A-ZU5B and DD show two clear peaks, the first peak, eluting at 15.4 mL, corresponds to a protein of approximately 36.5 kDa identified as ZU5AB. The second peak, eluting shortly after ZU5A-ZU5B at 16.8 mL, corresponds to a protein of approximately 21.9 kDa and identified as DD. Increasing the amount of DD in the protein mix, to a molar ration of 1:4, did not alter the elution profile. No interaction could be detected between ZU5A-ZU5B and DD by either of these two approaches. The DD construct of Ankyrin-R is 12 kDa in size, thus the DD must be forming a homodimer that obscures the putative ZU5B binding interface and prevents ZU5B/DD interaction. Another possibility for the observed missing interaction between ZU5B and DD may be the absence of the UPA domain. The UPA domain could play a role mediating the interactions between the ZU5B and DD domains.
The structure of the ZU5 domain of the tight junction protein Zona Occludens 1 (ZO-1) is known alone and in complex with a short peptide derived from the glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (GRINL1A)32. The structure shows that ZO-1 ZU5 forms an incomplete ZU5 repeat, as was anticipated from the structure of Ank-R ZU523. The ZO-1 ZU5 domain has been compared previously to Ank-R ZU5A and UNC5b ZU5. Comparison with Ank-R ZU5B reveals that the ZO-1 repeat is similar to both ZU5B and ZU5A (Figure 4C). Comparing ZO-1 and ZO-1/GRINL1A against ZU5B results in RMSDs of 1.3 Å and 1.3 Å for 67 and 71 common Cα carbons, respectively. It also confirms that the N-terminal region of the ZO-1 construct and the C-terminal GRINL1A peptide form β strands that completes the central β sheet core of the ZU5 domain32. Overall, the ZU5 β-barrel core is similar in all the structures, however in ZO-1 there are different number of β-strands on each side of the β-barrel core. The core itself is different as not all strands are formed and a large central section is not well-defined. A β-strand formed by residues 1632–1640 in the ZU5/GRINL1A structure corresponds to a much shorter strand at the N-terminus of the Ank-R domains, although the longer strand in ZO-1 appears to be an artifact of the construct used. Finally, the C-terminus of the ZO-1/GRINL1A construct corresponds to a short segment of GRINL1A. This fragment forms a β strand that corresponds to the last strand of ZU5A. In this way, the GRINL1A peptide interacts with ZO-1 by incorporating itself to the β core. In addition, the ZO-1 ZU5 domain is also much smaller and seems to comprise a minimal β-barrel core, while the other ZU5 domains have additional structural elements.
As the ZU5A domain displays a conserved, positively-charged surface patch involved in spectrin binding23; 24, the electrostatic charge on the surface of ZU5A-ZU5B was calculated33. The positive patch present in the ZU5A domain is still present and unobstructed (Figure 3B), as expected since ZU5A retains its spectrin binding function even in the presence of the ZU5B domain. The corresponding region in ZU5B is not positively charged. Instead, it shows an electrostatic surface charge similar to that observed in the ZU5 domain of UNC5b (Figure 3C). Interestingly, ZU5B and ZU5 in UNC5b both show a negatively charged region. In the case of UNC5b, this region is in direct interaction with the UPA domain. The suggestion of a possible UPA - ZU5B interaction is based on the observed electrostatic surface charge, as the residues involved in the ZU5/UPA interface in the UNC5b supramodule are not conserved in ZU5B. The residues in the UPA domain, however, are conserved.
Structure-based sequence alignment of the ZU5A and ZU5B domains of Ankyrin-R, the ZU5 domain of the UNC5b netrin receptor, and the ZU5 domain of ZO-1 show the position of conserved regions across these ZU5 domains (Figure 4D). The structure-based sequence alignment presented here is different from the one reported based on the ZO-1 structure32. Some regions clearly have no structural equivalent and the sequences cannot be aligned based on the structures, even though a sequence-based alignment is possible. In addition, some regions are structurally very different among all of the structures. In general, there are few strictly conserved residues amongst the four structures, however, some of these conserved residues are not conserved in other related proteins21. The alignment shows that there is a preponderance of conserved glycines and prolines and that many of the conserved residues are at the beginning or end of the strands forming the core.
