Abstract
The most significant advance in the medical management of HIV-1 infection has been the treatment of patients with antiviral drugs, which can suppress HIV-1 replication to undetectable levels. The discovery of HIV-1 as the causative agent of AIDS together with an ever-increasing understanding of the virus replication cycle have been instrumental in this effort by providing researchers with the knowledge and tools required to prosecute drug discovery efforts focused on targeted inhibition with specific pharmacological agents. To date, an arsenal of 24 Food and Drug Administration (FDA)-approved drugs are available for treatment of HIV-1 infections. These drugs are distributed into six distinct classes based on their molecular mechanism and resistance profiles: (1) nucleoside-analog reverse transcriptase inhibitors (NNRTIs), (2) non–nucleoside reverse transcriptase inhibitors (NNRTIs), (3) integrase inhibitors, (4) protease inhibitors (PIs), (5) fusion inhibitors, and (6) coreceptor antagonists. In this article, we will review the basic principles of antiretroviral drug therapy, the mode of drug action, and the factors leading to treatment failure (i.e., drug resistance).
Highly active antiretroviral therapy (HAART) dramatically suppresses viral replication. It uses three antiretroviral agents directed against at least two distinct molecular targets and may forestall the evolution of drug resistance.
BASIC PRINCIPLES OF ANTIRETROVIRAL THERAPY
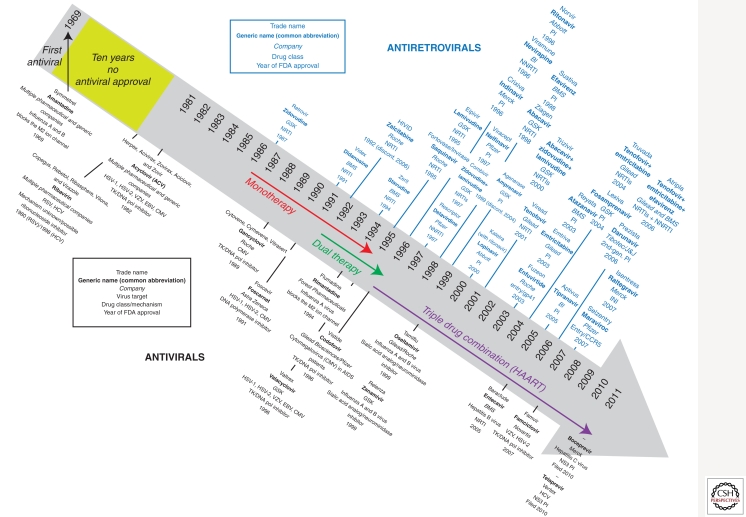
Before 1996, few antiretroviral treatment options for HIV-1 infection existed. The clinical management of HIV-1 largely consisted of prophylaxis against common opportunistic pathogens and managing AIDS-related illnesses. The treatment of HIV-1 infection was revolutionized in the mid-1990s by the development of inhibitors of the reverse transcriptase and protease, two of three essential enzymes of HIV-1, and the introduction of drug regimens that combined these agents to enhance the overall efficacy and durability of therapy. A timeline of antiretroviral drug development and approval for human use is described in Figure 1.
Figure 1.
Timeline for FDA approval for current antiviral and antiretroviral drugs.
Since the first HIV-1 specific antiviral drugs were given as monotherapy in the early 1990s, the standard of HIV-1 care evolved to include the administration of a cocktail or combination of antiretroviral agents (ARVs). The advent of combination therapy, also known as HAART, for the treatment of HIV-1 infection was seminal in reducing the morbidity and mortality associated with HIV-1 infection and AIDS (Collier et al. 1996; D’Aquila et al. 1996; Staszewski et al. 1996). Combination antiretroviral therapy dramatically suppresses viral replication and reduces the plasma HIV-1 viral load (vLoad) to below the limits of detection of the most sensitive clinical assays (<50 RNA copies/mL) resulting in a significant reconstitution of the immune system (Autran et al. 1997; Komanduri et al. 1998; Lederman et al. 1998;) as measured by an increase in circulating CD4+ T-lymphocytes. Importantly, combination therapy using three antiretroviral agents directed against at least two distinct molecular targets is the underlying basis for forestalling the evolution drug resistance.
In an untreated individual, on average there are 104–105 or more HIV-1 particles per mL of plasma, which turn over at a rate of ∼1010/d (Ho et al. 1995; Wei et al. 1995; Perelson et al. 1996). Owing to the error-prone reverse transcription process, it is estimated that one mutation is introduced for every 1000–10,000 nucleotides synthesized (Mansky and Temin 1995; O’Neil et al. 2002; Abram et al. 2010). As the HIV-1 genome is ∼10,000 nucleotides in length, one to 10 mutations may be generated in each viral genome with every replication cycle. With this enormous potential for generating genetic diversity, HIV-1 variants with reduced susceptibility to any one or two drugs will often preexist in the viral quasispecies before initiating therapy (Coffin 1995). The success of HAART results in part from using drug combinations that decrease the probability of selecting virus clones (from an intrapatient HIV-1 population) bearing multiple mutations and conferring resistance to a three-antiretroviral-drug regimen.
Given the rate of HIV-1 turnover and the size of the virus population, mathematical modeling studies have suggested that any combinations in which at least three mutations are required should provide durable inhibition (Frost and McLean 1994; Coffin 1995; Nowak et al. 1997; Stengel 2008). In the simplest interpretation of these models, three drug combinations should be more advantageous than two drug regimens, and in fact, this was the precedent established in early clinical trials of combination antiretroviral therapy. However, this interpretation assumes that all drugs have equal activity, that they require the same number of mutations to engender resistance, and that resistance mutations impact viral replication capacity or viral fitness to a similar degree. Trial and error with early antiretroviral agents helped to establish the basic principles for effective drug combinations in HAART. Since these early days, therapies have evolved, with the introduction of newer drugs with greater potency and higher barriers to the development of resistance. Moreover, some antiretroviral agents have been shown to select for mutations which are either incompatible with or engender hypersensitivity to other antiretroviral drugs, suggesting certain ARVs may offer an advantage with respect to resistance barrier when used in the context of specific combinations (Larder et al. 1995; Kempf et al. 1997; Hsu et al. 1998). Therefore, whether HIV-1 treatment can be simplified to two or even one potent drug(s) remains an open question that can only be answered with future clinical studies.
In 2010, HIV-1 treatment guidelines in the United States and European Union recommend the initiation of HAART with three fully active antiretroviral agents when CD4 cells in peripheral blood decline to 350 per cubic mm, a stage at which viral levels can often reach 10,000–100,000 copies per mL (as measured by RNA in the blood) (see http://aidsinfo.nih.gov/Guidelines/). With proper adherence, HAART can suppress viral replication for decades, dramatically increasing the life expectancy of the HIV-infected individual. However, HAART alone cannot eliminate HIV-1 infection. HIV-1 is a chronic infection for which there is currently no cure—the prospect of maintaining therapy for the lifetime of a patient presents major challenges. The potential for persistent viral replication in compartments and reservoirs may continue to drive pathogenic disease processes (Finzi et al. 1997, 1999). The effect of therapy can be impaired by nonadherence, poor drug tolerability, and drug interactions among antiretroviral agents and other medications that decrease optimal drug levels. Each of these can lead to virologic failure and the evolution of drug resistance.
For all antiretroviral drug classes, drug resistance has been documented in patients failing therapy as well as in therapy-naïve patients infected with transmitted, drug-resistant viruses. Resistance testing is therefore recommended before initiating HAART in therapy-naïve patients as well as when reoptimizing antiretroviral therapy after treatment failure. Given the number of agents and distinct classes of antiretroviral drugs available today, most patients, even those with a history of failure, can be successfully treated. However, as the virus continues to evolve and escape, with even the most effective therapies, new HIV-1 treatments will always be needed.
THE HIV REPLICATION CYCLE AND DRUG TARGETS
Antiretroviral agents for the treatment of HIV-1 are a relatively new addition to the armamentarium of antiviral drugs. In the 1960s, amantadine and rimantidine were the first approved antiviral drugs for treatment of a human influenza virus infection (Davies et al. 1964; Wingfield et al. 1969), but more than 20 years passed before the elucidation of their mechanism of action (Hay et al. 1985). With the advent of modern molecular biology, such serendipitous approaches to antiviral drug discovery have been largely replaced by mechanistic-based approaches, which include (1) high throughput compound screens with virus-specific replication or enzymatic assays, (2) optimization of inhibitors using lead compounds based on homologous enzymes or targets, and/or (3) rational drug design modeled on the structures of viral proteins. These methods together with advances in the corresponding enabling technologies greatly accelerated the development of antiretroviral drugs in the early 1990s. The highly divergent evolution of HIV-1 genes from the human host provided the basis for implementing targeted screening efforts and/or designing and optimizing inhibitors with minimal off-target activities, thus capitalizing on these technological advances. A full timeline in the development of antiviral and antiretroviral drugs for human use is described in Figure 1.
