Abstract
Great progress has been made toward understanding the pathogenesis of Parkinson’s disease (PD) during the past two decades, mainly as a consequence of the discovery of specific gene mutations contributing to the onset of PD. Recently, dysregulation of the autophagy pathway has been observed in the brains of PD patients and in animal models of PD, indicating the emerging role of autophagy in this disease. Indeed, autophagy is increasingly implicated in a number of pathophysiologies, including various neurodegenerative diseases. This article will lead you through the connection between autophagy and PD by introducing the concept and physiological function of autophagy, and the proteins related to autosomal dominant and autosomal recessive PD, particularly α-synuclein and PINK1-PARKIN, as they pertain to autophagy.
Proteins associated with inherited forms of Parkinson’s disease (PD) (e.g., α-synuclein) are involved in autophagy. Basal, constitutive autophagy may be essential for neuronal survival; its dysregulation may lead to neurodegeneration.
There seem to be various causes of Parkinson’s disease (PD), yet the pathogenesis of this disease appears to be converging on common themes—oxidative stress, mitochondrial dysfunction, and protein aggregation—all of which are tightly linked to autophagy, a highly conserved cellular homeostatic process essential for bulk degradation of cytoplasmic contents. In particular, the recent identification of autosomal dominant and autosomal recessive mutations in familial PD has revealed the involvement of the corresponding gene products in autophagy. Although autophagy has commonly been regarded as an adaptive response to nutrient deprivation, increasing evidence indicates that basal, constitutive autophagy is essential for neuronal survival and that its dysregulation leads to neurodegeneration.
AUTOPHAGY
Main Types of Autophagy
Autophagy is an evolutionarily conserved catabolic process that mediates the degradation of long-lived proteins and dysfunctional or superfluous organelles in eukaryotic cells. Autophagy is induced by various adverse conditions such as limited nutrients, low oxygen levels, and decreased energy supply, and its action results in the release of degradation products, especially amino acids, back into the cytoplasm to be used in essential biosynthetic pathways.
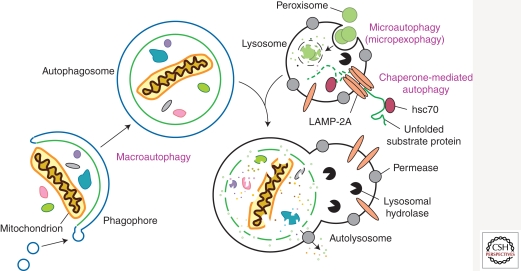
According to the different pathways by which cargo is delivered to the lysosome or vacuole, autophagy can be divided into three main types (Fig. 1): macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). CMA involves direct translocation of unfolded proteins across the lysosome membrane. Chaperone proteins mediate this process by binding to cytosolic substrates that enter the lysosome through interaction with a receptor/channel on the lysosomal membrane (Majeski and Dice 2004). Microautophagy describes the process of direct uptake of cytoplasmic materials at the lysosome surface by invagination of the lysosome membrane. After vesicles containing the cytosolic substrates pinch off into the lysosomal lumen, they are rapidly degraded (Kunz et al. 2004). In contrast, during macroautophagy, portions of the cytoplasm are engulfed by a double-membrane phagophore that expands into a cytosolic vesicle called an autophagosome; the completed autophagosome is targeted to the lysosome in mammalian cells or the vacuole in yeast (Klionsky 2005). The outer membrane of the autophagosome subsequently fuses with the lysosomal/vacuolar membrane, allowing hydrolases access to the inner autophagosome membrane and its cargo, which is degraded and recycled. In contrast to the ubiquitin-26S proteasome system, macroautophagy can mediate nonselective and bulk degradation of cytoplasmic contents, including entire organelles (Shintani et al. 2002; Kanki et al. 2009). Among the three main types of autophagy, macroautophagy is the best characterized process and will hereafter be referred to as autophagy.
Figure 1.
Schematic model of the three main types of autophagy. The modes of autophagy differ depending on the nature of the substrate and the site of sequestration. In chaperone-mediated autophagy, the substrates contain a KFERQ-consensus motif, are unfolded by HSC70 chaperones, and translocate directly across the lysosome membrane via interaction with a LAMP-2A oligomer. There are various types of microautophagy-like processes including micropexophagy and micromitophagy, the selective degradation of peroxisomes and mitochondria, respectively. Again, sequestration occurs at the lysosome-limiting membrane, but the substrates do not have to be unfolded. Macroautophagy uses a double-membrane phagophore to sequester the cargo. Essentially any cytoplasmic component can be enwrapped by a phagophore, which expands into an autophagosome. Fusion with the lysosome allows the cargo to be degraded, and the resulting macromolecules are released into the cytosol through permeases, allowing them to be reused for anabolic processes.