Discussion
The ZU5A-ZU5B structure presented here is the first structure of tandem ZU5 repeats, revealing the general arrangement of the two domains and demonstrating the structural similarity between them. Overall, both ZU5A and ZU5B fold into compact β-strand rich cores with variable numbers of surface loops and helices. In the fragment, the ZU5B domain extends away from the ZU5A domain and is connected by a short loop in between the two (Figure 3A). The region of ZU5A that forms the spectrin binding surface23; 24 is completely exposed and is away from the ZU5B domain. As there are few interactions between the two ZU5 domains, the ZU5B domain does not appear capable to interfere directly with the spectrin binding ability of the ZU5A domain, even if the relative arrangements of the two domains were to change. This was further confirmed by native gel shift assays and SPR data, where the ZU5A-ZU5B protein fragment was shown to form a complex with β-spectrin with the same binding characteristics as the ZU5A domain alone. No changes in binding parameters were observed in the presence of the ZU5B domain, suggesting that binding only involves interactions between ZU5A and spectrin and that the ZU5B domain does not contribute to binding. Placing these biophysical data in the context of the structure, superposition of ZU5A-ZU5B model with the ZU5A/β-spectrin complex structure also indicates that the presence of ZU5B would not interfere with the ZU5A/spectrin interaction (Figure 5A).
Figure 5. The ZU5 domain is a versatile protein interaction module.
A. Superposed structures of the ZU5A domain in ZU5A-ZU5B on the ZU5A in complex with spectrin. Ribbon diagram of the ZU5A-ZU5B (turquoise, green) structure superposed on the ZU5A domain (red) of the complex with β-spectrin (blue)24. The diagram shows that ZU5B is unlikely to interact with spectrin as it is found away from the spectrin/ankyrin interface. This finding is supported by biochemical experiments that show that the ZU5B domain does not alter the binding characteristics significantly. B. Superposed structures of the ZU5A-ZU5B fragment of ankyrin-R on the UNC5b supramodule. Ribbon diagram of the ZU5A-ZU5B fragment (turquoise, green) superposed on the UNC5b supramodule comprising the ZU5, UPA and DD domains25. The ZU5 domain of UNC5b (pink) was superposed on the ankyrin-R ZU5B domain. The superposition shows that the ankyrin DD domain could interact in a similar manner without any major conformational changes. The UPA domain could also interact with ZU5B in a similar manner, although there are some potential clashes in the superposed structures. Some of these potential clashes may not exist as they involve a region in the UNC5b UPA domain that is not present in ankyrin. Finally, the ankyrin ZU5A domain clashes with the UNC5b UPA domain, suggesting that a small reorientation of the ZU5A domain may be needed for proper interaction. Overall, the superposition shows that a similar supramodule could be formed in ankyrin and that this supramodule is not likely to interact directly with spectrin. C. The ZU5 domain has several distinct binding surfaces. The ZU5 domain can use different regions to interact with different proteins. The known areas of interaction are marked, showing the different, non-overlapping areas of the ZU5 domain involved in different complexes. Furthermore, the incorporation of additional β strands can also serve as a mechanism for interaction, as observed in the interaction with the DD domain in UNC5b25 and the GRINL1A peptide in ZO-132.
The ZU5 domain has been found in a wide-range of proteins and has been implicated in interactions with different other domains. To understand the role of the ZU5 domain as an interaction module, all known ZU5 domain structures were compared. Comparison of the four ZU5 domains with known structures (ZU5A and ZU5B of ankyrin-R, ZU5 of UNC5b, and ZU5 of ZO-1) shows that all of these domains share a similar fold, with a compact β-sandwich core with a variable number of surface loops and helices (Figure 5A, 5B, 5C). This variability may, in fact, signal to the variable function(s) of these domains and the usage of ZU5 as an adaptable protein-protein binding module.