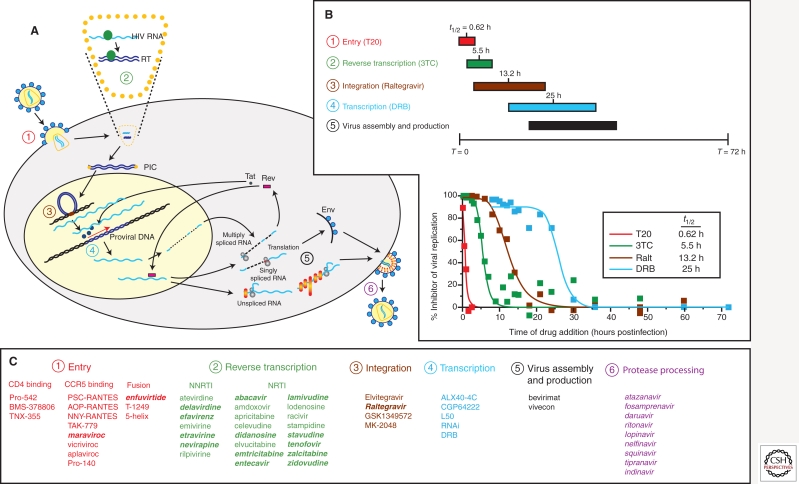
Whereas the HIV-1 life cycle presents many potential opportunities for therapeutic intervention, only a few have been exploited. The replication scheme of HIV-1 is shown in Figure 2, marked with the steps blocked by approved inhibitors (numbers in panel 2A). A timing of the retroviral lifecycle is described in panel B based on the specific time window of inhibition by a specific drug class. In panel 2C, the inhibitors in development (normal text) or FDA approved (italic/bold text) are listed by inhibition of a specific retroviral replication event. The first step in the HIV-1 replication cycle, viral entry (Doms and Wilen 2011), is the target for several classes of antiretroviral agents: attachment inhibitors, chemokine receptor antagonists, and fusion inhibitors. The HIV-1 envelope gp120/gp41 has affinity for the CD4 receptor and directs HIV-1 to CD4+ immune cells (Dalgleish et al. 1984; Klatzmann et al. 1984). Interaction of the gp120 subunit of the HIV-1 envelope with CD4 is followed by binding to an additional coreceptor, either the CC chemokine receptor CCR5 or the CXC chemokine receptor CXCR4 (Alkhatib et al. 1996; Deng et al. 1996; Doranz et al. 1996; Feng et al. 1996). The disposition of these coreceptors on the surface of lymphocytes and monocyte/macrophages, and coreceptor recognition by the viral envelope, are major determinants of tropism for different cell types. These sequential receptor-binding events trigger conformational changes in the HIV-1 envelope, exposing a hydrophobic domain on gp41 that mediates fusion with the cellular membrane. The entire entry process is completed within 1 h of virus contact with the cell (Fig. 2B). Gp120 and CD4 are targets for small-molecule and antibody-based attachment inhibitors BMS-378806 and TNX-355, each of which have shown some clinical promise, although neither is approved for use in HIV-1 patients (Reimann et al. 1997; Lin et al. 2003; Kuritzkes et al. 2004). BMS-378806 binds to a pocket on gp120 important for binding CD4 and alters the conformation of the envelope protein such that it cannot recognize CD4 (Lin et al. 2003). TNX-355 is a humanized anti-CD4 monoclonal antibody that binds to CD4 and inhibits HIV-1 envelope docking, but does not inhibit CD4 function in immunological contexts (Reimann et al. 1997). Gp41 and the coreceptor CCR5 are the targets for the two approved entry agents that will be discussed in more detail below: the peptide-based fusion inhibitor, fuzeon, and the small-molecule CCR5 chemokine receptor antagonist, maraviroc.
Figure 2.
Identifying distinct steps in HIV-1 life cycle as potential or current target for antiretroviral drugs. (A) Schematic of the HIV-1 life cycle in a susceptible CD4+ cell. (B) Time frame for antiretroviral drug action during a single-cycle HIV-1 replication assay. In this experiment, HIV-1 inhibitors are added following a synchronized inhibition. The addition of drug following the HIV-1 replication step targeted by the drug will result in a lack of inhibition. The time window of drug inhibition provides an estimate for the time required for these replication steps. For example, T30 or enfuvirtide (T20) only inhibits within 1–2 h of infection, whereas lamivudine (3TC) inhibits within a 2- to 10-h time frame, which coincides with reverse transcription. (C) Preclinical, abandoned (normal text), or FDA-approved (bold italic text) inhibitors are listed in relation to specificity of action and drug target.
Viral entry and fusion of the HIV-1 envelope with the host cell membrane allow for uncoating of the viral core and initiate a slow dissolution process that maintains protection of the viral RNA genome while permitting access to deoxyribonucleoside triphosphates (dNTPs) necessary for reverse transcription and proviral DNA synthesis (Fig. 1). Reverse transcription is a process extending over the next 10 h of infection (Fig. 2A,B). Reverse transcriptase (RT) was the first HIV-1 enzyme to be exploited for antiretroviral drug discovery (Fig. 1). RT is a multifunctional enzyme with RNA-dependent DNA polymerase, RNase-H, and DNA-dependent DNA polymerase activities, all of which are required to convert the single-stranded HIV-1 viral RNA into double-stranded DNA (Hughes and Hu 2011). RT is the target for two distinct classes of antiretroviral agents: the NRTIs (Fig. 2C), which are analogs of native nucleoside substrates, and the NNRTIs (Fig. 2C), which bind to a noncatalytic allosteric pocket on the enzyme. Together, the 12 licensed agents in these two classes account for the nearly half of all approved antiretroviral drugs. Although the NRTIs and NNRTIs differ with respect to their site of interaction on the enzyme and molecular mechanism, both affect the DNA polymerization activity of the enzyme and block the generation of full-length viral DNA.
The completion of reverse transcription is required to form the viral preintegration complex, or PIC. The PIC, comprised of viral as well as cellular components, is transported to the nucleus where the second essential HIV-1 enzyme, integrase, catalyzes the integration of the viral DNA with the host DNA (Craigie and Bushman 2011). Integrase orchestrates three sequence-specific events required for integration, assembly with the viral DNA, endonucleolytic processing of the 3′ ends of the viral DNA, and strand transfer or joining of the viral and cellular DNA. In the context of HIV-1 infection, the process occurs in a stepwise manner, with the rate-limiting event being strand transfer and the stable integration of the viral genome into the human chromosome occurring within the first 15–20 h of infection (Fig. 2B). The newest class of approved ARVs, integrase inhibitors (INIs or InSTIs) (Fig. 2C), specifically inhibit strand transfer and block integration of the HIV-1 DNA into the cellular DNA.
Integration of the HIV-1 DNA is required to maintain the viral DNA in the infected cell and is essential for expression of HIV-1 mRNA and viral RNA. Following integration, the cellular machinery can initiate transcription; however, transcript elongation requires binding of the HIV-1 regulatory protein Tat to the HIV-1 RNA element (TAR) (Karn and Stoltzfus 2011). This mechanism is unique to HIV-1 and is thus considered a highly desirable therapeutic target. A variety of candidate small-molecule inhibitors of either HIV transcription, or more specifically, the Tat–TAR interaction, have been identified during the last 15 yrs (Fig. 2A,C, section 4) (Hsu et al. 1991; Cupelli and Hsu 1995; Hamy et al. 1997; Hwang et al. 2003). Unfortunately, none of these compounds were sufficiently potent and/or selective to progress beyond phase I clinical trials. Recent reports describe a new cyclic Tat peptidomimetic that binds to TAR with high affinity and shows broad and potent HIV-1 inhibition (Davidson et al. 2009; Lalonde 2011). Surprisingly, this drug inhibits both HIV-1 reverse transcription and Tat-mediated mRNA transcription (Lalonde 2011).
The assembly and maturation of HIV-1 on the inner plasma membrane is also an active area for drug discovery. Inhibitors such as betulinic acid have been shown to block HIV-1 maturation by interacting with the viral capsid (Fig. 2A,C, section 5) (Fujioka et al. 1994; Li et al. 2003). Although a promising new mechanism of action, insufficient antiviral activity precluded the development beyond early phase clinical trials (Smith et al. 2007).
The context of the HIV-1 life cycle, the final class of approved ARVs, is the HIV-1 PIs. PIs block proteolysis of the viral polyprotein, a step required for the production of infectious viral particles (Sundquist and Kräusslich 2011). PIs are among the most potent agents developed to date, but are large, peptidelike compounds that generally require the coadministration of a “boosting” agent to inhibit their metabolism and enhance drug levels. Therefore, PI-containing regimens contain a fourth drug, albeit one that does not directly contribute to overall antiviral activity. To date, ritonavir (RTV) is the only boosting agent or pharmacokinetic enhancer (PKE) available for use (Kempf et al. 1997; Hsu et al. 1998), although other compounds are in early stages of clinical development.
This description of the HIV-1 replication cycle (Fig. 2) provides a cursory overview of the most advanced antiretroviral drug targets with a focus on the approved agents that will be covered in more detail below. However, it should be noted that nearly all viral processes that are distinct from the cellular life cycle are potentially suitable for screening/designing inhibitors. Enhancing or modulating the activities of cellular restriction factors (Malim and Bieniasz 2011) could also potentially provide an approach to inhibiting HIV-1 replication and/or modulate pathogenesis and transmission, but this topic is not covered further here.
