Abstract
We analysed 67 short tandem repeat polymorphisms from the non-recombining part of the Y-chromosome (Y-STRs), including 49 rarely-studied simple single-copy (ss)Y-STRs and 18 widely-used Y-STRs, in 590 males from 51 populations belonging to 8 worldwide regions (HGDP-CEPH panel). Although autosomal DNA profiling provided no evidence for close relationship, we found 18 Y-STR haplotypes (defined by 67 Y-STRs) that were shared by two to five men in 13 worldwide populations, revealing high and widespread levels of cryptic male relatedness. Maximal (95.9%) haplotype resolution was achieved with the best 25 out of 67 Y-STRs in the global dataset, and with the best 3-16 markers in regional datasets (89.6-100% resolution). From the 49 rarely-studied ssY-STRs, the 25 most informative markers were sufficient to reach the highest possible male lineage differentiation in the global (92.2% resolution), and 3-15 markers in the regional datasets (85.4-100%). Considerably lower haplotype resolutions were obtained with the three commonly-used Y-STR sets (Minimal Haplotype, PowerPlex Y®, and AmpFlSTR® Yfiler®). Six ssY-STRs (DYS481, DYS533, DYS549, DYS570, DYS576 and DYS643) were most informative to supplement the existing Y-STR kits for increasing haplotype resolution, or – together with additional ssY-STRs - as a new set for maximizing male lineage differentiation. Mutation rates of the 49 ssY-STRs were estimated from 403 meiotic transfers in deep-rooted pedigrees, and ranged from ~4.8×10−4 for 31 ssY-STRs with no mutations observed to 1.3×10−2 and 1.5×10−2 for DYS570 and DYS576, respectively, the latter representing the highest mutation rates reported for human Y-STRs so far. Our findings thus demonstrate that ssY-STRs are useful for maximizing global and regional resolution of male lineages, either as a new set, or when added to commonly-used Y-STR sets, and support their application to forensic, genealogical and anthropological studies.
Keywords: Y-STR; microsatellites; Y-chromosome; haplotype resolution; lineage differentiation; HGDP-CEPH, mutation rates
Introduction
Human Y-chromosomal short tandem repeat polymorphisms (Y-STRs) or microsatellites, usually in combination as haplotypes, are widely used to resolve and relate male lineages in forensic, genealogical, evolutionary and anthropological studies [1-4]. Haplotype resolution is central to most applications of Y-STRs and depends not only on the number of markers used but also on their independent mutability. Differences in Y-STR diversity and allelic spectra between geographic regions are well known [5,6], and can sometimes be used to infer the geographic region of paternal ancestry of a person [7,8]. However, they can also result in differences in haplotype resolution between loci and geographic regions [9]. Also, at a more local level, higher frequencies of indistinguishable Y-chromosomes can sometimes be found due to members of the same male lineage living in the same geographic region, a phenomenon usually referred to as (male) population substructure. It has been observed that 7-16 highly polymorphic Y-STRs are insufficient for differentiating male lineages when applied to populations that underwent a strong (male) bottleneck in their history: for example, identical Y-STR haplotypes were found in two populations from Pakistan with a frequency of 14% (16 Y-STRs) [10], in Finns at 13% (16 Y-STRs) [11], or even over entire geographic regions such as Polynesia at 16% (7 Y-STRs) [12]. Reduced Y-STR diversity leading to a large number of indistinguishable Y-STR haplotypes can also be caused by cultural effects such as patrilocal residence pattern and polygyny as previously observed in New Guinea [13], or by strongly biased male expansion due to male occupation history and privilege as in Central and Eastern Asia [14,15]. The resulting lack of resolution can be especially problematic when a limited number of Y-STRs is used in forensic applications of male lineage identification.
Therefore, more Y-STRs than included in the three commonly-used sets (the 9 Y-STRs comprising the so-called Minimal Haplotype, the 12 Y-STRs included in the PowerPlex Y® System [Promega], or the 17 Y-STRs from the AmpFlSTR® Yfiler® PCR Amplification Kit [Applied Biosystems]) are needed for improving the resolution of male lineage differentiation in particular populations, and also for differentiating male relatives in any population. A large number of additional Y-STRs (166) have been described previously [16]; however, population-genetic data are still scarce for most of these additional markers. In this study, we analysed 67 Y-STRs in 590 unrelated males from 51 globally distributed populations covering eight geographic regions from all inhabited continents except Australia, (the HGDP-CEPH panel [17]). These 67 Y-STRs comprise 18 previously-used Y-STRs including all of those that are part of commercially-available Y-STR kits, as well as 49 additional Y-STRs described recently [16,18]. From the latter, we chose simple loci that exist in a single copy on the non-recombining part of the human Y-chromosome and contain only one uninterrupted variable stretch of repeats to avoid the problems of length homoplasy (as with complex Y-STRs) and allele-locus assignment (as with multi-copy Y-STRs). Simple Y-STRs also have a great advantage over complex markers due to a more direct relationship between mutation rate and length variation [16] as relevant in evolutionary studies. Previous analysis suggests that ssY-STRs may lead to more precise time estimates when applied to male lineage dating in anthropological and evolutionary studies due to a higher correlation between repeat count and repeat variance compared with complex Y-STRs [18]. Single-copy Y-STRs do not suffer from the problem of equivocal allele-locus assignment usually associated with multi-copy Y-STRs, which may result in an underestimation of the haplotype resolution, and additionally can cause problems in correctly inferring the number of males who contributed to a crime scene sample in forensic studies.
Here, we investigate male lineage differentiation, both on a global level and regional levels, considering a total of 67 Y-STRs as well as a set of 49 rarely-studied simple single-copy Y-STRs (ssY-STRs) alone. In addition, we studied improvements of global and regional haplotype resolution by adding the most informative ssY-STRs to the three commonly-used sets of Y-STRs. Finally, we estimated mutation rates for all 49 ssY-STRs by analyzing deep-rooted pedigrees to understand the basis of their value in resolving male lineages and to stimulate future uses of these markers in forensic, genealogical, and anthropological studies where the knowledge of mutation rates is crucial.
Material and Methods
DNA samples from the Human Genome Diversity Panel (HGDP) [17] were provided by The Centre d’Etude du Polymorphisme Humain (CEPH). Samples where identity, first and second degree biological relationship, origin mix-up or duplicated samples were identified previously based on autosomal 783 STRs [19] have been excluded from the analysis and only males from the H952 set [19] were used. Altogether 590 males from 51 populations of 8 worldwide geographic regions were studied: 163 from East Asia (18 populations), 84 from Europe (8), 48 from the Middle East (3), 20 from North Africa (1), 16 from Oceania (2), 20 from the Americas (5), 162 from South and West Asia (8), and 77 from Sub-Saharan Africa (6). In addition, DNA samples from 104 members of 28 deep-rooted pedigrees from Canada [20], Germany [4], and China [21] covering all together 403 meiotic transfers were analysed.