ZU5A from ankyrin-R has a large positive patch on its surface where it interacts with a negative patch on the β-spectrin23; 24. Not surprisingly, a similar large positive patch is not found on ZU5B, UNC5b ZU5, or ZO-1 ZU5. In particular, it is interesting to note that a positive electrostatic charge found on the surface of ZU5 UNC5b, corresponding to the ZU5/UPA interface, is also found on the ZU5B surface (Figure 3C). This could be a potential site of interaction between the ZU5B and UPA domains of ankyrin-R. UNC5b ZU5 also binds a Death domain using another interface and also expanding one of the β sheets by adding a short strand to it. As noted before, ZO-1 has an incomplete domain that is completed through the interaction with another protein32. Taken together, these observations suggest that the ZU5 domain is a versatile protein/protein interaction domain with more than one interaction surface (Figure 5C) that acts in different modes to interact with a variety of partners. In the case of Ank-R ZU5A, it interacts with spectrin through a positively charged surface area24. In the case of UNC5b ZU5, and probably ankyrin ZU5B, the same domain can interact with the UPA and DD domains using completely different areas of the protein25. Finally, DD and ZO-1 illustrate the case where the interaction involves expansion or completion of a β sheet32.
The involvement of the ZU5 domain of the UNC5b netrin receptor in supramodule formation and functional regulation25 suggests that the equivalent ankyrin region, spanning the ZU5B, UPA and DD domains, could also be forming a similar supramodule and be involved in auto-regulating ankyrin function. Structural and sequence comparison of the UNC5b ZU5 and Ank-R ZU5A-ZU5B fragment shows that over the common structural regions the sequence of the ZU5 domain of UNC5b is more similar to the one in ZU5A. However, structurally, the ZU5B domain resembles the UNC5b ZU5 domain, as both have the same overall shape and with equivalent exposed surfaces. This supports the possibility that ZU5B may be capable of forming a supramodule. Detailed analysis of the potential site for a ZU5B/DD interaction reveals that the ankyrin DD domain could be accommodated by the ZU5B domain in the same manner as in UNC5b without any major structural changes. However, gel filtration analysis of ZU5A-ZU5B/DD (Supplemental Figure S2) suggests that DD forms a homodimer that interferes with the ZU5B/DD interaction. It is important to note that homodimerization has also been observed in the UNC5b DD domain25. The UNC5b DD domains form homodimers when separated from the ZU5-UPA domains of UNC5b, suggesting that DD dimerization competes with binding to the ZU5 interface (Supplemental Figure S3B). In addition, the structure of Ankyrin DD (PDB: 2YVI) shows dimer formation utilizing a different binding region than the one involved in the potential ZU5B interaction (Supplemental Figure S3A). Thus, our experiments, together with previous data on UNC5b, suggest competition between DD homodimerization and DD/ZU5B dimerization. It also appears that DD binding to ZU5B is enhanced by having both domains linked through the UPA domain. It thus appears that DD homodimerization prevents ZU5 binding in the absence of the UPA domain.
On the other hand, comparison of the ZU5-UPA interface reveals that ZU5B has an extra helix (residues 1114–1125) that would interfere with UPA binding as this helix would collide with a long loop in UPA (~778–790 in UNC5b). Sequence analysis of the UPA region in ankyrin reveals that this region contains a large deletion. It is thus possible that the UPA of ankyrin has a shorter loop and in this way ZU5B could be accommodated. Without a structure of the ankyrin ZU5B-UPA region it is not possible to ascertain whether this is the case, but it is possible that in ankyrin the UPA domain has a shorter loop to allow proper interaction with the Ank-R ZU5B domain.
Mapping of known clinical mutations to the ZU5B region of ankyrin-R shows two mutants related to hereditary spherocytosis34; 35. The Tubarao mutation, a missense mutation, changes isoleucine1075 into a threonine36. Isoleucine 1075 is one of the first residues in the first β strand of ZU5B region and faces towards the solvent in the wildtype ZU5A-ZU5B structure. Since it is one of the amino acids of the first β strand comprising the β core of ZU5B, this residue may be crucial in completing the ZU5 fold (Supplemental Figure S4A, S4B). A second mutation, Trp1185Arg, maps to the hydrophobic interior of the protein. Placing a charged residue at this position may interfere with the proper folding of the ZU5B domain (Supplemental Figure S4C).34; 35; 37. Other mutants in the ZU5B region, such as Thr1127, cause either a frameshift or introduce premature stop codons. These mutations would lead to incomplete or nonsensical proteins.