NUCLEOSIDE/NUCLEOTIDE REVERSE TRANSCRIPTASE INHIBITORS
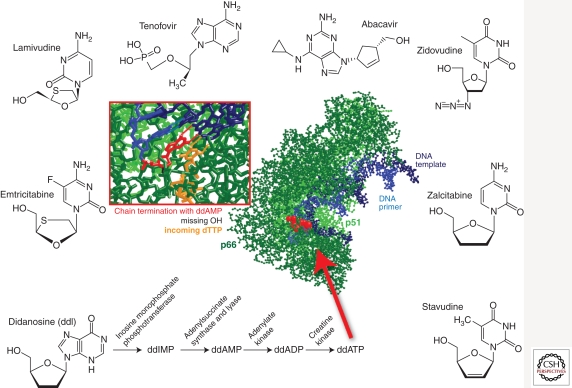
NRTIs were the first class of drugs to be approved by the FDA (Fig. 1) (Young 1988). NRTIs are administered as prodrugs, which require host cell entry and phosphorylation (Mitsuya et al. 1985; Furman et al. 1986; Mitsuya and Broder 1986; St Clair et al. 1987; Hart et al. 1992) by cellular kinases before enacting an antiviral effect (Fig. 3). Lack of a 3′-hydroxyl group at the sugar (2′-deoxyribosyl) moiety of the NRTIs prevents the formation of a 3′-5′-phosphodiester bond between the NRTIs and incoming 5′-nucleoside triphosphates, resulting in termination of the growing viral DNA chain. Chain termination can occur during RNA-dependent DNA or DNA-dependent DNA synthesis, inhibiting production of either the (−) or (+) strands of the HIV-1 proviral DNA (Cheng et al. 1987; Balzarini et al. 1989; Richman 2001). Currently, there are eight FDA-approved NRTIs: abacavir (ABC, Ziagen), didanosine (ddI, Videx), emtricitabine (FTC, Emtriva), lamivudine (3TC, Epivir), stavudine (d4T, Zerit), zalcitabine (ddC, Hivid), zidovudine (AZT, Retrovir), and Tenofovir disoprovil fumarate (TDF, Viread), a nucleotide RT inhibitor (Fig. 3).
Figure 3.
Nucleos(t)ide reverse transcriptase inhibitors and X-ray crystal structure of HIV-1 RT in complex with DNA primer/template chain terminated with ddAMP and with an incoming dTTP. The cartoon of the crystal structure data was adapted from coordinates deposited by Huang et al. (1998) (1RTD).
As with all antiretroviral therapies, treatment with any of these agents often results in the emergence of HIV-1 strains with reduced drug susceptibility. Resistance to NRTIs is mediated by two mechanisms: ATP-dependent pyrophosphorolysis, which is the removal of NRTIs from the 3′ end of the nascent chain, and reversal of chain termination (Arion et al. 1998; Meyer et al. 1999; Boyer et al. 2001) and increased discrimination between the native deoxyribonucleotide substrate and the inhibitor. NRTI mutations occur in RT and are classified as nucleoside/nucleotide associated mutations (NAMs) or thymidine analog mutations (TAMs). TAMs promote pyrophosphorolysis and are involved in the excision of AZT and d4T (Arion et al. 1998; Meyer et al. 2002; Naeger et al. 2002). TAM amino acid changes in HIV-1 RT include two distinct pathways: the TAM1 pathway (M41L, L210W, T215Y, and occasionally D67N) and the TAM2 pathway (D67N, K70R, T215F, and 219E/Q) (Larder and Kemp 1989; Boucher et al. 1992; Kellam et al. 1992; Harrigan et al. 1996; Bacheler et al. 2001; Marcelin et al. 2004; Yahi et al. 2005).
A second mechanism of NRTI resistance is the prevention of NRTI incorporation into the nascent chain. Mutations associated with this mechanism include the M184V/I and the K65R. The M184V mutation emerges with 3TC or FTC therapy (Schinazi et al. 1993; Quan et al. 1996), whereas treatment with Tenofovir, ddC, ddI, d4T, and ABC can select K65R (Wainberg et al. 1999; Margot et al. 2002; Garcia-Lerma et al. 2003; Shehu-Xhilaga et al. 2005). In general, K65R rarely emerges in patients receiving any AZT-containing regimen because this mutation is phenotypically antagonistic to the TAMs (Parikh et al. 2006; White et al. 2006). M184V restores Tenofovir susceptibility in the presence of K65R (Deval et al. 2004), thus K65R viruses are also infrequent in patients on Tenofivir who fail 3TC or emtricitabine (FTC) with M184V.
Many primary and secondary NRTI mutations (or combinations of these) have been shown to decrease RT function and viral replicative fitness (Quinones-Mateu and Arts 2002, 2006). Although several studies have suggested a potential for a clinical benefit associated with reduced replicative fitness of NRTI-resistant variants, it is important to note that additional mutations can accumulate in the presence of ongoing treatment resulting in higher levels of resistance. The loss in replicative fitness owing to drug resistance mutations (in the absence of drug) can also be compensated by accumulating secondary mutations.
NON–NUCLEOSIDE REVERSE TRANSCRIPTASE INHIBITORS
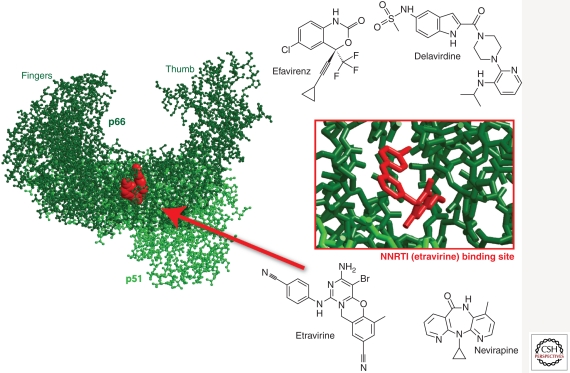
NNRTIs inhibit HIV-1 RT by binding and inducing the formation of a hydrophobic pocket proximal to, but not overlapping the active site (Fig. 4) (Kohlstaedt et al. 1992; Tantillo et al. 1994). The binding of NNRTIs changes the spatial conformation of the substrate-binding site and reduces polymerase activity (Kohlstaedt et al. 1992; Spence et al. 1995). The NNRTI-binding pocket only exists in the presence of NNRTIs (Rodgers et al. 1995; Hsiou et al. 1996) and consists of hydrophobic residues (Y181, Y188, F227, W229, and Y232), and hydrophilic residues such as K101, K103, S105, D192, and E224 of the p66 subunit and E138 of the p51 subunit (Fig. 4) (Sluis-Cremer et al. 2004). Unlike NRTIs, these non/uncompetitive inhibitors do not inhibit the RT of other lentiviruses such as HIV-2 and simian immunodeficiency virus (SIV) (Kohlstaedt et al. 1992; Witvrouw et al. 1999). Currently, there are four approved NNRTIs: etravirine, delavirdine, efavirenz, and nevirapine, and several in development, including rilpivirine in phase 3 (Fig. 4).
Figure 4.
Non–nucleoside RT inhibitors and the X-ray crystal structure of HIV-1 RT complexed with etravirine (Lansdon et al. 2010) (3MEE).
NNRTI resistance generally results from amino acid substitutions such as L100, K101, K103, E138, V179, Y181, and Y188 in the NNRTI-binding pocket of RT (Tantillo et al. 1994). The most common NNRTI mutations are K103N and Y181C (Bacheler et al. 2000, 2001; Demeter et al. 2000; Dykes et al. 2001). As with NRTI resistance, complex patterns of NNRTI-resistant mutations can arise and alternative pathways have been observed in nonsubtype B infected individuals (Brenner et al. 2003; Spira et al. 2003; Gao et al. 2004). Most NNRTI mutations engender some level of cross resistance among different NNRTIs, especially in the context of additional secondary mutations (Antinori et al. 2002).
In contrast to the significant reductions in replicative fitness observed with resistance to other drug classes, with NNRTIs, single nucleotide changes can result in high-level resistance with only a slight loss of replicative fitness (Deeks 2001; Dykes et al. 2001; Imamichi et al. 2001). A lower genetic barrier, minimal impact on replicative fitness, and the slow reversion of these mutations in patients in the absence of drug contribute to transmission and stability of NNRTI-resistant HIV-1 in the population. Interestingly, the majority of NNRTI-resistance mutations selected under NNRTI treatment are commonly found as wild-type sequence in HIV-1 group O and HIV-2. HIV-1 group O can actually be subdivided into lineages based on a C181 or Y181 amino acid in RT (Tebit et al. 2010). Furthermore, nearly all primate lentiviruses can be phylogenetically classified into different lineages based on signature sequences in NNRTI-binding pocket and linked to a Cys, Ile, or Tyr at position 181, i.e., the primary codon-conferring resistance to NNRTIs (Tebit et al. 2010). Given the intrinsic resistance in most primate lentiviruses, aside from HIV-1 group M, it is not surprising that acquired resistance to NNRTIs has the least fitness impact.