We genotyped 49 simple single-copy Y-STRs ascertained from 166 recently described Y-STRs [16] (see Supplementary Table S1 or Table 3 for the list of all markers) using 15 small multiplex reactions as described elsewhere [18] with the following modifications. PCRs on HGDP-CEPH samples were carried out in 384-well microtiter plates using 10 μl volumes containing 1× PCR buffer including 1.5 mM MgCl2 (Applied Biosystems Inc., Foster City, CA), 1 mM dNTPs (Roche Diagnostics GmbH, Mannheim, Germany), 0.25 units of AmpliTaqGold DNA polymerase (Applied Biosystems), 0.25–1 ng DNA and the primer pairs and concentrations described in Supplementary Table S1, using a GeneAmp PCR Systems 9700 machine (Applied Biosystems). Initial denaturation was at 95°C for 15 min, followed by 20 cycles of touchdown PCR: 94°C for 30 s, 70°C for 45 s, 72°C for 1 min, with a 1°C decrease in annealing temperature every cycle, and then 15 cycles of standard PCR (94°C for 30 s, 50°C for 45 s, 72°C for 1 min), and finishing with extension at 60°C for 45 min and storage at 4°C. PCRs of deep-rooting pedigree samples were carried out in 96-well microtiter plates in 20 μl volumes using a MJ Research PTC-200 (Bio-Rad, Hercules, CA). PCR fragment lengths were analyzed by mixing 1 μl of PCR product with 9.7 μl Hi–Di Formamide (Applied Biosystems) and 0.3 μl CXR 60–400 bases size marker (Promega, Madison, WI) and running on 36 cm×50 μm capillaries containing POP-4 polymer (Applied Biosystems) on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Capillary electrophoresis was carried out at 1 kV for 22 s followed by 15 kV for 25 min with a run temperature of 60°C. Allele sizes were measured using GeneMapper v3.7 software (Applied Biosystems). In addition, 18 previously used Y-STRs (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, Y-GATA-H4, and DYS388) of which all except DYS388 were genotyped using the AmpFlSTR® Yfiler® PCR amplification kit(Applied Biosystems) according to the instructions provided by the manufacturer. DYS388 was genotyped as described elsewhere [1]. Furthermore, 15 autosomal STRs were genotyped in the HGDP-CEPH samples using the PowerPlex 16 kit (Promega) according to the instructions provided by the manufacturer.
Table 3.
Mutation data of 49 simple single-copy Y-STRs from analyzing 104 members of 28 deep-rooted pedigrees covering 403 meiotic transfers at each of the 49 Y-STRs analyzed.
| Bayesian Mutation Rates | ||||
|---|---|---|---|---|
| Y-STR Locus | # Observed Mutations | Median | 2.5% Credible Interval | 97.5% Credible Interval |
|
|
||||
| DYS472 | 0 | 4.82E-04 | 1.97E-05 | 0.003265 |
| DYS476 | 0 | 4.68E-04 | 2.09E-05 | 0.003355 |
| DYS480 | 0 | 4.63E-04 | 1.99E-05 | 0.003305 |
| DYS481 | 3 | 0.004454 | 0.001014 | 0.01381 |
| DYS485 | 1 | 0.001363 | 1.57E-04 | 0.006591 |
| DYS487 | 1 | 0.001383 | 1.53E-04 | 0.006378 |
| DYS488 | 0 | 4.66E-04 | 2.07E-05 | 0.003314 |
| DYS490 | 0 | 4.74E-04 | 2.27E-05 | 0.003391 |
| DYS491 | 0 | 4.64E-04 | 2.01E-05 | 0.003213 |
| DYS492 | 0 | 4.68E-04 | 2.00E-05 | 0.003431 |
| DYS494 | 0 | 4.71E-04 | 2.25E-05 | 0.003457 |
| DYS495 | 0 | 4.77E-04 | 2.27E-05 | 0.00342 |
| DYS497 | 1 | 0.001351 | 1.70E-04 | 0.006358 |
| DYS505 | 0 | 4.69E-04 | 2.00E-05 | 0.003256 |
| DYS508 | 2 | 0.002722 | 4.85E-04 | 0.009925 |
| DYS511 | 1 | 0.001367 | 1.52E-04 | 0.00651 |
| DYS522 | 0 | 4.82E-04 | 2.17E-05 | 0.003332 |
| DYS525 | 0 | 4.87E-04 | 2.18E-05 | 0.003343 |
| DYS530 | 0 | 4.79E-04 | 2.16E-05 | 0.003356 |
| DYS531 | 0 | 4.84E-04 | 2.00E-05 | 0.003406 |
| DYS533 | 2 | 0.002731 | 4.86E-04 | 0.009715 |
| DYS537 | 0 | 4.77E-04 | 2.01E-05 | 0.003365 |
| DYS540 | 0 | 4.73E-04 | 2.23E-05 | 0.003331 |
| DYS549 | 1 | 0.001381 | 1.51E-04 | 0.006572 |
| DYS554 | 1 | 0.001393 | 1.64E-04 | 0.006736 |
| DYS556 | 0 | 4.68E-04 | 2.19E-05 | 0.003403 |
| DYS565 | 2 | 0.002761 | 4.96E-04 | 0.009897 |
| DYS567 | 0 | 4.80E-04 | 2.02E-05 | 0.003413 |
| DYS568 | 0 | 4.83E-04 | 2.26E-05 | 0.00338 |
| DYS569 | 0 | 4.84E-04 | 1.99E-05 | 0.003296 |
| DYS570 | 7 | 0.0127 | 0.004839 | 0.02724 |
| DYS572 | 1 | 0.001377 | 1.62E-04 | 0.006533 |
| DYS573 | 2 | 0.002786 | 4.81E-04 | 0.01005 |
| DYS575 | 1 | 0.001407 | 1.66E-04 | 0.006531 |
| DYS576 | 8 | 0.01492 | 0.006175 | 0.02935 |
| DYS578 | 0 | 4.76E-04 | 1.94E-05 | 0.003479 |
| DYS579 | 0 | 4.81E-04 | 2.39E-05 | 0.003286 |
| DYS580 | 0 | 4.69E-04 | 2.06E-05 | 0.003371 |
| DYS583 | 0 | 4.72E-04 | 2.17E-05 | 0.003352 |
| DYS589 | 0 | 4.75E-04 | 2.17E-05 | 0.003304 |
| DYS590 | 0 | 4.83E-04 | 2.08E-05 | 0.003286 |
| DYS594 | 0 | 4.79E-04 | 2.16E-05 | 0.003368 |
| DYS617 | 0 | 4.70E-04 | 1.99E-05 | 0.003222 |
| DYS618 | 0 | 4.61E-04 | 2.31E-05 | 0.003409 |
| DYS636 | 1 | 0.001385 | 1.56E-04 | 0.006328 |
| DYS638 | 1 | 0.001362 | 1.63E-04 | 0.006613 |
| DYS640 | 2 | 0.002787 | 4.95E-04 | 0.01017 |
| DYS641 | 0 | 4.81E-04 | 2.24E-05 | 0.003421 |
| DYS643 | 0 | 4.73E-04 | 2.22E-05 | 0.003388 |
| All 49 Y-STRs | 38 | 7.90E-04 | 2.85E-04 | 0.001537 |
Haplotype resolution was estimated by dividing the number of haplotypes identified in the sample by the total number of samples per global or regional dataset. Evaluation of Y-STR loci according to their contribution towards increasing haplotype resolution was achieved by applying a hill-climbing approach. The marker with the highest variability was selected first. The next marker was then chosen according to the maximal increase in the resolution of the haplotype containing the already-chosen markers and the additional marker. To reduce computing time, individuals that were already distinguished by the chosen markers were excluded from the next calculation. In this way, we obtained the minimal number of markers that provided the maximal increase in haplotype resolution in a given set of individuals. Haplotype resolution was preferred to rank the Y-STR loci according to their ability for maximizing male lineage differentiation as it is a more direct measure for resolving haplotypes than the often used haplotype diversity. However, we would like to point out that simple haplotype resolution as estimated here is sensitive to sample size effects, which shall be considered when interpreting large values from small sample sizes. Mutation rates were estimated using a Bayesian-based approach as described in detail elsewhere [22]. In brief, a hierarchical binomial model was used. Three different MCMC chains were run in parallel for each mutation rate estimation in order to improve chain mixing. Each MCMC chain consisted of 100,000 runs but only the last 50,000 were retained. Of the 50,000 retained simulations per parameter estimation, a thin of 15 was applied to reduce the amount of autocorrelation of close steps. Therefore, a final set of 10,000 simulations was used.