In conclusion, the ZU5A-ZU5B protein fragment of human erythrocyte ankyrin has been solved. Both the structure and biophysical experiments show that this second ZU5B domain is unlikely to interfere with or facilitate ZU5A binding to spectrin. In addition, the newly elucidated ZU5B domain shows that it is possible that this domain is involved in formation of an auto-regulating supramodule involving the UPA and DD domains of ankyrin. Finally, analysis of different ZU5 domains shows that the common feature is the presence of a conserved β core. The ZU5 domain can interact with many other partners using different regions of the protein and even by completing the core. Thus, the ZU5 domain represents a compact and very versatile protein interaction module.
Materials and Methods
Cloning, expression, and purification of ankyrin fragments
Preparation of ZU5A-ZU5B
A fragment spanning residues 911–1233 of human ankyrin-R was cloned from human erythrocyte cDNA Ank3738; 39; 40 into the vector pMCSG7 by ligation independent cloning41. Ank37 cDNA was kindly given by Dr. Patrick Gallagher38; 40. The resulting construct, ZU5A-ZU5B, was designed with an N-terminal His-tag and a TEV cleavage site for ease of purification. The DNA sequence was verified by the Northwestern University Sequencing Facility.
Preparation of DD
A fragment of spanning residues 1394–1497 of human ankyrin-R was cloned in a similar manner as the ZU5AB construct from human erythrocyte cDNA Ank1538; 39; 40 into the vector pMCSG7. The DNA sequence was verified by the Northwestern University Sequencing Facility.
Protein expression of ZU5A-ZU5B
For protein expression, E. coli BL21(DE3) competent cells were transformed using the verified plasmids by heat shock and plated on LB agar plates with 100 μg/mL ampicillin. After overnight incubation at 37 °C, single colonies were used to inoculate 100 mL of Luria-Bertrani (LB) media with 100 μg/mL ampicillin. These starter cultures were grown overnight at 37 °C with shaking then transferred to 1 L Terrific Broth (TB) media. The TB cultures were incubated at 37 °C with shaking until the OD600 had reached ~0.8. Protein expression was induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). Induced cultures were further incubated at 16 °C overnight with shaking. Cells were harvested via centrifugation at 3500 × g. The resulting cell pellet was flash frozen with liquid nitrogen and stored at −80 °C until needed. Selenomethionine protein was produced in the same manner as described for native ZU5A-ZU5B, except for the use of M9 media instead of TB, and addition of 50 mg selenomethionine to each liter of culture at the time of IPTG induction.
Protein expression of DD
Similar methods were used in expressing the DD construct in E. coli BL21(DE3) as for ZU5A-ZU5B, except for the use of LB media throughout instead of TB media.
Protein purification of ZU5A-ZU5B
Frozen cell pellets from 1 L of culture were thawed and resuspended in 20 mL of lysis buffer (50 mM sodium phosphate pH 8.0, 10 mM imidazole) and lysed by sonication. After lysis, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) was added to the cell lysate to prevent proteolysis. Cell debris was removed from the cell lysate via centrifugation at 66,000 × g at 4 °C for 40 mins. The resulting supernatant was purified using Ni-NTA resin (Qiagen), following the manufacturer's protocol. After elution from the Ni-NTA resin, TEV protease was added (1:20 mass ratio) to the peak protein fractions and dialyzed overnight at 4 °C in PEDP buffer (20 mM sodium phosphate pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF) with 0.3 M sodium chloride. The cleaved protein was loaded on a Sulfonyl methacrylate (Bio-Rad) column and eluted with a 0.1 – 1.0 M NaCl gradient in PEDP and further purified on a P-60 (Bio-Rad) size-exclusion column. The final purified protein fractions were concentrated by centrifugation in Amicon Ultra centrifugal filtration devices. Purity of the target protein was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5 % polyacrylamide gels. The final protein concentration was determined by Aλ=280 measurements using a Nanodrop spectrophotometer with extinction coefficient (Abs 1% = 8.64) calculated from UV absorbing amino acids in the sequence. Proper folding of the protein was checked via circular dichroism, and protein molecular mass was verified via Mass spectrometry at the Integrated Molecular Structure Education and Research Center at Northwestern University.