INTEGRASE INHIBITORS
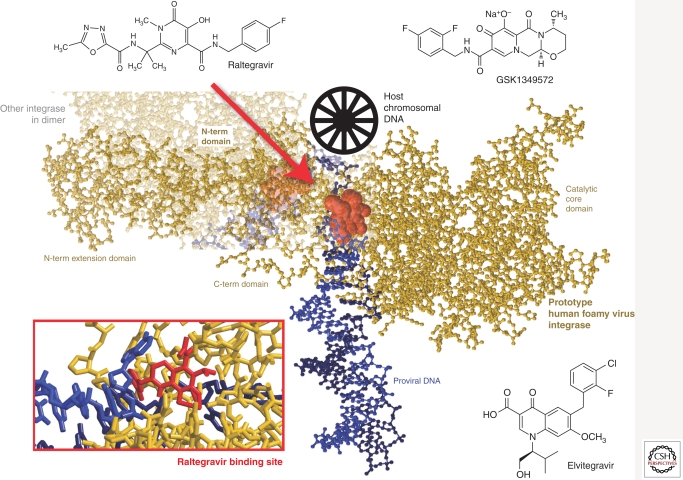
Integrase was the most recent HIV-1 enzyme to be successfully targeted for drug development (Espeseth et al. 2000; Hazuda et al. 2004a,b). Raltegravir (RAL), MK-0518 was FDA approved in 2007, and other integrase inhibitors, including Elvitegravir (EVG), GS-9137 are progressing through clinical development (Fig. 5) (Sato et al. 2006; Shimura et al. 2008). As mentioned above, integrase catalyzes 3′ end processing and viral DNA and strand transfer. All integrase inhibitors in development target the strand transfer reaction and are thus referred to as either INIs or more specifically, integrase strand transfer inhibitors (InSTIs) (Espeseth et al. 2000; Hazuda et al. 2004a, b; McColl and Chen 2010). The selective effect on strand transfer is a result of a now well-defined mechanism of action in which the inhibitor (1) binds only to the specific complex between integrase and the viral DNA and (2) interacts with the two essential magnesium metal ion cofactors in the integrase active site and also the DNA (Fig. 5). Therefore, all InSTIs are comprised of two essential components: a metal-binding pharmacophore, which sequestors the active site magnesiums, and a hydrophobic group, which interacts with the viral DNA as well as the enzyme in the complex (Grobler et al. 2002). InSTIs are therefore the only ARV class that interacts with two essential elements of the virus, the integrase enzyme as well as the viral DNA, which is the substrate for integration.
Figure 5.
Integrase strand transfer inhibitors and the crystal structure of prototype human foamy virus integrase (as a model of HIV-1 IN) complexed to dsDNA and Raltegravir (Hare et al. 2010) (3OYH). N-term, amino-terminal; C-term, carboxy-terminal.
The recent cocrystallization of the foamy virus integrase DNA complex or intasome with both RAL and EVG (Hare et al. 2010) corroborates the biochemical mechanism and provides a structural basis for understanding the unique breadth of antiviral activity that has been observed for InSTIs across all HIV-1 subtypes as well as other retroviruses, such as HIV-2 and XMRV (Fig. 5) (Maignan et al. 1998; Damond et al. 2008; Shimura et al. 2008; Van Baelen et al. 2008; Garrido et al. 2010; Singh et al. 2010). In the cocrystal structure, the general architecture and amino acids within the active site of the foamy virus intasome are highly conserved with other retroviral integrases, as are the immediate surrounding interactions with InSTIs. The common mechanism of action and conserved binding mode for InSTIs also has important implications for understanding resistance to the class. Mutations that engender resistance to InSTIs almost always map within the integrase active site near the amino acid residues that coordinate the essential magnesium cofactors (Hazuda et al. 2004a; Hare et al. 2010). Thus, these mutations have deleterious effects on enzymatic function and viral replicative capacity (Marinello et al. 2008; Quercia et al. 2009). In clinical studies, resistance to Raltegravir is associated with three independent pathways or sets of mutations in the integrase gene, as defined by primary or signature mutations at Y143, N155, or Q148 (Fransen et al. 2009). These primary mutations are generally observed with specific secondary mutations; for N155(H) these include E92Q, V151L, T97A, G163R, and L74M, whereas for Q148(K/R/H), G140S/A and E138K are common. Significant cross resistance is observed among the InSTIs almost regardless of the primary/secondary mutation sets (Goethals et al. 2008; Marinello et al. 2008). Although cross resistance is prevalent, different agents appear to preferentially select different patterns of mutations (Hazuda et al. 2004a).
PROTEASE INHIBITORS
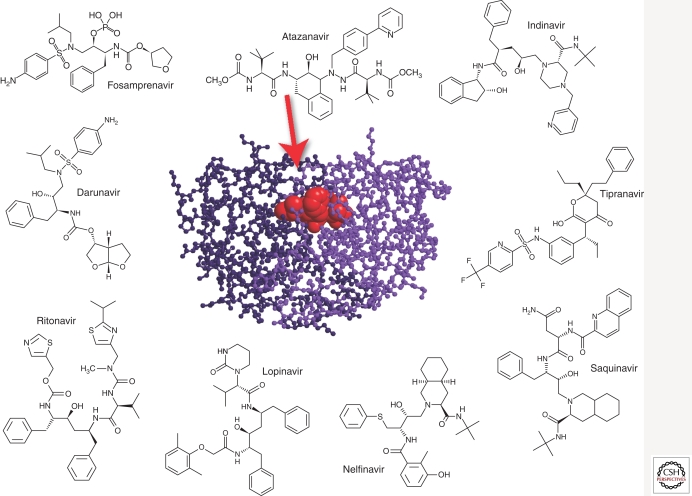
The HIV-1 protease is the enzyme responsible for the cleavage of the viral gag and gag-pol polyprotein precursors during virion maturation (Park and Morrow 1993; Miller 2001). Ten PIs are currently approved: amprenavir (APV, Agenerase), atazanavir (ATZ, Reyataz), darunavir (TMC114, Prezista), fosamprenavir (Lexiva), indinavir (IDV, Crixivan), lopinavir (LPV), nelfinavir (NFV, Viracept), ritonavir (RTV, Norvir), saquinavir (SQV, Fortovase/Invirase), and tipranavir (TPV, Aptivus) (Fig. 6).
Figure 6.
Protease inhibitors and the crystal structure of HIV-1 protease complexed with atazanavir (CA Schiffer, unpubl.) (3EKY).
Because of its vital role in the life cycle of HIV-1 and relatively small size (11 kDa), it was initially expected that resistance to protease inhibitors would be rare. However, the protease gene has great plasticity, with polymorphisms observed in 49 of the 99 codons, and more than 20 substitutions known to be associated with resistance (Shafer et al. 2000). The emergence of protease inhibitor resistance likely requires the stepwise accumulation of primary and compensatory mutations (Molla et al. 1996a) and each PI usually selects for certain signature primary mutations and a characteristic pattern of compensatory mutations. Unlike NNRTIs, primary drug-resistant substitutions are rarely observed in the viral populations in protease inhibitor-naïve individuals (Kozal et al. 1996).
All PIs share relatively similar chemical structures (Fig. 6) and cross resistance is commonly observed. For most PIs, primary resistance mutations cluster near the active site of the enzyme, at positions located at the substrate/inhibitor binding site (e.g., D30N, G48V, I50V, V82A, or I84V, among others). These amino acid changes usually have a deleterious effect on the replicative fitness (Nijhuis et al. 2001; Quinones-Mateu and Arts 2002; Quinones-Mateu et al. 2008). In addition to mutations in the protease gene, changes located within eight major protease cleavage sites (i.e., gag and pol genes), have been associated with resistance to protease inhibitors (Doyon et al. 1996; Zhang et al. 1997; Clavel et al. 2000; Miller 2001; Nijhuis et al. 2001). Cleavage site mutants are better substrates for the mutated protease, and thus partially compensate for the resistance-associated loss of viral fitness (Doyon et al. 1996; Mammano et al. 1998; Zennou et al. 1998; Clavel et al. 2000; Nijhuis et al. 2001). With PI resistance, HIV-1 appears to follows a “stepwise” pathway to overcome drug selection: (1) acquisition of primary resistance mutations in the protease gene, (2) selection of secondary/compensatory protease mutations to repair the enzymatic function and rescue viral fitness, and (3) selection of mutations in the major cleavage sites of the gag and gag-pol polyprotein precursors that restore protein processing and increase production of the HIV-1 protease itself (Condra et al. 1995; Molla et al. 1996b; Doyon et al. 1998; Berkhout 1999; Nijhuis et al. 2001).
ENTRY INHIBITORS
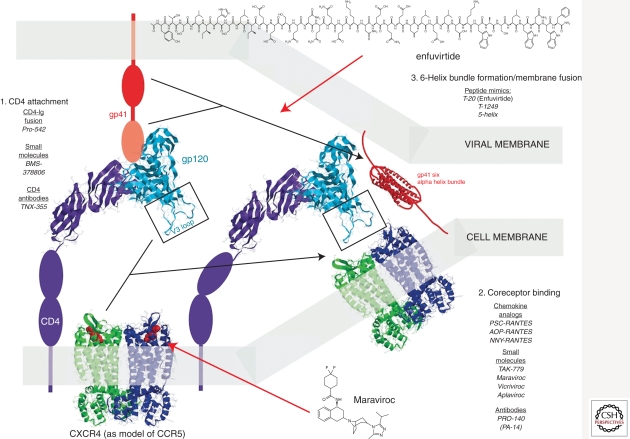
HIV-1 entry exploits several host proteins for a set of intricate events leading to membrane fusion and virus core release into the cytoplasm (Fig. 7). HIV-1 entry inhibitors can be subdivided into distinct classes based on disruption/inhibition of distinct targets/steps in the process.
Figure 7.