Results and Discussion
Male lineage differentiation in worldwide HGDP-CEPH samples using 67 Y-STRs
Male lineage differentiation was investigated in 590 HGDP-CEPH samples from the H952 set [19] using 67 Y-STRs, and identified 563 different haplotypes. Notably, we found 47 males who were involved in the sharing of 20 haplotypes representing 16 pairs, two trios, one quartet and one quintet of individuals (Table 1). All males who shared a complete 67-locus Y-STR haplotype were sampled from the same respective population. This finding is remarkable as based on the previous analysis of 783 autosomal STRs it was concluded that the H952 set contains no pairs of relatives closer than first cousins, with possible exceptions in the Karitiana and Surui from Brazil [19]. We additionally tested for close relationships between the individuals sharing a complete 67-locus Y-STR haplotype by inspecting the data of 15 autosomal STRs usually applied to forensic human identity testing (PowerPlex 16 System, Promega) that were originally generated for other purposes (PdK in preparation). We found that males who share an identical Y-chromosome differed in their autosomal STR profile by 19 (63.3%) to 28 (93.3%) of the 30 possible alleles (with the exception of the Karitiana and Surui, described below). This autosomal DNA finding excludes close biological relationships and is in agreement with Rosenberg’s conclusions [19]. However, two Karitiana males differed by only 50% of the 30 autosomal STR alleles and three Surui males by only 46.7% or 50%, respectively, indicating that a somewhat close relationship cannot be excluded in these cases also in agreement with Rosenberg [19]; thus, one of the Karitiana and two of the Surui were excluded from further analyses (Table 1). This left a total of 587 males in the global dataset, of which 24 could not be differentiated by the 67 Y-STRs analyzed.
Table 1.
HGDP-CEPH males from the H952 set* with respectively shared 67-locus Y-STR haplotypes and their autosomal STR differences from PowerPlex 16 analysis.
| HGDP-CEPH ID | Population | Subregion | Region | Pairwise allele differences from PowerPlex 16 Analysis** |
|---|---|---|---|---|
| 945 | Yakut | Siberia | East Asia | |
| 969 | Yakut | Siberia | East Asia | 24 (80%) |
|
| ||||
| 952 | Yakut | Siberia | East Asia | |
| 953 | Yakut | Siberia | East Asia | |
| 961 | Yakut | Siberia | East Asia | |
| 960 | Yakut | Siberia | East Asia | |
| 965 | Yakut | Siberia | East Asia | 19 (63.3%) to 26 (86.7%) |
|
| ||||
| 1317 | Lahu | China | East Asia | |
| 1318 | Lahu | China | East Asia | 23 (76.7%) |
|
| ||||
| 1327 | She | China | East Asia | |
| 1330 | She | China | East Asia | 21 (70%) |
|
| ||||
| 1348 | Tu | China | East Asia | |
| 1352 | Tu | China | East Asia | 27 (90%) |
|
| ||||
| 562 | Druze | Israel | Middle East | |
| 580 | Druze | Israel | Middle East | 22 (73.3%) |
|
| ||||
| 616 | Bedouin | Israel | Middle East | |
| 630 | Bedouin | Israel | Middle East | |
| 631 | Bedouin | Israel | Middle East | 20 (66.7) to 23 (76.7%) |
|
| ||||
| 628 | Bedouin | Israel | Middle East | |
| 642 | Bedouin | Israel | Middle East | 20 (66.7%) |
|
| ||||
| 727 | Palestinian | Israel | Middle East | |
| 733 | Palestinian | Israel | Middle East | 23 (76.7%) |
|
| ||||
| 546 | Papuan | New Guinea | Oceania | |
| 547 | Papuan | New Guinea | Oceania | 21 (70%) |
|
| ||||
| 15 | Brahui | Pakistan | South Asia | |
| 45 | Brahui | Pakistan | South Asia | 23 (76.7%) |
|
| ||||
| 109 | Hazara | Pakistan | South Asia | |
| 110 | Hazara | Pakistan | South Asia | 21 (70%) |
|
| ||||
| 116 | Hazara | Pakistan | South Asia | |
| 118 | Hazara | Pakistan | South Asia | 22 (73.3%) |
|
| ||||
| 102 | Hazara | Pakistan | South Asia | |
| 106 | Hazara | Pakistan | South Asia | |
| 121 | Hazara | Pakistan | South Asia | |
| 122 | Hazara | Pakistan | South Asia | 20 (66.7%) to 28 (93.3%) |
|
| ||||
| 267 | Kalash | Pakistan | South Asia | |
| 277 | Kalash | Pakistan | South Asia | 22 (73.3%) |
|
| ||||
| 302 | Kalash | Pakistan | South Asia | |
| 307 | Kalash | Pakistan | South Asia | 25 (83.3%) |
|
| ||||
| 454 | Biaka Pygmies | Central African Republic | Sub-Saharan Africa | |
| 472 | Biaka Pygmies | Central African Republic | Sub-Saharan Africa | 23 (76.7%) |
|
| ||||
| 462 | Mbuti Pygmies | Democratic Republic of Congo | Sub-Saharan Africa | |
| 478 | Mbuti Pygmies | Democratic Republic of Congo | Sub-Saharan Africa | 22 (73.3%) |
|
| ||||
| 998 | Karitiana | Brazil | South America | |
| 1015*** | Karitiana | Brazil | South America | 15 (50%) |
|
| ||||
| 837 | Surui | Brazil | South America | |
| 845*** | Surui | Brazil | South America | |
| 849*** | Surui | Brazil | South America | 14 (46.7%) to 15 (50%) |
see [19],
PowerPlex 16 Analysis consists of 15 STRs with 30 possible alleles,
not considered in further haplotype resolution analyses because close relationship cannot be excluded
Our Y-chromosomal and autosomal STR data together with Rosenberg’s extensive autosomal STR data [19] implies that substantial male relatedness in the respective populations exists that shall be further distant than the level of first cousins. Male-line common ancestors in these populations must have lived sufficiently long ago for the observed autosomal STR diversity to have accumulated, largely by random assortment and recombination, but not so long ago that Y-STR mutations would have accumulated. It also demonstrates that males who are not closely related based on autosomal DNA evidence may nevertheless be difficult to differentiate by Y-chromosomal DNA-analysis, even when large numbers of Y-STRs are used. The occurrence of this phenomenon in 13 of the 51 populations from five of the eight continental regions studied (East Asia, Middle East, Oceania, South Asia and sub-Saharan Africa) indicates that this may not be a rare phenomenon, at least in anthropological samples i.e. samples from indigenous populations of usually small population size. Additional effects that could have caused the observed Y-chromosome sharing may include strong male bottlenecks, preferential mating and polygynie, patrilocality and strongly biased male expansion due to male occupation history and privilege.