Protein purification of DD
DD construct was purified following similar methods to the ones used to purify ZU5AB. Q-sepharose (GE Healthcare) column was used instead of Sulfonyl methacrylate column after the Ni-NTA column purification. Protein was eluted off with a 0.1 – 1.0 M NaCl gradient in TEDP buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF) and further purified on a Sephadex G-25 (Pharmacia) size-exclusion column. Proper folding of the isolated protein was checked via circular dichroism, and protein molecular mass was verified with mass spectrometry at the Integrated Molecular Structure Education and Research Center at Northwestern University.
Gel-shift assay
Native gel shift assays were used to assess the ZU5A-ZU5B interaction with β-spectrin fragments. ZU5A-ZU5B and human erythrocyte spectrin repeats 13-or repeats 14–15 were mixed in a 1:1 molar ratio and incubated at room temperature for 20 minutes. Ten percent polyacrylamide gels at pH 7.4 were pre-run at 30 V for 30 minutes before loading. Protein samples were then mixed with 6 × loading buffer (40% sucrose) prior to loading. Gels were run at 30 V for 4 hours at 4 °C. Protein was visualized by Coomassie Blue staining.
Analytical Gel Filtration
Analytical gel filtration was conducted on an AKTA FPLC system (GE Healthcare). Protein mixes of ZU5A-ZU5B/DD were loaded onto Superdex 200 10/300 GL (GE Healthcare) equilibrated with a 20 mM sodium phosphate pH 8.0 buffer with 0.1 M NaCl.
Surface Plasmon Resonance (SPR)
Binding affinity and kinetics of the complex formed between the ZU5A-ZU5B and biotinylated spectrin fragments were measured with a Biacore 3000 equipped with a streptavidin biosensor at the University of Chicago Biophysics Core, as described previously21. For the measurements, purified ZU5A-ZU5B in HBS-P (10 mM HEPES pH 7.4, 0.15 M NaCl, 0.005 % (v/v) surfactant P20, Biacore) was diluted serially (final concentrations of 0, 2, 5, 10, 20, 50 nM) and maintained at 10 °C. Injections were performed in triplicate. Data for each concentration series were processed globally and simultaneously to obtain the ka and kd after reference cell and buffer subtraction (Table I, Figure 2B, C) using the included BIAevaluation software version 4.1.
Crystallization and characterization of ZU5A-ZU5B
Crystals of ZU5A-ZU5B Ankyrin-R were grown by vapor diffusion against 25% ethanol, 0.1 M Tris pH 7. One microliter of purified protein solution (~6.5 mg/mL) was mixed with an equal volume of reservoir solution in a sitting drop reservoir. Crystals grew as thin plates at 10 °C overnight. There were no differences in crystal size, growth rate, morphology, or space group between native crystals and selenomethionine (SeMet) ZU5A-ZU5B crystals. However, two slightly different but closely related crystal forms were identified under the same conditions for the native crystals. For data collection, crystals were cryo-protected by direct addition of either glycerol (final concentration of 10%) or MPD (final concentration of 30%) to the crystallization drops. Crystals were then harvested and flash-frozen in liquid nitrogen.