Structure predictions of various viral-host components involved in the HIV-1 entry process and the inhibitors. Section 1 describes the components involved for initial CD4 attachment, specifically the D1 domain of CD4 and the C4 domain of gp120. The gp120 structure is shown as an overlay of two structures (2NY2 and 3HI1) (Zhou et al. 2007; Chen et al. 2009). Inhibitors of this CD4 process are listed. Interactions between gp120 and CXCR4 are described in section 2. A rough model of maraviroc (MVC) binding to CCR5 in Figure 7 is based on data from a recently published structure of CXCR4 complexed to a small molecule, IT1t (Wu et al. 2010). The final step in the entry process involves the formation of the gp41 six alpha-helix bundle, which can be blocked by T20 (enfuvirtide). The structure for HIV-1 gp41 six alpha-helix bundle is based on that of SIV gp41 (Malashkevich et al. 1998) (2SIV).
Fusion Inhibitors
The crystal structure of the gp41 ectodomain and of the ectodomain partnered with an inhibitory peptide (C34) revealed that the fusion-active conformation of gp41 was a six-helix bundle in which three N helices form an interior, trimeric coiled coil onto which three antiparallel C helices pack (Doms and Wilen 2011). Peptide fusion inhibitors were designed based on the discovery that two homologous domains in the viral gp41 protein must interact with each other to promote fusion, and that mimicry of one of these domains by a heterologous protein can bind and disrupt the intramolecular interactions of the virus protein. Alpha-helical peptides homologous to the leucine zipper domain of gp41 had significant antiviral activity against HIV-1, and this activity depended on their ordered solution structure (Wild et al. 1993, 1994). Rational design of helical inhibitors ultimately produced a molecule (T-20, enfuvirtide) with potent antiviral activity in vivo (Fig. 7) (Kilby et al. 1998; Lalezari et al. 2003).
Resistance to early alpha-helical inhibitors was shown to be mediated by mutations in the amino-terminal heptad repeat region of gp41 (Rimsky et al. 1998), which provide further evidence for binding of these peptides to the virus. Monotherapy with enfuvirtide resulted in viral load rebounds after 14 days with resistance which mapped to determinants in the HR1 domain (G36D, I37T, V38A, V38M, N42T, N42D, N43K) (Wei et al. 2002). Mutations that confer resistance to enfuvirtide result in reduced replication capacity/replicative fitness presumably because mutations that reduce enfuvirtide binding also reduce the efficiency of six-helix bundle formation and overall fusion rates (Reeves et al. 2004, 2005). These mutations do not confer cross resistance to other entry inhibitors (attachment inhibitors or coreceptor inhibitors) (Ray et al. 2005) but can sensitize viruses to neutralization by monoclonal antibodies that target the gp41 domain by prolonging the exposure of fusion intermediates that are specifically sensitive to these antibodies (Reeves et al. 2005). Adaptation to enfuvirtide has even resulted in viruses that require enfuvirtide for fusion (Baldwin et al. 2004).
Resistance mutations in gp41 decrease fusion efficiency and reduce viral fitness (Labrosse et al. 2003). Nonetheless, studies of baseline susceptibility to enfuvirtide suggested that large variations in intrinsic susceptibility existed in diverse HIV-1 isolates, and that these variations mapped to regions outside the enfuvirtide-binding site (Derdeyn et al. 2000). Sequences associated with the V3 loop were correlated with intrinsic enfuvirtide susceptibility, suggesting that interactions with the coreceptor were important determinants of susceptibility of a drug that inhibits virus fusion. A seminal observation in the understanding of entry inhibitor susceptibility was the discovery that efficiency of the fusion process was the principal modulator of intrinsic enfuvirtide susceptibility (Reeves et al. 2002). Mutations in the coreceptor-binding site that reduced gp120 affinity for CCR5 resulted in viruses with reduced fusion kinetics (Reeves et al. 2004; Biscone et al. 2006). Engagement of CD4 by gp120 initiates a process of structural rearrangement in the envelope glycoprotein resulting in fusion. Completion of this process requires engagement of the coreceptor molecule, but enfuvirtide susceptibility is limited to the time between CD4 engagement and six-helix bundle formation. Any decrease in the rate of this entry process (e.g., reducing the levels of coreceptor expression) also increases susceptibility of the virus to inhibition by enfuvirtide. Consistent with this, ENF is synergistic with compounds that inhibit CD4 or coreceptor engagement (Tremblay et al. 2000; Nagashima et al. 2001).
Small-Molecule CCR5 Antagonists
Small-molecule CCR5 antagonists bind to hydrophobic pockets within the transmembrane helices of CCR5 (Dragic et al. 2000; Tsamis et al. 2003). This site does not overlap the binding sites of either CCR5 agonists or HIV-1 envelope. Instead, drug binding induces and stabilizes a receptor conformation that is not recognized by either. Thus, these molecules are considered allosteric inhibitors. Ideally, a small-molecule inhibitor of CCR5 would block binding by HIV-1 envelope but continue to bind native chemokines and allow signal transduction. Most small-molecule inhibitors, however, are pure antagonists of the receptor. Oral administration of small-molecule antagonists has been shown to inhibit viral replication in macaque models (Veazey et al. 2003) and to prevent vaginal transmission (Veazey et al. 2005). Thus far, three antagonists (VCV, MVC, and Aplaviroc) have been shown to inhibit virus replication in humans (Dorr et al. 2005). The compound MVC was approved for therapeutic use by the FDA in 2007 (Fig. 7).
MVC binds a hydrophobic transmembrane cavity of CCR5. Binding alters the conformation of the second extracellular loop of the receptor and prevents interaction with the V3 stem loop of gp120 (Dragic et al. 2000; Kondru et al. 2008). A rough model of MVC binding to CCR5 in Figure 7 is based on a recently published structure of CXCR4 complexed to a small molecule, IT1t (Wu et al. 2010). CXCR4 also serves as a coreceptor for HIV-1 but attempts at development of CXCR4 antagonists (e.g., AMD3100) fail in clinical studies (Hendrix et al. 2004). Because MVC binds to a host cell protein, resistance to MVC is unlike that of other ARVs. Potential resistance mechanisms include (1) tropism switching (utilization of CXCR4 instead of CCR5 for entry), (2) increased affinity for the coreceptor, (3) utilization of inhibitor-bound receptor for entry, and (4) faster rate of entry. Tropism switching has been a concern in the therapeutic administration of this class as primary infection with, or early emergence of CXCR4 tropic virus, although rare, typically leads to faster disease progression. Thus, the selection of CXCR4 tropic virus owing to CCR5 antagonist treatment could have a negative impact on HIV-1 pathogenesis.
Small-molecule CCR5 inhibitors have been used to select for drug resistance in peripheral blood mononuclear cell cultures (PBMC), which express CCR5 and CXCR4, as well as a variety of other chemokine receptors that could potentially substitute for HIV-1 coreceptors. In these experiments, inhibitor-resistant viruses continue to require CCR5 for entry (Trkola et al. 2002; Marozsan et al. 2005; Baba et al. 2007; Westby et al. 2007). Furthermore, evaluation of coreceptor tropism of viruses from patients who failed MVC therapy during clinical trials has suggested that tropism change occurred only when X4 tropic viruses were preexisting in the patient quasispecies before initiating treatment with MVC (Westby et al. 2006). Thus, it appears that de novo mutations conferring altered coreceptor usage is not the favored pathway for resistance in vitro or in vivo. It should be noted that in some treatment failures, the use of CCR5 was maintained even in the presence of MVC. These “resistant” HIV-1 isolates did not display the same shift in drug susceptibility, typically characterized by an increase in IC50 values, but were capable of using both the free and inhibitor-bound CCR5 for entry (Trkola et al. 2002; Tsibris et al. 2008). In such cases, resistance is reported as MPI (or maximum percent inhibition) for saturating concentrations of drug.
Although it is still early with respect to the clinical experience for CCR5 antagonists, there are documented cases of treatment failures that are not accounted for by either CXCR4 tropism switch or resistance owing to increased MPI. Recent studies suggest discrepancies in the sensitivity to CCR5 antagonists may be assay dependent. Susceptibility to entry CCR5 antagonists can be affected by cell type, state of cellular activation, and number of virus replication cycles (Kuhmann et al. 2004; Marozsan et al. 2005; Lobritz et al. 2007;Westby et al. 2007). Also, different primary HIV-1 isolates can vary in sensitivity by as much as 100-fold in IC50 values (Torre et al. 2000; Dorr et al. 2005; Lobritz et al. 2007), and this difference is much more demonstrable with infection assays using replication-competent primary HIV-1 isolates as compared with defective viruses limited to single-cycle replication. These complexities make it quite challenging to detect resistance at the time of treatment failure with routine resistance testing assays. Given the issues, the use of CCR5 antagonists in clinical practice is somewhat more complex than other classes of ARV agents.