Maximizing global and regional Y-STR haplotype resolution
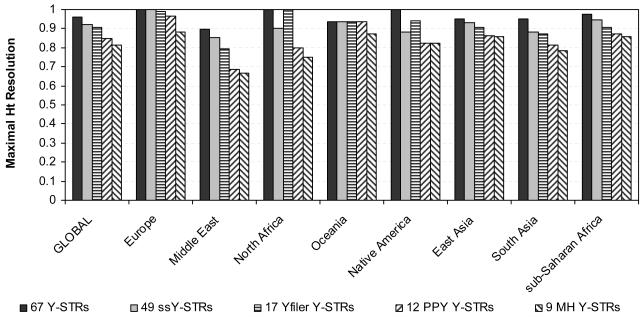
We investigated the maximal and highest possible differentiation of the 587 HGDP males that could be obtained, both on the global and various regional scales, and the minimal number of Y-STRs needed for this (Table 2, Figure 1 and 2, Supplementary Table S2). We considered five marker sets: i) all 67 Y-STRs analysed, ii) 49 rarely-studied ssY-STRs, and for comparative reasons the three commonly-used Y-STR sets iii) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems) (Yfiler), iv) 12 Y-STRs included in the PowerPlex Y® System (Promega) (PPY), and v) 9 Y-STRs comprising the so called Minimal Haplotype (MH). In the analysis of all 67 Y-STRs, the best 25 markers were sufficient to obtain the highest possible haplotype resolution of 95.9% in the global dataset (Table 2), which also marked the maximal resolution in this dataset (see above for why this is lower than 100%). From the 49 ssY-STRs, 25 best markers were enough to obtain the highest possible haplotype resolution of 92.2% in the global dataset (Table 2), 3.7% lower than the maximal value. In comparison, the highest possible haplotype resolution obtainable from the global sample set using Yfiler markers, which was already reached with 15 of the 17 markers (only one of GATAH4, DYS438 and DYS391 was needed to improve resolution), was 90.5%. In particular, this value is 1.7% lower than the resolution obtained with the most informative 25 ssY-STR, and 5.4% lower than the maximal resolution in the global dataset obtained with the best 25 of all 67 markers (although only about half of the number of markers were used with Yfiler). This shows that the male lineage differentiation obtained with the commonly-used Yfiler kit is fairly high, but also that it does not always reflect the maximum resolution, and can be further increased using additional Y-STRs (see below). However, considerably lower resolutions were obtained with the PPY Y-STRs at 84.7% (for which all 12 markers were needed), which is 11.2% lower than maximum in the global dataset, and especially with the MH Y-STRs at 81.3% (for which all 9 markers were needed), which is 14.6% lower than the maximal resolution in the global dataset. This may be expected from the smaller number of markers involved, but does clearly illustrate the limitation of both marker sets, in particular of the Minimal Haplotype traditionally used in forensic and anthropological studies, and the need of supplementation with additional Y-STRs (see below).
Table 2.
Most informative Y-STRs for maximizing global haplotype resolution using the males from the H952 set of the HGDP-CEPH samples (for regional analyses see Supplementary Table S2).
| 67 Y-STRs | 49 ssY-STRs | 49 ssY-STRs suppl. Yfiler* | 49 ssY-STRs suppl. PowerPlex-Y** | 49 ssY-STRs suppl. Min Ht*** | |||||
|---|---|---|---|---|---|---|---|---|---|
| HR | Marker | Marker | HR | Marker | HR | Marker | HR | Marker | HR |
| DYS458 | 0.027 | DYS481 | 0.022 | DYS576 | 0.922 | DYS576 | 0.876 | DYS576 | 0.857 |
| DYS385 | 0.152 | DYS570 | 0.133 | DYS549 | 0.930 | DYS549 | 0.893 | DYS549 | 0.876 |
| DYS481 | 0.455 | DYS576 | 0.382 | DYS533 | 0.935 | DYS533 | 0.903 | DYS533 | 0.889 |
| DYS576 | 0.687 | DYS643 | 0.622 | DYS569 | 0.940 | DYS570 | 0.910 | DYS570 | 0.901 |
| DYS570 | 0.816 | DYS549 | 0.729 | DYS522 | 0.944 | DYS497 | 0.915 | DYS556 | 0.908 |
| DYS389AB | 0.860 | DYS533 | 0.792 | DYS556 | 0.947 | DYS522 | 0.920 | DYS522 | 0.913 |
| DYS439 | 0.888 | DYS522 | 0.828 | DYS487 | 0.949 | DYS569 | 0.925 | DYS569 | 0.918 |
| DYS456 | 0.901 | DYS525 | 0.850 | DYS495 | 0.951 | DYS556 | 0.928 | DYS497 | 0.922 |
| DYS533 | 0.911 | DYS495 | 0.865 | DYS511 | 0.952 | DYS589 | 0.932 | DYS617 | 0.925 |
| DYS549 | 0.920 | DYS572 | 0.876 | DYS531 | 0.954 | DYS487 | 0.934 | DYS589 | 0.928 |
| DYS19 | 0.925 | DYS497 | 0.882 | DYS572 | 0.956 | DYS617 | 0.935 | DYS487 | 0.930 |
| DYS392 | 0.928 | DYS589 | 0.889 | DYS636 | 0.957 | DYS505 | 0.937 | DYS481 | 0.932 |
| DYS390 | 0.932 | DYS511 | 0.894 | DYS5703 | 0.9593 | DYS508 | 0.939 | DYS505 | 0.934 |
| DYS522 | 0.935 | DYS487 | 0.898 | DYS511 | 0.940 | DYS508 | 0.935 | ||
| DYS556 | 0.939 | DYS508 | 0.901 | DYS531 | 0.942 | DYS511 | 0.937 | ||
| DYS569 | 0.942 | DYS540 | 0.905 | DYS572 | 0.944 | DYS531 | 0.939 | ||