Selective Autophagy
In some cases, autophagy displays substrate specificity, even though autophagy is often considered to be a nonselective pathway for the degradation of bulk cytoplasmic components. Indeed, the unique feature of the autophagy process where the initial sequestering compartment expands into an autophagosome allows for flexible cargo selection. For example, in the cytoplasm to vacuole targeting (Cvt) pathway, autophagy fulfills a biosynthetic role by delivering three vacuolar hydrolases, α-mannosidase (Ams1) (Hutchins and Klionsky 2001), aminopeptidase I (Ape1) (Klionsky et al. 1992), and aspartyl aminopeptidase (Ape4) (Yuga et al. 2011) to their final destination, the vacuole. In addition, superfluous or damaged organelles and misfolded or aggregated proteins are selectively targeted for degradation by autophagy. Different terms are used depending on the cargo, for example, “mitophagy” for selective autophagic degradation of mitochondria, “pexophagy” for peroxisomes, “reticulophagy” for the endoplasmic reticulum (ER), and “ribophagy” for ribosomes (Klionsky et al. 2007). Among them, mitophagy has been increasingly implicated in the pathogenesis of PD through the PINK1-PARKIN-mediated pathway.
Molecular Mechanisms of Autophagy
The molecular understanding of autophagy was initiated by pioneering work in yeast utilizing genetic screens that led to the discovery of autophagy-related (ATG) genes, followed by the identification of homologs in higher eukaryotes. The corresponding Atg proteins can be divided into four major groups: the Atg1/unc-51-like kinase (ULK) complex, two ubiquitin-like protein (Atg12 and Atg8/LC3) conjugation systems, the class III phosphatidylinositol 3-kinase (PtdIns3K)/Vps34 complex I, and the Atg9/mATG9 transmembrane protein system (Xie and Klionsky 2007). The target of rapamycin (TOR), a master regulator of nutrient and growth factor signaling, is one of the critical components involved in controlling the induction of autophagy (He and Klionsky 2009). In most cell types, TOR activity is necessary and sufficient to suppress autophagy under favorable growth conditions, primarily in response to nitrogen. Other kinases, including protein kinase A, AMPK/Snf1, and Pho85, modulate autophagy in response to various types of stress.
Both the Atg1/ULK complex and the membrane protein Atg9 function early in the process of autophagosome formation. Atg1 is a target of the Tor signaling pathway and acts in part by regulating the localization of other Atg proteins such as Atg9, an integral membrane protein that cycles back and forth between the site of phagophore nucleation/autophagosome formation, termed the phagophore assembly site (PAS), and other peripheral locations (Reggiori et al. 2004). Due to its subcellular itinerary and its characterization as a membrane protein, Atg9 is thought to be responsible for the transport of donor membrane that contributes to autophagosome formation.
The main product of the ubiquitin-like conjugation systems is the covalent attachment of Atg8 to phosphatidylethanolamine (PE) (Geng and Klionsky 2008). Atg8–PE becomes associated with both the inner and outer membrane of the phagophore. Once the autophagosome is complete, Atg8 is cleaved off of PE from the outer membrane, whereas the Atg8 on the inner membrane remains associated with PE and is degraded in the vacuole. Atg8 controls the size of the forming autophagosome (Xie et al. 2008), and is also involved in cargo recognition during selective autophagy (Okamoto et al. 2009; Shintani et al. 2002).
Physiological Functions and Connections to PD
Although autophagy is primarily a starvation response in yeast, in higher eukaryotic organisms, autophagy is involved in a wide range of physiological and pathological processes, including responses to nutrient deprivation, development, intracellular clearance, suppression of tumor formation, aging, cell death and survival, and immunity (Huang and Klionsky 2007). As a primary protective mechanism that maintains nutrient and energy homeostasis in response to stress, dysregulation of autophagy underlies the pathophysiologies of many diseases. Increasing evidence suggests that dysregulation of autophagy results in the accumulation of abnormal proteins and/or damaged organelles, which is commonly observed in neurodegenerative diseases, such as Alzheimer, Huntington’s, and Parkinson’s diseases (Banerjee et al. 2010). Of note, autophagy is the only known mechanism that eukaryotic cells possess to degrade protein aggregates and damaged organelles that cannot be processed by the proteasome. Recent studies from transgenic mice, animal, and cell models of PD suggest the involvement of proteins genetically linked to autosomal dominant PD, particularly α-synuclein and LRRK2, in the autophagy pathway (Bandyopadhyay and Cuervo 2007; Alegre-Abarrategui and Wade-Martins 2009). In addition, proteins related to recessive PD, such as PINK1 and PARKIN, have an important role in the process of mitophagy.