CONCLUSIONS
The breadth and depth of the HIV-1 therapy pipeline may arguably be among the most successful for treating any single human disease, infection, or disorder as illustrated by the number of antiretroviral agents and unique drug classes available. In reviewing the history of ARV drug development, however, there are some key lessons and parallels that need to be kept in mind as we consider the development of small-molecule prevention strategies for HIV-1 and evolving treatment strategies for other viral infections, including hepatitis C virus (HCV). The road to successful HIV-1 treatment was hard, and in the early days many patients were inadequately treated with suboptimal regimens that rapidly led to failure and drug resistance. Although it is unknown whether the prevention of HIV-1 transmission will require the same number of agents, the inherent plasticity of HIV-1 would suggest erring on the side of caution and focusing early on combination products that would mitigate this risk. In the case of HCV, the breadth of genetic diversity appears to be greater than that observed in an HIV-1 infected individual. Anti-HCV drugs in the most advanced stages for approval inhibit a small number of targets (e.g., the NS5b polymerase and NS3 protease) and each class appears to share significant cross resistance; when tested as single agents, the emergence of HCV drug resistance is rapid. The success of HAART should provide the benchmark for HCV drug development and a roadmap for the development of novel prevention strategies in HIV-1 to avoid potential risk to both the individual patient and the population by preventing the acquisition and transmission of drug resistance.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol 84: 9864–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA 1996. CC CKR5: A RANTES, MIP-1a, MIP-1b receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272: 1955–1958 [DOI] [PubMed] [Google Scholar]
- Antinori A, Zaccarelli M, Cingolani A, Forbici F, Rizzo MG, Trotta MP, Di Giambenedetto S, Narciso P, Ammassari A, Girardi E, et al. 2002. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res Hum Retroviruses 18: 835–838 [DOI] [PubMed] [Google Scholar]
- Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): Increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37: 15908–15917 [DOI] [PubMed] [Google Scholar]
- Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J 1997. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277: 112–116 [DOI] [PubMed] [Google Scholar]
- Baba M, Miyake H, Wang X, Okamoto M, Takashima K 2007. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob Agents Chemother 51: 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, et al. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob Agents Chemother 44: 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L, Jeffrey S, Hanna G, D’Aquila R, Wallace L, Logue K, Cordova B, Hertogs K, Larder B, Buckery R, et al. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol 75: 4999–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin CE, Sanders RW, Deng Y, Jurriaans S, Lange JM, Lu M, Berkhout B 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J Virol 78: 12428–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini J, Herdewijn P, De Clercq E 1989. Differential patterns of intracellular metabolism of 2′,3′-didehydro-2′,3′-dideoxythymidine and 3′-azido-2′,3′-dideoxythymidine, two potent anti-human immunodeficiency virus compounds. J Biol Chem 264: 6127–6133 [PubMed] [Google Scholar]
- Berkhout B 1999. HIV-1 evolution under pressure of protease inhibitors: Climbing the stairs of viral fitness. J Biomed Sci 6: 298–305 [DOI] [PubMed] [Google Scholar]
- Biscone MJ, Miamidian JL, Muchiri JM, Baik SS, Lee FH, Doms RW, Reeves JD 2006. Functional impact of HIV coreceptor-binding site mutations. Virology 351: 226–236 [DOI] [PubMed] [Google Scholar]
- Boucher CA, O’Sullivan E, Mulder JW, Ramautarsing C, Kellam P, Darby G, Lange JM, Goudsmit J, Larder BA 1992. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis 165: 105–110 [DOI] [PubMed] [Google Scholar]
- Boyer PL, Sarafianos SG, Arnold E, Hughes SH 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol 75: 4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, Marlink RG, Schapiro J, Roger M, Wainberg MA 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17: F1–F5 [DOI] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326: 1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC, Dutschman GE, Bastow KF, Sarngadharan MG, Ting RY 1987. Human immunodeficiency virus reverse transcriptase. General properties and its interactions with nucleoside triphosphate analogs. J Mol Biol 262: 2187–2189 [PubMed] [Google Scholar]
- Clavel F, Race E, Mammano F 2000. HIV drug resistance and viral fitness. Adv Pharmacol 49: 41–66 [DOI] [PubMed] [Google Scholar]
- Coffin JM 1995. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 267: 483–489 [DOI] [PubMed] [Google Scholar]
- Collier AC, Coombs RW, Schoenfeld DA, Bassett RL, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe VJ, et al. 1996. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. AIDS Clinical Trials Group. N Engl J Med 334: 1011–1017 [DOI] [PubMed] [Google Scholar]
- Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature (London) 374: 569–571 [DOI] [PubMed] [Google Scholar]
- *.Craigie R, Bushman FD 2011. HIV DNA integration. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupelli LA, Hsu MC 1995. The human immunodeficiency virus type 1 Tat antagonist, Ro 5-3335, predominantly inhibits transcription initiation from the viral promoter. J Virol 69: 2640–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aquila RT, Hughes MD, Johnson VA, Fischl MA, Sommadossi JP, Liou SH, Timpone J, Myers M, Basgoz N, Niu M, et al. 1996. Nevirapine, zidovudine, and didanosine compared with zidovudine and didanosine in patients with HIV-1 infection. A randomized, double-blind, placebo-controlled trial. National Institute of Allergy and Infectious Diseases AIDS Clinical Trials Group Protocol 241 Investigators. Ann Intern Med 124: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312: 763–767 [DOI] [PubMed] [Google Scholar]
- Damond F, Lariven S, Roquebert B, Males S, Peytavin G, Morau G, Toledano D, Descamps D, Brun-Vezinet F, Matheron S 2008. Virological and immunological response to HAART regimen containing integrase inhibitors in HIV-2-infected patients. AIDS 22: 665–666 [DOI] [PubMed] [Google Scholar]
- Davidson A, Leeper TC, Athanassiou Z, Patora-Komisarska K, Karn J, Robinson JA, Varani G 2009. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc Natl Acad Sci 106: 11931–11936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WL, Grunert RR, Haff RF, Mcgahen JW, Neumayer EM, Paulshock M, Watts JC, Wood TR, Hermann EC, Hoffmann CE 1964. Antiviral activity of 1-adamantanamine (amantadine). Science 144: 862–863 [DOI] [PubMed] [Google Scholar]
- Deeks SG 2001. International perspectives on antiretroviral resistance. Nonnucleoside reverse transcriptase inhibitor resistance. J Acquir Immune Defic Syndr 26: S25–S33 [DOI] [PubMed] [Google Scholar]
- Demeter LM, Shafer RW, Meehan PM, Holden-Wiltse J, Fischl MA, Freimuth WW, Para MF, Reichman RC 2000. Delavirdine susceptibilities and associated reverse transcriptase mutations in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdine monotherapy (ACTG 260). Antimicrob Agents Chemother 44: 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature (London) 381: 661–666 [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74: 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem 279: 509–516 [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85: 1149–1158 [DOI] [PubMed] [Google Scholar]
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, et al. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49: 4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol 70: 3763–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon L, Payant C, Brakier-Gingras L, Lamarre D 1998. Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors. J Virol 72: 6146–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, et al. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci 97: 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes C, Fox K, Lloyd A, Chiulli M, Morse E, Demeter LM 2001. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology 285: 193–203 [DOI] [PubMed] [Google Scholar]
- Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, et al. 2000. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci 97: 11244–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA 1996. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272: 872–877 [DOI] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278: 1295–1300 [DOI] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5: 512–517 [DOI] [PubMed] [Google Scholar]
- Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W 2009. Loss of Raltegravir susceptibility of HIV-1 is conferred by multiple non-overlapping genetic pathways. J Virol 83: 11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SD, McLean AR 1994. Quasispecies dynamics and the emergence of drug resistance during zidovudine therapy of HIV infection. AIDS 8: 323–332 [DOI] [PubMed] [Google Scholar]
- Fujioka T, Kashiwada Y, Kilkuskie RE, Cosentino LM, Ballas LM, Jiang JB, Janzen WP, Chen IS, Lee KH 1994. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J Nat Prod 57: 243–247 [DOI] [PubMed] [Google Scholar]
- Furman PA, Fyfe JA, St Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc Natl Acad Sci 83: 8333–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Paxinos E, Galovich J, Troyer R, Baird H, Abreha M, Kityo C, Mugyenyi P, Petropoulos C, Arts EJ 2004. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J Virol 78: 5390–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lerma JG, MacInnes H, Bennett D, Reid P, Nidtha S, Weinstock H, Kaplan JE, Heneine W 2003. A novel genetic pathway of human immunodeficiency virus type 1 resistance to stavudine mediated by the K65R mutation. J Virol 77: 5685–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C, Geretti A, Zahonero N, Booth C, Strang A, Soriano V, De Mendoza C, et al. 2010. Integrase variability and susceptibility to HIV integrase inhibitors: Impact of subtypes, antiretroviral experience and duration of HIV infection. J Antimicrob Chemother 65: 320–326 [DOI] [PubMed] [Google Scholar]