| DYS589 | 0.945 | DYS556 | 0.908 | DYS4814 | 0.9454 | DYS537 | 0.940 | ||
| DYS487 | 0.947 | DYS488 | 0.910 | DYS540 | 0.942 | ||||
| DYS393 | 0.949 | DYS505 | 0.911 | DYS572 | 0.944 | ||||
| DYS511 | 0.951 | DYS531 | 0.913 | DYS6365 | 0.9455 | ||||
| DYS531 | 0.952 | DYS537 | 0.915 | ||||||
| DYS572 | 0.954 | DYS565 | 0.917 | ||||||
| DYS636 | 0.956 | DYS568 | 0.918 | ||||||
| DYS448 | 0.957 | DYS569 | 0.920 | ||||||
| DYS3851 | 0.9591 | DYS6362 | 0.9222 | ||||||
HR: haplotype resolution (number of haplotypes divided by number of individuals), ssY-STRs: simple single-copy Y-chromosomal short tandem repeat polymorphisms
AmpFlSTR® Yfiler®: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS348, DYS439, DYS448, DYS456, DYS458, DYS635, and Y-GATA-H4
PowerPlex Y®: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS438, and DYS439
Minimal Haplotype: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS385a/b
or alternatively any one of DYS388,-472,-488,-497,-491,-617,-618,-476,-492,-480,-485,-490,-495,-494,-389CD,-391,-437,- 635,-505,-508,-525,-530,-537,-540,-554,-565,-567,-568,-573,-575,-578,-579,-580,-583,-638,-640,-641,-438,-590,-594,-643, GATA_H4
or alternatively any one of DYS472,-491,-617,-618,-476,-492,-480,-485,-490,-494,-530,-554,-567,-573,-575,-578,-579,-580,- 583,-638,-640,-641,-590,-594
or alternatively any one of DYS472,-488,-497,-491,-617,-618,-476,-492,-480,-481,-485,-490,-494,-505,-508,-525,-530,-537,-540,-554,-565,-567,-568,-573,-575,-578,-579,-580,-589,-583,-638,-640,-641,-590,-594,-643
or alternatively any one of DYS472,-488,-491,-618,-476,-492,-480,-485,-490,-495,-494,-525,-530,-537,-540,-554,-565,-567,-568,-573,-575,-578,-579,-580,-583,-636,-638,-640,-641,-590,-594,-643
or alternatively anyone of DYS472,-488,-491,-618,-476,-492,-480,-485,-490,-495,-494,-525,-530,-554,-565,-567,-568,-573,- 575,-578,-579,-580,-583,-638,-640,-641,-590,-594,-643
Figure 1.
Maximal Y-STR haplotype resolution in global and regional analyses using males from the H952 set of the worldwide HGDP-CEPH samples considering i) 67 Y-STRs, ii) 49 simple single copy Y-STRs, iii) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems), iv) 12 Y-STRs included in the PowerPlex Y® System (Promega), and v) 9 Y-STRs comprising the Minimal Haplotype.
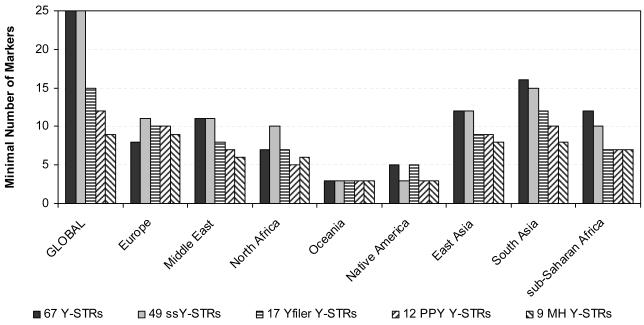
Figure 2.
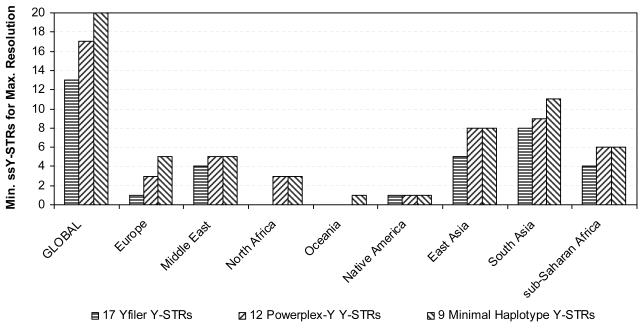
Minimal number of markers needed to reach the maximum Y-STR haplotype resolution in global and regional analyses using males from the H952 set of the worldwide HGDP-CEPH samples considering i) 67 Y-STRs, ii) 49 simple single copy Y-STRs, iii) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems), iv) 12 Y-STRs included in the PowerPlex Y® System (Promega), and v) 9 Y-STRs comprising the Minimal Haplotype.
The separate analyses of the eight geographic regions (Figure 1 and 2, Supplementary Table S2) using the complete set of 67 Y-STRs revealed maximal haplotype resolutions ranging from 89.6% (with 11 markers) in the Middle East to 100% in Europe, North Africa and Native Americans (with eight, seven and five markers, respectively). Complete individualization in the latter three regions were observed because they did not include males with identical 67-locus Y-STR haplotypes (or potentially closely related males were excluded on the basis of autosomal DNA profiling as with Native Americans, see above). For the regional analysis using the 49 ssY-STRs, the highest possible resolution ranged from 85.4% (with 11 markers) in the Middle East, to 100% in Europe (with 11 markers). The Yfiler analysis provided highest possible regional haplotype resolutions ranging from 79.2% in the Middle East (with eight markers) to 100% in North Africa (with seven markers). As expected, and also seen in the global dataset, male differentiation was more limited when using the PPY Y-STRs (68.8% with seven markers in the Middle East to 96.4% in Europe with 10 markers) and even more so with the MH Y-STRs (from 66.6% in the Middle East with six markers to 88.1% in Europe with all nine markers), clearly demonstrating the limitations of these markers sets for male lineage differentiation in egional analysis in addition to global analyses as described above, and the need for supplementing with additional Y-STR markers (see below).