EARLY DISCOVERIES: α-SYNUCLEIN AND AUTOPHAGY
α-Synuclein and PD
α-Synuclein was found to localize to the presynaptic terminals in the central nervous system and is involved in vesicular release (Clayton and George 1998, 1999; Spencer et al. 2009). It is a natively unfolded protein, but can be found in several aberrant conformational states including an oligomer, a protofibril, and an amyloid fibril (Martinez-Vicente et al. 2008). α-Synuclein was identified as a component of Lewy bodies, cytosolic inclusions that are a pathological trait of PD (Spillantini et al. 1997, 1998). Studies of familial cases of autophagy reveal two separate autosomal dominant mutations in the α-synuclein gene: A53T and A30P (Polymeropoulos et al. 1997). In addition to the point mutations, several posttranslational modifications such as phosphorylation, ubiquitination, nitration, oxidation, and dopamine-dependent adduct formation also create toxic forms of the protein (Martinez-Vicente et al. 2008).
Chaperone-Mediated Autophagy
α-Synuclein in its native form is degraded by chaperone-mediated autophagy (Cuervo et al. 2004). The protein contains a 15 amino acid sequence that consists of imperfect yet overlapping variations of the KFERQ CMA recognition motif. The chaperone protein HSC70 recognizes the pentapeptide sequence motif and binds to α-synuclein. α-Synuclein then binds to the lysosomal-associated membrane protein type 2A (LAMP-2A) at the lysosomal membrane. This CMA receptor with the aid of a lysosomal lumenal HSC70 transports α-synuclein into the lysosome where it is degraded by proteases. Mutant forms of the protein prevent its degradation by the CMA pathway resulting in toxic aggregation in the cytoplasm as was seen in cell culture and postmortem tissues (Martinez-Vicente et al. 2008). Autophagy can partially compensate for the lack of CMA-mediated degradation, but may cause autophagic cell death under stress conditions. Cells expressing mutant α-synuclein can be characterized by an increase in cell death, accumulation of autophagosomes, and a loss of ability to store catecholamine along with a failure to release dopamine (Stefanis et al. 2001).
All mutant forms of α-synuclein vary in the degree to which they hamper the lysosomal/CMA degradation pathway and thus have different levels of toxicity. In cell culture studies, the A53T and A30P mutants of the protein bind more strongly to the LAMP-2A receptor than the wild-type form, but fail to be transported across the lysosomal membrane (Martinez-Vicente et al. 2008). The mutants act as receptor inhibitors, preventing other CMA targets from binding. This leads to a complete block in CMA resulting in a higher degree of toxicity. Overexpression of the wild-type protein is matched by an increase in expression of the CMA receptor protein LAMP-2A, but high levels of protein expression lead to the formation of oligomeric forms that cannot be degraded by CMA (Vogiatzi et al. 2008; Xilouri et al. 2009). This results in a toxicity level that is lower than the familial point mutant forms. Similar intermediate levels of toxicity are seen in certain posttranslationally modified versions of α-synuclein as was illustrated in cell and post-mortem tissues. Phosphorylated, ubiquitinated, nitrated, and oxidized forms are less susceptible to CMA degradation than the nonmodified protein, but they do not block CMA in its entirety. It is thought that these modifications promote higher-order oligomers that cannot be broken down and degraded. This is reflected in the observation of all of these modifications in cytosolic aggregates.
Modification of the protein with dopamine gives a phenotype that more closely resembles the point mutations. α-Synulcein can be modified with oxidized dopamine through a noncovalent interaction. Dopamine α-synuclein (DA-α-syn) inhibits not only its own degradation, but it also blocks CMA activity in general. This defect can be seen in a variety of experimental conditions including isolated lysosomes, dopaminergic cell lines, and visceral motor neurons. The CMA defect results from DA-α-syn expression because it is not seen in α-synuclein deletion cells, it is not observed with the dopamine insensitive form of α-synuclein, and it is reproduced with isolated lysosomes when presented with DA-α-syn. The complete blockage of CMA creates a high level of toxicity (Martinez-Vicente et al. 2008). What is interesting about the DA-α-syn modification is that it is the dopaminergic substantia nigra and the norepinephrine-releasing locus coeruleus neurons that are killed first in the progression of PD. Both of these types of neurons contain cytosolic dopamine and produce neuromelanin (a product of dopamine modifications). The toxic DA-α-syn form of the protein may explain why those two types of neurons are particularly sensitive.