- Goethals O, Clayton R, Van Ginderen M, Vereycken I, Wagemans E, Geluykens P, Dockx K, Strijbos R, Smits V, Vos A, et al. 2008. Resistance mutations in HIV type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J Virol 82: 10366–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, Espeseth AS, Wolfe A, Egbertson M, Bourgeois M, Melamed J, et al. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: Implications for metal binding in the active site of phosphotransferase enzymes. Proc Natl Acad Sci 99: 6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T 1997. An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 replication. Proc Natl Acad Sci 94: 3548–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci 107: 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan PR, Kinghorn I, Bloor S, Kemp SD, Najera I, Kohli A, Larder BA 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol 70: 5930–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GJ, Orr DC, Penn CR, Figueiredo HT, Gray NM, Boehme RE, Cameron JM 1992. Effects of (–)–2′-deoxy-3′-thiacytidine (3TC) 5′-triphosphate on human immunodeficiency virus reverse transcriptase and mammalian DNA polymerases alpha, beta, and gamma. Antimicrob Agents Chemother 36: 1688–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J 4: 3021–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, Guare JP Jr, Egbertson MS, et al. 2004a. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc Natl Acad Sci 101: 11233–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, et al. 2004b. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science 305: 528–532 [DOI] [PubMed] [Google Scholar]
- Hendrix CW, Collier AC, Lederman MM, Schols D, Pollard RB, Brown S, Jackson JB, Coombs RW, Glesby MJ, Flexner CW, et al. 2004. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J Acquir Immune Defic Syndr 37: 1253–1262 [DOI] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature (London) 373: 123–126 [DOI] [PubMed] [Google Scholar]
- Hsiou Y, Ding J, Das K, Clark AD Jr, Hughes SH, Arnold E 1996. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: Implications of conformational changes for polymerization and inhibition mechanisms. Structure 4: 853–860 [DOI] [PubMed] [Google Scholar]
- Hsu MC, Schutt AD, Holly M, Slice LW, Sherman MI, Richman DD, Potash MJ, Volsky DJ 1991. Inhibition of HIV replication in acute and chronic infections in vitro by a Tat antagonist. Science 254: 1799–1802 [DOI] [PubMed] [Google Scholar]
- Hsu A, Granneman GR, Cao G, Carothers L, El-Shourbagy T, Baroldi P, Erdman K, Brown F, Sun E, Leonard JM 1998. Pharmacokinetic interactions between two human immunodeficiency virus protease inhibitors, ritonavir and saquinavir. Clin Pharmacol Ther 63: 453–464 [DOI] [PubMed] [Google Scholar]
- *.Hu W-S, Hughes SH 2011. HIV-1 reverse transcription. Cold Spring Harb Perspect Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 282: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Hwang S, Tamilarasu N, Kibler K, Cao H, Ali A, Ping YH, Jeang KT, Rana TM 2003. Discovery of a small molecule Tat-trans-activation-responsive RNA antagonist that potently inhibits human immunodeficiency virus-1 replication. J Biol Chem 278: 39092–39103 [DOI] [PubMed] [Google Scholar]
- Imamichi T, Murphy MA, Imamichi H, Lane HC 2001. Amino acid deletion at codon 67 and Thr-to-Gly change at codon 69 of human immunodeficiency virus type 1 reverse transcriptase confer novel drug resistance profiles. J Virol 75: 3988–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karn J, Stoltzfus CM 2011. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 10.1101/cshperspect. a006916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellam P, Boucher CA, Larder BA 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci 89: 1934–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf DJ, Marsh KC, Kumar G, Rodrigues AD, Denissen JF, McDonald E, Kukulka MJ, Hsu A, Granneman GR, Baroldi PA, et al. 1997. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob Agents Chemother 41: 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med 4: 1302–1307 [DOI] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312: 767–768 [DOI] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256: 1783–1790 [DOI] [PubMed] [Google Scholar]
- Komanduri KV, Viswanathan MN, Wieder ED, Schmidt DK, Bredt BM, Jacobson MA, McCune JM 1998. Restoration of cytomegalovirus-specific CD4+ T-lymphocyte responses after ganciclovir and highly active antiretroviral therapy in individuals infected with HIV-1. Nat Med 4: 953–956 [DOI] [PubMed] [Google Scholar]
- Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol 73: 789–800 [DOI] [PubMed] [Google Scholar]
- Kozal MJ, Shah N, Shen N, Yang R, Fucini R, Merigan TC, Richman DD, Morris D, Hubbel E, Chee M, et al. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nature Med 2: 753–759 [DOI] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, et al. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol 78: 2790–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes DR, Jacobson J, Powderly WG, Godofsky E, DeJesus E, Haas F, Reimann KA, Larson JL, Yarbough PO, Curt V Jr, et al. 2004. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. J Infect Dis 189: 286–291 [DOI] [PubMed] [Google Scholar]
- Labrosse B, Labernardiere JL, Dam E, Trouplin V, Skrabal K, Clavel F, Mammano F 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J Virol 77: 1610–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalezari JP, Henry K, O’Hearn M, Montaner JS, Piliero PJ, Trottier B, Walmsley S, Cohen C, Kuritzkes DR, Eron JJ Jr, et al. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N Engl J Med 348: 2175–2185 [DOI] [PubMed] [Google Scholar]
- Lalonde M, Lobritz M, Ratcliff A, Chaminian M, Athanassiou Z, Tyagi M, Karn J, Robinson JA, Varani G, Arts EJ 2011. Inhibition of both HIV-1 reverse transcription and gene expression by a cyclic peptide that binds the Tat-transactivating response element (TAR) RNA. PLoS Pathog 7: e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdon EB, Brendza KM, Hung M, Wang R, Mukund S, Jin D, Birkus G, Kutty N, Liu X 2010. Crystal structures of HIV-1 reverse transcriptase with etravirine (TMC125) and rilpivirine (TMC278): Implications for drug design. J Med Chem 53: 4295–4299 [DOI] [PubMed] [Google Scholar]
- Larder BA, Kemp SD 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Larder BA, Kemp SD, Harrigan PR 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269: 696–699 [DOI] [PubMed] [Google Scholar]
- Lederman MM, Connick E, Landay A, Kuritzkes DR, Spritzler J, St Clair M, Kotzin BL, Fox L, Chiozzi MH, Leonard JM, et al. 1998. Immunologic responses associated with 12 weeks of combination antiretroviral therapy consisting of zidovudine, lamivudine, and ritonavir: Results of AIDS Clinical Trials Group Protocol 315. J Infect Dis 178: 70–79 [DOI] [PubMed] [Google Scholar]
- Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, et al. 2003. PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci 100: 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, et al. 2003. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci 100: 11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobritz MA, Marozsan AJ, Troyer RM, Arts EJ 2007. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J Virol 81: 8258–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: High level of similarity of the active site with other viral integrases. J Mol Biol 282: 359–68 [DOI] [PubMed] [Google Scholar]
- Malashkevich VN, Chan DC, Chutkowski CT, Kim PS 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: Conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci 95: 9134–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Malim MH, Bieniasz PD 2011. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F, Petit C, Clavel F 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: Phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol 72: 7632–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky LM, Temin HM 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 69: 5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin AG, Delaugerre C, Wirden M, Viegas P, Simon A, Katlama C, Calvez V 2004. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J Med Virol 72: 162–165 [DOI] [PubMed] [Google Scholar]
- Marinello J, Marchand C, Mott B, Bain A, Thomas CJ, Pommier Y 2008. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry 47: 9345–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margot NA, Isaacson E, McGowan I, Cheng AK, Schooley RT, Miller MD 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16: 1227–1235 [DOI] [PubMed] [Google Scholar]
- Marozsan AJ, Moore DM, Lobritz MA, Fraundorf E, Abraha A, Reeves JD, Arts EJ 2005. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J Virol 79: 7121–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl DJ, Chen X 2010. Strand transfer inhibitors of HIV-1 integrase: Bringing IN a new era of antiretroviral therapy. Antiviral Res 85: 101–118 [DOI] [PubMed] [Google Scholar]
- Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA 1999. A mechanism of AZT resistance: An increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell Biol 4: 35–43 [DOI] [PubMed] [Google Scholar]
- Meyer PR, Matsuura SE, Tolun AA, Pfeifer I, So AG, Mellors JW, Scott WA 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 46: 1540–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V 2001. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J Acquir Immune Defic Syndr 26 Suppl 1: S34–S50 [DOI] [PubMed] [Google Scholar]
- Mitsuya H, Broder S 1986. Inhibition of the in vitro infectivity and cytopathic effect of human T-lymphotrophic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) by 2′,3′-dideoxynucleosides. Proc Natl Acad Sci 83: 1911–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya H, Weinhold KJ, Furman PA, St Clair MH, Lehrman SN, Gallo RC, Bolognesi D, Barry DW, Broder S 1985. 3′-Azido-3′-deoxythymidine (BW A509U): An antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci 82: 7096–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper PJ, Mo HM, Markowitz M, Chernyavskiy T, Niu P, Lyons N, et al. 1996a. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med 2: 760–766 [DOI] [PubMed] [Google Scholar]
- Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper PJ, Mo HM, Markowitz M, Chernyavskiy T, Niu P, Lyons N, et al. 1996b. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med 2: 760–766 [DOI] [PubMed] [Google Scholar]
- Naeger LK, Margot NA, Miller MD 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 46: 2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima KA, Thompson DA, Rosenfield SI, Maddon PJ, Dragic T, Olson WC 2001. Human immunodeficiency virus type 1 entry inhibitors PRO 542 and T-20 are potently synergistic in blocking virus-cell and cell-cell fusion. J Infect Dis 183: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Nijhuis M, Deeks S, Boucher C 2001. Implications of antiretroviral resistance on viral fitness. Curr Opin Infect Dis 14: 23–28 [DOI] [PubMed] [Google Scholar]
- Nowak MA, Bonhoeffer S, Shaw GM, May RM 1997. Anti-viral drug treatment: Dynamics of resistance in free virus and infected cell populations. J Theor Biol 184: 203–217 [DOI] [PubMed] [Google Scholar]