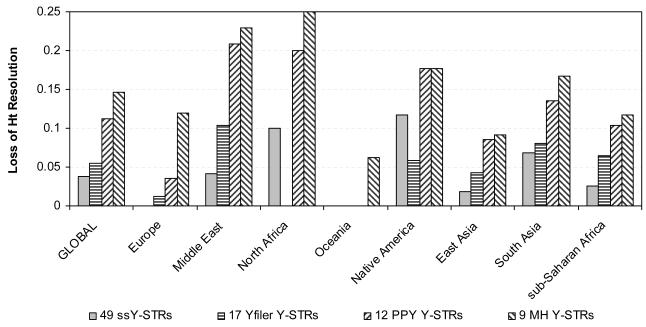
Figure 3 illustrates the loss of resolution with the various Y-STR marker sets in relation to the maximal male lineage differentiation as obtained with the most informative 25 of all 67 Y-STRs studied. Loss of resolution obtained with the minimal set of ssY-STRs ranged from zero (three markers) for Oceania to 11.8% (three markers) for Native Americans; from zero in Oceania (three markers) and North Africa (seven markers) to 10.4% in the Middle East (eight markers) for the minimal set of the Yfiler markers; from zero in Oceania (three markers) to 20.8% in the Middle East (seven markers) for the minimal set of the PPY markers; and from 6.3% in Oceania (three markers) to 25% in North Africa (six markers) for the minimal set of the MH markers. We found that the highest possible haplotype resolution usually decreased with decreasing numbers of markers initially considered in the marker sets (Figure 1), and, consequently, loss of resolution increased (Figure 3), both in the global as well as the regional datasets. This may be expected as more markers considered initially provide more possibilities to find the minimal number with maximal resolution, but again demonstrates the limitation of the commonly-used Y-STR sets. Exceptions were Oceania where the highest possible haplotype resolution was obtained with all minimal markers sets (except for MH); North Africa as well as in Native Americans, where the highest possible haplotype resolution provided by the minimal set of Yfiler markers was higher than that obtained with the minimal set of ssY-STRs; and again in Native Americans where the same maximal haplotype resolution was obtained with the minimal sets of PPY and MH markers. We also found that the minimal number of Y-STRs needed to reach highest possible haplotype resolution usually decreased with decreasing numbers of markers initially included in the marker sets, although some exceptions were observed (Figure 2). Most notable were the findings in Oceania, where only three Y-STRs from all five sets (although not always the same ones) were sufficient to provide maximal possible haplotype resolution, which additionally was identical for four of the five sets. This can be explained by strong bottleneck / founder effects in the regional population history and can be additionally influenced by the low sample size of 16 individuals from the two populations analysed.
Figure 3.
Loss of haplotype resolution in global and regional analyses using males from the H952 set of the worldwide HGDP-CEPH samples in relation to the maximal haplotype resolution from analyzing 67 Y-STRs considering i) 49 simple single copy Y-STRs, ii) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems), iii) 12 Y-STRs included in the PowerPlex Y® System (Promega), and iv) 9 Y-STRs comprising the Minimal Haplotype.
Most informative Y-STRs for maximizing male lineage differentiation
Furthermore, we investigated how many and which of the Y-STRs from the five marker sets were most informative for maximizing male lineage differentiation in the global and regional datasets (Table 2, Figure 2, Supplementary Table S2). From the 67 Y-STRs, three (Oceania) to 26 (South Asia) were needed to reach maximal resolution in the regional datasets, compared with the 25 best markers that provided maximal resolution in the global dataset. Highest possible differentiation was reached with three (Oceania) to 15 (South Asia) ssY-STR markers compared with the 25 best ssY-STRs that achieved highest possible resolution in the global dataset. Of the 17 Yfiler markers, between three (Oceania) and 12 (South Asia) were needed for the highest possible resolution (15 markers were needed in the global analysis), as well as between three (Oceania) and 10 (South Asia) for the 12 PPY markers (all 12 markers were also needed in the global analysis), and between three (Oceania) and all nine (Europe) for the MH markers (all nine markers were also needed in the global analysis).
The results from the global analysis suggest that three rarely-studied ssY-STRs (DYS570, DYS576, and DYS481) partly in addition to two commonly-used Y-STRs (DYS385, DYS458) contributed most to haplotype resolution (Table 2). However, the regional analyses revealed more heterogeneity in the most resolving markers (see Supplementary Table S2), as may be expected. The contribution of DYS385 may be overestimated since the two loci were not separated experimentally; instead, allele-locus assignment was performed artificially by assigning the smaller of the two alleles to one locus and the larger to the other, which has been shown to not always represent the true assignment when experimentally separating both alleles [23]. Moreover, two other ssY-STRs DYS549 and DYS533 (also DYS643 to a certain extent) scored high in the haplotype resolution ranking list of the 49 ssY-STRs, but somewhat less so in the 67 Y-STR analyses (Table 2). The three most informative ssY-STRs DYS570, DYS576, and DYS481 as identified here in the worldwide analysis were also highlighted previously as three of the five Y-STRs that comprised the smallest set of loci in supplementing the nine MH Y-STRs leading to 100% resolution in six regional German, one Dutch and one Turkish population sample respectively [9]. Four highly resolving ssY-STRs DYS481, DYS570, DYS576 and DYS549 from our study were among the 14 Y-STRs that provided 99.5% and 99.7% resolution respectively in an European American and an African American population sample [24]. Not surprisingly, four Y-STRs that showed highest degree of variation in a previous study of 76 worldwide individuals (DYS481, DYS570, DYS576, and DYS643) [18], also showed the highest values in maximizing global haplotype resolution in our study (perhaps somewhat less so for DYS643, see below for the limited value of this marker in supplementing commonly-used Y-STR sets).
Simple single-copy Y-STRs for maximizing resolution of commonly-used Y-STR sets
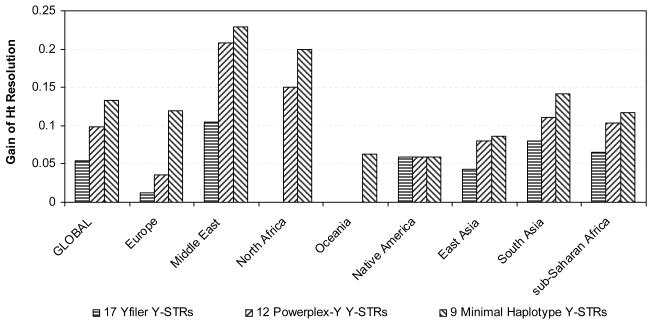
Finally, we extracted the minimal set of ssY-STRs that provided highest possible haplotype resolution when supplementing the three commonly-used Y-STR sets: Yfiler, PPY and MH. This revealed the most informative ssY-STRs that can be used on top of the commonly-used Y-STR sets to further increase the resolution of male lineage resolution in forensic or anthropological cases where males in question could not be differentiated with the conventional Y-STRs. In the global dataset, we achieved the overall maximal haplotype resolution of 95.9% with 13 ssY-STRs on top of the Yfiler markers, an increase of 5.4% (Figures 4 and 5, Table 2). For the PPY and the MH analyses, 17 and 20 ssY-STRs, respectively, were needed to reach the highest possible resolution of 94.5% (which marks 98.5% of the overall maximal resolution), representing an increase of 9.9 and 13.3% respectively (Figure 4 and 5, Table 2). Also in the regional analyses we could considerably increase by supplementing with ssY-STRs the haplotype resolution achieved by all three commonly-used Y-STR sets (except where the maximal value was reached by the original marker sets), mostly only with a few ssY-STR markers (Figure 4 and 5, Supplementary Table S2). Gain of resolution ranged from 1.2% in Europe to 10.4% in the Middle East for the Yfiler supplementary analysis; from 3.6% in Europe to 20.8% in the Middle East for the PPY supplementary analysis; and from 5.9% in Native Americans to 22.9% in the Middle East for the MH supplementary analysis. The number of ssY-STRs required to reach the highest possible haplotype resolution on top of the three commonly-used Y-STR sets ranged from one in Europe to eight in South Asia supplementing Yfiler, from one in Native Americans to nine in South Asia supplementing PPY, and from one in Oceania and Native Americans to 11 in South Asia supplementing MH (Figure 5). The most informative ssY-STR marker to supplement commonly-used Y-STR sets for maximizing male lineage differentiation was DYS576 followed by DYS549, DYS533 and DYS570 in the global analyses of all three commonly-used marker sets (Table 2), whereas other ssY-STRs were more informative in some of the various regional analyses (Supplementary Table S2).