The blockage of CMA activity with mutant forms of α-synuclein not only results in the direct buildup of toxicity in the neuron through the formation of aggregates, but it also prevents the protective activity of the protein myocyte enhancer factor 2D (MEF2D). MEF2D, a transcription factor, is an important player in neuronal survival. Patients with PD show an increase of this protein in brain neurons, and a genetic polymorphism of a related protein (MEF2A) has been linked to Alzheimer disease. CMA-dependent degradation regulates MEF2D activity. MEF2D is continuously shuttled to the cytosol from the nucleus where it interacts with hsc70. In cells, when CMA is inhibited, an inactive form of the protein accumulates in the cytosol and the amount of protein in the nucleus drops. This inactive form can no longer bind DNA. Wild-type and mutant forms of α-synuclein prevent binding between HSC70 and MEF2D (Yang et al. 2009). This suggests that not only does α-synuclein promote neuronal death through the formation of aggregates, but it also promotes cell death by inhibiting cell survival proteins.
α-Synuclein and Autophagy
As noted previously, inhibition of CMA by aberrant α-synuclein leads to an increase in autophagy. This appears to be a compensatory response, but rather than leading to cell survival, the induction of autophagy can be detrimental causing autophagic cell death. Blocking autophagy by knocking down the autophagy protein Atg5 in cells expressing the A53T α-synuclein mutant can rescue the cell from toxicity-induced cell death (Xilouri et al. 2009). However, autophagy-induced neuronal death is not always the outcome. One study suggests that the signaling pathway for activation of autophagy may be important as to whether or not autophagy will be protective or detrimental. Autophagy is mainly initiated through the mTOR signaling pathway either directly or indirectly through the autophagy protein Atg1. An additional signaling pathway for initiation of autophagy is the Vps34-Beclin 1 complex. It is this secondary signaling pathway that appears to promote cell survival. For example, a reduction in α-synuclein accumulation is seen when Beclin 1 is overexpressed. In addition, Beclin 1 overexpression decreases cell death and increases autophagy activity observed through enhanced lysosomal degradation (Spencer et al. 2009).
Not only does aberrant α-synuclein inhibit CMA, but it also inhibits autophagy through RAB1A and omegasome formation as seen in both cell and mouse models (Winslow et al. 2010). RAB1A is a GTPase involved in the early secretory pathway, specifically ER-to-Golgi transport. The early secretory pathway is important for autophagy, and inhibition of the secretory pathway blocks autophagy. Furthermore, RAB proteins can play a role in autophagy independent of the secretory pathway, as seen with Ypt1 in yeast (Lynch-Day et al. 2010) and RAB1A in mammalian cells (Huang et al. 2011). α-Synuclein overexpression blocks autophagosome formation, inhibits secretion, and increases Golgi fragmentation. Overexpression of RAB1A rescues this defect. The block in autophagy due to α-synuclein overexpression occurs early in the pathway, before autophagosome formation, suggesting an effect on ATG9, which is the only transmembrane protein required for autophagy. It is thought that ATG9 is responsible for the transport of membrane to the site of autophagosome formation and thus acts early in the process. ATG9 normally forms puncta at a perinuclear location (the site of autophagosome formation) and at the trans-Golgi network in mammalian cells. When α-synuclein is overexpressed, ATG9 is mislocalized and is diffuse throughout the cytoplasm of the cell. The same phenotype is seen with the knockdown of RAB1A. One preautophagosome structure that branches off of the ER is the omegasome, which generates an autophagosome, at least under some circumstances. Omegasome formation is reduced in cells that overexpress α-synuclein and in cells that have reduced RAB1A protein levels. α-Synuclein blocks autophagy by inhibiting the activity of RAB1A, which results in the mislocalization of ATG9 and inhibition of autophagosome formation (Winslow et al. 2010).