ZU5A-ZU5B Data collection, analysis, refinement, and model building
ZU5A-ZU5B Native 1 crystals belong to space group C2, with unit cell constants a=277.8 Å, b=40.5 Å, c=94.7 Å, α=γ=90°, β=92.3°. Native 2 crystals also belong to space group C2, with unit cell constants, a=160.1 Å, b=40.5 Å, c=178.1 Å, α=γ=90°, b=112.7°. ZU5A-ZU5B SeMet crystal were most similar to the Native 1 crystals with unit cell constants a=279.5 Å, b=40.9 Å, c=95.3 Å, α=γ=90°, β=92.0°. Data from Native 1 crystals were collected to 2.6 Å resolution whereas Native 2 data were collected up to 2.0 Å resolution. Data for SeMet ZU5A-ZU5B crystals were collected up to 2.2 Å resolution. All diffraction data were collected using synchrotron radiation at the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). Data were processed with XDS42 and scaled with SCALA43. Further processing were completed with various programs available from the CCP4 suite43.
The ZU5A-ZU5B structure was solved by a combination of Molecular Replacement (MR) and Single-wavelength Anomalous Dispersion (SAD) phasing29. An MR solution obtained using the ZU5A structure (PDB ID: 3F59)23 as a search model showed negligible density for the ZU5B domain. Nonetheless, these phasing data were able to assist in locating the Se atoms in the SeMet datasets. SAD phasing with the program AutoSHARP30 using the Se positions, followed by density modification with DM produced an electron density map with both domains clearly visible. The experimental map was of excellent quality and automated model building with PHENIX44 produced the initial model. The model was refined with Refmac5 using the Native 1 and SeMet data sets45. This model was further refined using BUSTER46 against the higher resolution Native 2 dataset. Manual rebuilding and inspection were done with Coot47. The final models were validated using MolProbity48. The final high resolution model includes 1,233 amino acids, 686 water molecules, and 18 ethanol molecules; and has a final Rfactor and Rfree of 17.3% and 20.6% respectively, with RMSD values of 0.010 Å and 1.05° for bond lengths and bond angles (Table I).
Figures
Figures were made with the Pymol molecular visualization system49. Superpositions of molecules were generated with the program lsqkab available in the CCP4 suite43; 50.
Supplemental data
Supplemental data are available.
Protein data bank accession codes
The final structure factors and coordinates have been deposited in the Protein Data Bank with accession codes 3UD1 and 3UD2 for the Native and SeMet proteins.
Supplementary Material
Highlights
Structure of tandem ZU5 repeats of human ankyrin reveals arrangement of the domains.
The ankyrin ZU5B domain is not directly involved in spectrin binding.
Structural comparisons of ZU5B suggest a possible ZU5B function.
The ZU5 domain has several different binding areas.
Acknowledgements
We thank Fernando Bazan for pointing out the presence of the second ZU5 domain to us before publication of the UNC5b structure. We thank the Pinkett laboratory for use of their AKTA FPLC instrument. We acknowledge staff and instrumentation support from the University of Chicago Biophysics Core Facility, the Keck Biophysics Facility at Northwestern University, the Center for Structural Biology at Northwestern University, and LS-CAT at the Advanced Photon Source (APS) at Argonne National Laboratory. Support from the R.H. Lurie Comprehensive Cancer Center of Northwestern University to the Structural Biology Facility is also acknowledged. LS-CAT was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the APS is supported by the Department of Energy. Research was supported by NIH grant GM057692 (to AM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bennett V, Healy J. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harbor Perspectives in Biology. 2009;1 doi: 10.1101/cshperspect.a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohandas N, Evans E. Mechanical properties of the red cell membrane in relation to molecular structure and genetic defects. Annu Rev Biophys Biomol Struct. 1994;23:787–818. doi: 10.1146/annurev.bb.23.060194.004035. [DOI] [PubMed] [Google Scholar]