Figure 4.
Gain of haplotype resolution obtained with simple single copy Y-STRs when supplementing commonly-used Y-STR sets in global and regional analyses using males from the H952 set of the worldwide HGDP-CEPH samples relative to the resolution provided by the initial analysis of commonly-used Y-STR sets: i) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems), ii) 12 Y-STRs included in the PowerPlex Y® System (Promega), and iii) 9 Y-STRs comprising the Minimal Haplotype.
Figure 5.
Minimal number of simple single copy Y-STRs needed to reach the maximum haplotype resolution when supplementing commonly-used Y-STR sets in global and regional analyses using males from the H952 set of the worldwide HGDP-CEPH samples: i) 17 Y-STRs included in the AmpFlSTR® Yfiler® PCR Amplification Kit (Applied Biosystems), ii) 12 Y-STRs included in the PowerPlex Y® System (Promega), and iii) 9 Y-STRs comprising the Minimal Haplotype.
Mutation rates of 49 ssY-STRs
To understand the discriminatory power of ssY-STRs and to stimulate their application to forensic and evolutionary studies, we estimated the mutation rates of the 49 rarely-studied ssY-STRs for which no mutation data were previously available. For this purpose, 104 members of 28 deep-rooted pedigrees covering 403 meiotic transfers per locus were genotyped. Bayesian-based estimates of median locus-specific mutation rates ranged from 4.8×10−4 (95% credible interval: 2×10−5-3×10−3) for 31 loci where no mutation was observed, to 1.3×10−2 (95% CI: 4.8×10−3-2.7×10−2) for DYS570 (7/403) and 1.5×10−2 (95% CI: 6.2×10−3-2.9×10−2) for DYS576 (8/403) (Table 3). The median mutation rate across all 49 ssY-STRs was 7.9×10−4 (95% CI: 2.8×10−4-1.5×10−3), which is somewhat smaller than the median mutation rate of 2.2×10−3 (95% CI: 1.9×10−3-2.6×10−3) previously estimated across 16 Yfiler Y-STRs in a comprehensive family-based study based on more than 135,000 meiotic transfers [22]. This difference may simply reflect stochastic fluctuation caused by the small number of meiotic transfers covered in this preliminary mutation analysis. Alternatively, this finding may be influenced by the ascertainment of the ssY-STRs used here, which was less biased towards highly-variable ones than for the commonly-used Y-STRs. More pedigree and/or family data are needed to establish a reliable estimate of the mutation rates and characteristics of these ssY-STRs. However, the mutation rates estimated here for DYS570 and DYS576 at >1% are the highest reported for any Y-STR locus studied thus far [22], and confirmation in a large number of father-son pairs is currently underway (M.K. et al. in preparation). Notably, the three markers with the highest mutation rate estimates here: DYS570, DYS576 and DYS481 (0.4×10−2; 95% CI: 1×10−3 −1.4×10−2) were also the three ssY-STRs that contributed most to the haplotype resolution in the global and some regional analyses (Table 2, Supplementary Table S2); and additionally, DYS576 (and to a lesser extent also DYS570) was most informative in supplementing commonly-used Y-STR sets in global analyses (Table 2, Supplementary Table S2). Also notable, although no mutation was observed among the 403 meiotic transfers considered here for DYS643, this locus was found to be similarly polymorphic as DYS576, DYS570, and DYS481 in a previous study of 76 worldwide individuals [18], and also was the number four in the list of the 25 most informative ssY-STRs (but did not appear as highly informative when supplementing commonly-used Y-STR sets, see Table1).
Conclusions
Simple single-copy Y-STRs have several advantages since their simple repetitive structure and unique copy number make fragment length analysis and allele-locus assignment unequivocal. The ability to provide very high resolution male lineage differentiation when used as new sets, or to markedly improve the haplotype resolution achieved by the three commonly-used Y-STR sets when used as supplementary sets, provide clear arguments in favour of using ssY-STRs in forensic and anthropological studies. This is especially so when performing Y-STR typing for forensic applications in populations with reduced Y-chromosome diversity, or for male relative differentiation in any population. Our results provide recommendations on how to supplement commonly-used Y-STR marker sets with additional ssY-STRs for maximizing male lineage differentiation, with DYS576, DYS570, DYS549 and DYS533 being suggested as most promising markers. Alternatively, our study may lead to the application of new sets of ssY-STRs avoiding complications introduced by complex and multi-copy Y-STRs, in particular the best 25 ssY-STR markers identified here that achieved 96.1% of the maximal haplotype resolution in the global dataset, with DYS570, DYS576, DYS481, DYS549, DYS533, and DYS643 recommended as most promising markers for this. The mutation data of the ssY-STRs generated here suggest that many of the ssY-STRs studied may mutate somewhat slower compared to most of the currently-used Y-STRs, providing advantages for paternity testing and anthropological studies. However, two ssY-STRs (DYS570 and DYS576) are suggestive for having a higher than usual mutation rate, providing advantages in resolving male lineages including differentiation of male relatives in forensic and anthropological studies.
Supplementary Material
Acknowledgements
We thank the original donors and CEPH for providing the HGDP DNA samples. We are grateful to the contributing members of the deep-rooting pedigrees. We thank Si-Keun Lim for providing information on genotyping assays before publication. Kaye Ballantyne is acknowledged for useful comments on the manuscript. AW was supported by the Deutsche Forschungsgemeinschaft (SFB 680 to MK), QY by a Joint Project from the Natural Scientific Foundation of China and the Royal Society, as well as YX and CTS were by The Wellcome Trust. This study was supported by funds from the Netherlands Forensic Institute (to MK) and received additional support by a grant from the Netherlands Genomics Initiative / Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (to MK and PdK).