- O’Neil PK, Sun G, Yu H, Ron Y, Dougherty JP, Preston BD 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J Biol Chem 277: 38053–38061 [DOI] [PubMed] [Google Scholar]
- Parikh UM, Bacheler L, Koontz D, Mellors JW 2006. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J Virol 80: 4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Morrow CD 1993. Mutations in the protease gene of human immunodeficiency virus type 1 affect release and stability of virus particles. Virology 194: 843–850 [DOI] [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD 1996. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. 1582–1586 [DOI] [PubMed] [Google Scholar]
- Quan Y, Gu Z, Li X, Li Z, Morrow CD, Wainberg MA 1996. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (–)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J Virol 70: 5642–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quercia R, Dam E, Perez-Bercoff D, Clavel F 2009. Selective-advantage profile of human immunodeficiency virus type 1 integrase mutants explains in vivo evolution of raltegravir resistance genotypes. Virol J 83: 10245–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Arts EJ 2002. Fitness of drug resistant HIV-1: Methodology and clinical implications. Drug Resist Updat 5: 224–233 [DOI] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Arts EJ 2006. Virus fitness: Concept, quantification, and application to HIV population dynamics. Curr Top Microbiol Immunol 299: 83–140 [DOI] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Moore-Dudley DM, Jegede O, Weber J, Arts J 2008. Viral drug resistance and fitness. Adv Pharmacol 56: 257–296 [DOI] [PubMed] [Google Scholar]
- Ray PE, Soler-Garcia AA, Xu L, Soderland C, Blumenthal R, Puri A 2005. Fusion of HIV-1 envelope-expressing cells to human glomerular endothelial cells through an CXCR4-mediated mechanism. Pediatr Nephrol 20: 1401–1409 [DOI] [PubMed] [Google Scholar]
- Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, et al. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci 99: 16249–16254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Miamidian JL, Biscone MJ, Lee FH, Ahmad N, Pierson TC, Doms RW 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol 78: 5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW 2005. Enfuvirtide resistance mutations: Impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol 79: 4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann KA, Lin W, Bixler S, Browning B, Ehrenfels BN, Lucci J, Miatkowski K, Olson D, Parish TH, Rosa MD, et al. 1997. A humanized form of a CD4-specific monoclonal antibody exhibits decreased antigenicity and prolonged plasma half-life in rhesus monkeys while retaining its unique biological and antiviral properties. AIDS Res Hum Retroviruses 13: 933–943 [DOI] [PubMed] [Google Scholar]
- Richman DD 2001. HIV chemotherapy. Nature 410: 995–1001 [DOI] [PubMed] [Google Scholar]
- Rimsky LT, Shugars DC, Matthews TJ 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol 72: 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci 92: 1222–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Motomura T, Aramaki H, Matsuda T, Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka K, et al. 2006. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J Med Chem 49: 1506–1508 [DOI] [PubMed] [Google Scholar]
- Schinazi RF, Lloyd RM Jr, Nguyen MH, Cannon DL, McMillan A, Ilksoy N, Chu CK, Liotta DC, Bazmi HZ, Mellors JW 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother 37: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer RW, Dupnik K, Winters MA, Eshleman SH 2000. A guide to HIV-1 reverse transcriptase and protease sequencing for drug resistance studies. In HIV sequence compendium 2000 (ed. Kuiken CL, et al. ). Los Alamos National Laboratory, Los Alamos, NM: [PMC free article] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Tachedjian G, Crowe SM, Kedzierska K 2005. Antiretroviral compounds: Mechanisms underlying failure of HAART to eradicate HIV-1. Curr Med Chem 12: 1705–1719 [DOI] [PubMed] [Google Scholar]
- Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, et al. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82: 764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Gorzynski J, Drobysheva D, Bassit L, Schinazi R 2010. Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome. PLoS One 5: e9948 10.1371/journal.pone.0009948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis-Cremer N, Temiz NA, Bahar I 2004. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr HIV Res 2: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PF, Ogundele A, Forrest A, Wilton J, Salzwedel K, Doto J, Allaway GP, Martin DE 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother 51: 3574–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence RA, Kati WM, Anderson KS, Johnson KA 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267: 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother 51: 229–240 [DOI] [PubMed] [Google Scholar]
- Staszewski S, Miller V, Rehmet S, Stark T, De CJ, De BM, Peeters M, Andries K, Moeremans M, De RM, et al. 1996. Virological and immunological analysis of a triple combination pilot study with loviride, lamivudine and zidovudine in HIV-1-infected patients. AIDS 10: F1–F7 [DOI] [PubMed] [Google Scholar]
- St Clair MH, Richards CA, Spector T, Weinhold KJ, Miller WH, Langlois AJ, Furman PA 1987. 3′-Azido-3′-deoxythymidine triphosphate as an inhibitor and substrate of purified human immunodeficiency virus reverse transcriptase. Antimicrob Agents Chemother 31: 1972–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel RF 2008. Mutation and control of the human immunodeficiency virus. Math Biosci 213: 93–102 [DOI] [PubMed] [Google Scholar]
- *.Sundquist WI, Kräusslich H-G 2011. Assembly, budding, and maturation. Cold Spring Harb Perspect Med 10.1101/cshperspect.1006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo C, Ding J, Jacobo-Molina A, Nanni RG, Boyer PL, Hughes SH, Pauwels R, Andries K, Janssen PA, Arnold EA et al. 1994. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol 243: 369–387 [DOI] [PubMed] [Google Scholar]
- Tebit DM, Lobritz M, Lalonde M, Immonen T, Singh K, Sarafianos S, Herchenroder O, Krausslich HG, Arts EJ 2010. Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: Impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. J Virol 84: 9817–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre VS, Marozsan AJ, Albright JL, Collins KR, Hartley O, Offord RE, Quinones-Mateu ME, Arts EJ 2000. Variable sensitivity of CCR5-tropic human immunodeficiency virus type 1 isolates to inhibition by RANTES analogs. J Virol 74: 4868–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay CL, Kollmann C, Giguel F, Chou TC, Hirsch MS 2000. Strong in vitro synergy between the fusion inhibitor T-20 and the CXCR4 blocker AMD-3100. J Acquir Immune Defic Syndr 25: 99–102 [DOI] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, et al. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci 99: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, et al. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J Virol 77: 5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris AM, Sagar M, Gulick RM, Su Z, Hughes M, Greaves W, Subramanian M, Flexner C, Giguel F, Leopold KE, et al. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol 82: 8210–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baelen K, Van Eygen V, Rondelez E, Stuyver LJ 2008. Clade-specific HIV-1 integrase polymorphisms do not reduce raltegravir elvitegravir phenotypic susceptibility. AIDS 22: 1877–80 [DOI] [PubMed] [Google Scholar]
- Veazey RS, Klasse PJ, Ketas TJ, Reeves JD, Piatak M Jr, Kunstman K, Kuhmann SE, Marx PA, Lifson JD, Dufour J, et al. 2003. Use of a small molecule CCR5 inhibitor in macaques to treat simian immunodeficiency virus infection or prevent simian-human immunodeficiency virus infection. J Exp Med 198: 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Klasse PJ, Schader SM, Hu Q, Ketas TJ, Lu M, Marx PA, Dufour J, Colonno RJ, Shattock RJ, et al. 2005. Protection of macaques from vaginal SHIV challenge by vaginally delivered inhibitors of virus-cell fusion. Nature (London) 438: 99–102 [DOI] [PubMed] [Google Scholar]
- Wainberg MA, Miller MD, Quan Y, Salomon H, Mulato AS, Lamy PD, Margot NA, Anton KE, Cherrington JM 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antiviral Therapy 4: 87–94 [DOI] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature (London) 373: 117–122 [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46: 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, van der Ryst E 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 80: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol 81: 2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KL, Chen JM, Feng JY, Margot NA, Ly JK, Ray AS, Macarthur HL, McDermott MJ, Swaminathan S, Miller MD 2006. The K65R reverse transcriptase mutation in HIV-1 reverses the excision phenotype of zidovudine resistance mutations. Antiviral Therapy 11: 155–163 [DOI] [PubMed] [Google Scholar]
- Wild C, Greenwell T, Matthews T 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses 9: 1051–1053 [DOI] [PubMed] [Google Scholar]
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci 91: 9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wilen CB, Tilton JC, Doms RW 2011. HIV: Cell binding and entry. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield WL, Pollack D, Grunert RR 1969. Therapeutic efficacy of amantadine HCl and rimantadine HCl in naturally occurring influenza A2 respiratory illness in man. N Engl J Med 281: 579–584 [DOI] [PubMed] [Google Scholar]
- Witvrouw M, Pannecouque C, Van Laethem K, Desmyter J, De Clercq E, Vandamme AM 1999. Activity of non-nucleoside reverse transcriptase inhibitors against HIV-2 and SIV. AIDS 13: 1477–1483 [DOI] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. 2010. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330: 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahi N, Fantini J, Henry M, Tourres C, Tamalet C 2005. Structural analysis of reverse transcriptase mutations at codon 215 explains the predominance of T215Y over T215F in HIV-1 variants selected under antiretroviral therapy. J Biomed Sci 12: 701–710 [DOI] [PubMed] [Google Scholar]
- Young FE 1988. The role of the FDA in the effort against AIDS. Public Health Rep 103: 242–245 [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Mammano F, Paulous S, Mathez D, Clavel F 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol 72: 3300–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Imamichi H, Imamichi T, Lane HC, Falloon J, Vasudevachari MB, Salzman NP 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol 71: 6662–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van RD, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445: 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]