- 3.Bodine D. M. t., Birkenmeier CS, Barker JE. Spectrin deficient inherited hemolytic anemias in the mouse: characterization by spectrin synthesis and mRNA activity in reticulocytes. Cell. 1984;37:721–9. doi: 10.1016/0092-8674(84)90408-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu SC, Derick LH, Agre P, Palek J. Alteration of the erythrocyte membrane skeletal ultrastructure in hereditary spherocytosis, hereditary elliptocytosis, and pyropoikilocytosis. Blood. 1990;76:198–205. [PubMed] [Google Scholar]
- 5.Mohler PJ, Bennett V. Ankyrin-based cardiac arrhythmias: a new class of channelopathies due to loss of cellular targeting. Curr Opin Cardiol. 2005;20:189–93. doi: 10.1097/01.hco.0000160372.95116.3e. [DOI] [PubMed] [Google Scholar]
- 6.Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, Priori SG, Keating MT, Bennett V. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A. 2004;101:9137–42. doi: 10.1073/pnas.0402546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogne K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–9. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–46. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett V, Stenbuck PJ. Identification and partial purification of ankyrin, the high affinity membrane attachment site for human erythrocyte spectrin. J Biol Chem. 1979;254:2533–41. [PubMed] [Google Scholar]
- 10.Cunha SR, Mohler PJ. Ankyrin protein networks in membrane formation and stabilization. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–9. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- 12.Cunha SR, Mohler PJ. Cardiac ankyrins: Essential components for development and maintenance of excitable membrane domains in heart. Cardiovasc Res. 2006;71:22–9. doi: 10.1016/j.cardiores.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Hall TG, Bennett V. Regulatory domains of erythrocyte ankyrin. J Biol Chem. 1987;262:10537–45. [PubMed] [Google Scholar]
- 14.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–48. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Rio M, Imam A, DeLeon M, Gomez G, Mishra J, Ma Q, Parikh S, Devarajan P. The death domain of kidney ankyrin interacts with Fas and promotes Fas-mediated cell death in renal epithelia. J Am Soc Nephrol. 2004;15:41–51. doi: 10.1097/01.asn.0000104840.04124.5c. [DOI] [PubMed] [Google Scholar]
- 16.Davis LH, Davis JQ, Bennett V. Ankyrin regulation: an alternatively spliced segment of the regulatory domain functions as an intramolecular modulator. J Biol Chem. 1992;267:18966–72. [PubMed] [Google Scholar]
- 17.Abdi KM, Mohler PJ, Davis JQ, Bennett V. Isoform specificity of ankyrin-B: a site in the divergent C-terminal domain is required for intramolecular association. J Biol Chem. 2006;281:5741–9. doi: 10.1074/jbc.M506697200. [DOI] [PubMed] [Google Scholar]
- 18.Mohler PJ, Hoffman JA, Davis JQ, Abdi KM, Kim CR, Jones SK, Davis LH, Roberts KF, Bennett V. Isoform specificity among ankyrins. An amphipathic alpha-helix in the divergent regulatory domain of ankyrin-b interacts with the molecular co-chaperone Hdj1/Hsp40. J Biol Chem. 2004;279:25798–804. doi: 10.1074/jbc.M401296200. [DOI] [PubMed] [Google Scholar]
- 19.Ackerman S, Kozak L, Przyborski S, Rund L, Boyer B, Knowles B. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature. 1997:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- 20.Leonardo E, Hinck L, Masu M, KeinoMasu K, Ackerman S, TessierLavigne M. Vertebrate homologues of C-elegans UNC-5 are candidate netrin receptors. Nature. 1997:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- 21.Ipsaro JJ, Huang L, Gutierrez L, MacDonald RI. Molecular epitopes of the ankyrin-spectrin interaction. Biochemistry. 2008;47:7452–64. doi: 10.1021/bi702525z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohler P, Yoon W, Bennett V. Ankyrin-B targets beta(2)-spectrin to an intracellular compartment in neonatal cardiomyocytes. Journal of Biological Chemistry. 2004:40185–40193. doi: 10.1074/jbc.M406018200. [DOI] [PubMed] [Google Scholar]