References
- [1].Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, Herrmann S, Herzog B, Hidding M, Honda K, Jobling M, Krawczak M, Leim K, Meuser S, Meyer E, Oesterreich W, Pandya A, Parson W, Penacino G, Perez-Lezaun A, Piccinini A, Prinz M, Schmitt C, Roewer L, et al. Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med. 1997;110:125–133. 141–129. doi: 10.1007/s004140050051. [DOI] [PubMed] [Google Scholar]
- [2].Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- [3].de Knijff P. Messages through bottlenecks: on the combined use of slow and fast evolving polymorphic markers on the human Y chromosome. Am J Hum Genet. 2000;67:1055–1061. doi: 10.1016/s0002-9297(07)62935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kayser M, Vermeulen M, Knoblauch H, Schuster H, Krawczak M, Roewer L. Relating two deep-rooted pedigrees from Central Germany by high-resolution Y-STR haplotyping. Forensic Sci Int Genet. 2007;1:125–128. doi: 10.1016/j.fsigen.2007.02.004. [DOI] [PubMed] [Google Scholar]
- [5].Kayser M, Krawczak M, Excoffier L, Dieltjes P, Corach D, Pascali V, Gehrig C, Bernini LF, Jespersen J, Bakker E, Roewer L, de Knijff P. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet. 2001;68:990–1018. doi: 10.1086/319510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].de Knijff P, Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, Herrmann S, Herzog B, Hidding M, Honda K, Jobling M, Krawczak M, Leim K, Meuser S, Meyer E, Oesterreich W, Pandya A, Parson W, Penacino G, Perez-Lezaun A, Piccinini A, Prinz M, Roewer L, et al. Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Legal Med. 1997;110:134–149. doi: 10.1007/s004140050052. [DOI] [PubMed] [Google Scholar]
- [7].Kayser M, Lao O, Anslinger K, Augustin C, Bargel G, Edelmann J, Elias S, Heinrich M, Henke J, Henke L, Hohoff C, Illing A, Jonkisz A, Kuzniar P, Lebioda A, Lessig R, Lewicki S, Maciejewska A, Monies DM, Pawlowski R, Poetsch M, Schmid D, Schmidt U, Schneider PM, Stradmann-Bellinghausen B, Szibor R, Wegener R, Wozniak M, Zoledziewska M, Roewer L, Dobosz T, Ploski R. Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis. Hum Genet. 2005;117:428–443. doi: 10.1007/s00439-005-1333-9. [DOI] [PubMed] [Google Scholar]
- [8].Roewer L, Croucher PJ, Willuweit S, Lu TT, Kayser M, Lessig R, de Knijff P, Jobling MA, Tyler-Smith C, Krawczak M. Signature of recent historical events in the European Y-chromosomal STR haplotype distribution. Hum Genet. 2005;116:279–291. doi: 10.1007/s00439-004-1201-z. [DOI] [PubMed] [Google Scholar]
- [9].Rodig H, Roewer L, Gross A, Richter T, de Knijff P, Kayser M, Brabetz W. Evaluation of haplotype discrimination capacity of 35 Y-chromosomal short tandem repeat loci. Forensic Sci Int. 2008;174:182–188. doi: 10.1016/j.forsciint.2007.04.223. [DOI] [PubMed] [Google Scholar]
- [10].Mohyuddin A, Ayub Q, Qamar R, Zerjal T, Helgason A, Mehdi SQ, Tyler-Smith C. Y-chromosomal STR haplotypes in Pakistani populations. Forensic Sci Int. 2001;118:141–146. doi: 10.1016/s0379-0738(01)00382-6. [DOI] [PubMed] [Google Scholar]
- [11].Hedman M, Pimenoff V, Lukka M, Sistonen P, Sajantila A. Analysis of 16 Y STR loci in the Finnish population reveals a local reduction in the diversity of male lineages. Forensic Sci Int. 2004;142:37–43. doi: 10.1016/j.forsciint.2003.07.003. [DOI] [PubMed] [Google Scholar]
- [12].Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA, Moyse-Faurie C, Rutledge RB, Schiefenhoevel W, Gil D, Lin AA, Underhill PA, Oefner PJ, Trent RJ, Stoneking M. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- [13].Kayser M, Brauer S, Weiss G, Schiefenhovel W, Underhill P, Shen P, Oefner P, Tommaseo-Ponzetta M, Stoneking M. Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet. 2003;72:281–302. doi: 10.1086/346065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, Qamar R, Ayub Q, Mohyuddin A, Fu S, Li P, Yuldasheva N, Ruzibakiev R, Xu J, Shu Q, Du R, Yang H, Hurles ME, Robinson E, Gerelsaikhan T, Dashnyam B, Mehdi SQ, Tyler-Smith C. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xue Y, Zerjal T, Bao W, Zhu S, Lim SK, Shu Q, Xu J, Du R, Fu S, Li P, Yang H, Tyler-Smith C. Recent spread of a Y-chromosomal lineage in northern China and Mongolia. Am J Hum Genet. 2005;77:1112–1116. doi: 10.1086/498583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kayser M, Kittler R, Erler A, Hedman M, Lee AC, Mohyuddin A, Mehdi SQ, Rosser Z, Stoneking M, Jobling MA, Sajantila A, Tyler-Smith C. A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet. 2004;74:1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- [18].Lim SK, Xue Y, Parkin EJ, Tyler-Smith C. Variation of 52 new Y-STR loci in the Y Chromosome Consortium worldwide panel of 76 diverse individuals. Int J Legal Med. 2007;121:124–127. doi: 10.1007/s00414-006-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- [20].Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P. Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet. 1997;6:799–803. doi: 10.1093/hmg/6.5.799. [DOI] [PubMed] [Google Scholar]
- [21].Wang QJ, Lu CY, Li N, Rao SQ, Shi YB, Han DY, Li X, Cao JY, Yu LM, Li QZ, Guan MX, Yang WY, Shen Y. Y-linked inheritance of non-syndromic hearing impairment in a large Chinese family. J Med Genet. 2004;41:e80. doi: 10.1136/jmg.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Goedbloed M, Vermeulen M, Fang RN, Lembring M, Wollstein A, Lao O, Brauer S, Krüger C, Roewer L, Lessig R, Ploski R, Dobosz T, Henke L, Henke J, Furtado MR, Kayser M. Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR® Yfiler® PCR amplification kit. Int J Legal Med. 2008 doi: 10.1007/s00414-009-0342-y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kittler R, Erler A, Brauer S, Stoneking M, Kayser M. Apparent intrachromosomal exchange on the human Y chromosome explained by population history. Eur J Hum Genet. 2003;11:304–314. doi: 10.1038/sj.ejhg.5200960. [DOI] [PubMed] [Google Scholar]
- [24].Hanson EK, Ballantyne J. An ultra-high discrimination Y chromosome short tandem repeat multiplex DNA typing system. PLoS ONE. 2007;2:e688. doi: 10.1371/journal.pone.0000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.