- 23.Ipsaro JJ, Huang L, Mondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood. 2009;113:5385–93. doi: 10.1182/blood-2008-10-184358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ipsaro JJ, Mondragon A. Structural basis for spectrin recognition by ankyrin. Blood. 2010 doi: 10.1182/blood-2009-11-255604. blood-2009-11-255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Wei ZY, Jin H, Wu H, Yu C, Wen WY, Chan LN, Wen ZL, Zhang MJ. Autoinhibition of UNC5b Revealed by the Cytoplasmic Domain Structure of the Receptor. Molecular Cell. 2009;33:692–703. doi: 10.1016/j.molcel.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Leunghagesteijn CY, Spence AM, Stern BD, Zhou YW, Su MW, Hedgecock EM, Culotti JG. Unc-5, a Transmembrane Protein with Immunoglobulin and Thrombospondin Type-1 Domains, Guides Cell and Pioneer Axon Migrations in C-Elegans. Cell. 1992;71:289–299. doi: 10.1016/0092-8674(92)90357-i. [DOI] [PubMed] [Google Scholar]
- 27.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 28.Davis JQ, Bennett V. Brain ankyrin. A membrane-associated protein with binding sites for spectrin, tubulin, and the cytoplasmic domain of the erythrocyte anion channel. J Biol Chem. 1984;259:13550–9. [PubMed] [Google Scholar]
- 29.Mccoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. Journal of Applied Crystallography. 2007:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vonrhein C, Blanc E, Bricogne G. Automated structure solution with autoSHARP. Methods Molec Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Huo L, Wen W, Wang R, Kam C, Xia J, Feng W, Zhang M. Cdc42-dependent formation of the ZO-1/MRCKbeta complex at the leading edge controls cell migration. EMBO J. 2011;30:665–78. doi: 10.1038/emboj.2010.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker N, Sept D, Joseph S, Holst M, McCammon J. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher PG, Forget BG. Hematologically important mutations: spectrin and ankyrin variants in hereditary spherocytosis. Blood Cells Mol Dis. 1998;24:539–43. doi: 10.1006/bcmd.1998.0217. [DOI] [PubMed] [Google Scholar]
- 35.Gallagher P. Hematologically important mutations: Ankyrin variants in hereditary spherocytosis. Blood Cells Molecules and Diseases. 2005:345–347. doi: 10.1016/j.bcmd.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Gallagher P, Ferreira J, Saad S, Kerbally J, Costa F, Forget B. A recurring frameshift mutation of the ankyrin-1 gene associated with severe hereditary spherocytosis in Brazil. Blood. 1996:11–11. doi: 10.1046/j.1365-2141.2000.02441.x. [DOI] [PubMed] [Google Scholar]
- 37.Eber S, Gonzalez J, Lux M, Scarpa A, Tse W, Dornwell M, Herbers J, Kugler W, Ozcan R, Pekrun A, Gallagher P, Schroter W, Forget B, Lux S. Ankyrin-1 mutations are a major cause of dominant and recessive hereditary spherocytosis. Nature Genetics. 1996:214–218. doi: 10.1038/ng0696-214. [DOI] [PubMed] [Google Scholar]
- 38.Lambert S, Yu H, Prchal JT, Lawler J, Ruff P, Speicher D, Cheung MC, Kan YW, Palek J. cDNA sequence for human erythrocyte ankyrin. Proc Natl Acad Sci U S A. 1990;87:1730–4. doi: 10.1073/pnas.87.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 40.Gallagher P, Tse W, Scarpa A, Lux S, Forget B. Structure and organization of the human ankyrin-1 gene - Basis for complexity of pre-mRNA processing. Journal of Biological Chemistry. 1997:19220–19228. doi: 10.1074/jbc.272.31.19220. [DOI] [PubMed] [Google Scholar]
- 41.Aslanidis C, De Jong P. LIGATION-INDEPENDENT CLONING OF PCR PRODUCTS (LIC-PCR) Nucleic Acids Research. 1990:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl. Crystallog. 1993;26:795–800. [Google Scholar]
- 43.Collaborative-Computational-Project-4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994:750–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Murshadov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Chen V, Arendall W, Headd J, Keedy D, Immormino R, Kapral G, Murray L, Richardson J, Richardson D. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D-Biological Crystallography. 2010:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano W. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 50.Kabsch W. Solution for Best Rotation to Relate 2 Sets of Vectors. Acta Crystallographica Section A. 1976;32:922–923. [Google Scholar]
- 51.Diederichs K, Karplus P. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature Structural Biology. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.