α-Synuclein and Mitophagy
More recent studies of α-synuclein in PD have shown a relationship between its aberrant expression and mitophagy. Mitochondrial dysfunction is another characteristic of PD and will be described in more detail later in this article. However, a connection has been made between the activation of autophagy by aberrant α-synuclein expression and mitochondrial dysfunction. In cells expressing the A53T α-synuclein mutant, there is an observed increase in colocalization between autophagosomes and normal, polarized mitochondria. In addition, there is a decrease in the number and length of mitochondria in these cells. Similar results are seen when wild-type α-synuclein is overexpressed; however, the phenotype is not as severe. The increase in mitochondria clearance in these cell lines is dependent on mitochondrial fragmentation and on the protein PARKIN (Choubey et al. 2011). PARKIN, currently another large area of focus for autophagy and PD research, will be discussed in the next section of this article. However, the role of α-synuclein in the promotion of mitophagy of polarized mitochondria suggests that there may be a connection between it and PARKIN in the promotion of the disease.
RECENT STUDIES: CONTROVERSIES ABOUND
In this section, we will discuss new yet controversial areas of research with regard to PD and autophagy. Recent studies have focused on the hypothesis of mitochondrial dysfunction as a cause of the disease. These studies have resulted in some interesting data, but to date there is no clear indication as to whether mitochondrial dysfunction is a cause of Parkinson or is rather correlated with the progression of the disease.
The Hypothesis: Mitochondria Dysfunction in PD
Mitochondria are essential organelles that provide >90% of the energy in all eukaryotic cells through oxidative phosphorylation (McBride et al. 2006). Mitochondria are also involved in various other processes such as calcium homeostasis (Celsi et al. 2009) and regulation of apoptosis (Keeble and Gilmore 2007). However, mitochondria are also the major source of cellular reactive oxygen species (ROS). Normal levels of ROS can be tolerated because of cellular antioxidants, whereas in pathological situations of mitochondrial respiratory defect, dramatic ROS production exceeds the capability of antioxidant protection and causes severe damage to a wide range of cellular components including mitochondria. Accumulation of this damage is related to aging, cancer, and recently to neurodegenerative diseases such as PD (Wallace 2005).
PD is characterized primarily by the selective loss of dopaminergic neurons in the substantia nigra pars compacta leading to a dopamine deficit in the striatum. Recent evidence suggests that mitochondria dysfunction may play a role in the pathogenesis of both sporadic PD and familial Parkinsonism. One current model suggests that mitochondrial dysfunction results from damage to complex I of the mitochondrial electron transport chain (Schuler and Casida 2001). Indeed, some studies have shown complex I activities to be significantly reduced in post-mortem substantia nigra of PD patients (Schapira et al. 1989; Schapira 1993). There are several lines of evidence that suggest that increased oxidative damage and ATP depletion may cause dopaminergic neuronal cell death, but the hypothesis linking PD and complex I deficiency is still highly debatable, and the question of causation versus correlation remains to be answered.
Familial variants of PD account for up to 10% of all cases (Gasser 2009). In familial PD, several genes have been linked to autosomal recessive (PARK2, PARK6, PARK7) or dominant (LRRK2) Parkinson (Hardy et al. 2009). These genes have been linked to mitochondrial function and several very recent studies have demonstrated that the corresponding gene products are involved in the selective removal of damaged mitochondria through autophagy (Narendra et al. 2008, 2010b). Thus, these proteins may provide a link between mitophagy and PD.
Mitophagy: Autophagic Mitochondrial Removal
As discussed above, autophagy can be highly specific. During autophagy, the phagophore gradually expands and engulfs a portion of the cytoplasm, or specific cargos, to form the double-membrane autophagosome (Nair and Klionsky 2005). The diameter of a typical autophagosome is approximately 500 nm (Xie et al. 2008); however, the mechanism of autophagosome formation, involving the sequential expansion of the phagophore, provides autophagy with the capacity to sequester essentially any cellular components, including entire organelles, and deliver them into the lysosome for degradation.
Pioneering studies in yeast have demonstrated that autophagic degradation of mitochondria, mitophagy, can be a highly selective and tightly regulated process (Kanki et al. 2009; Mao et al. 2011). In yeast cells, mitophagy fits with the common model of a receptor-adaptor system for the selective degradation of a specific cargo by autophagy; a tag on the cargo is recognized by a receptor and/or adapter, which links the cargo with the autophagy machinery via interaction with Atg8 (Shintani et al. 2002; Wang and Klionsky 2011). In the case of mitophagy, yeast genetic screens discovered a mitochondrial outer membrane resident protein, Atg32, which functions as the receptor for the sequestration of mitochondria into an autophagosome (Kanki et al. 2009; Okamoto et al. 2009). During mitophagy, Atg32 is recognized by an adaptor protein, Atg11, which is proposed to play a role in mediating cargo recognition and transport to the phagophore assembly site (PAS), the nucleating structure for generation of the phagophore (Yorimitsu and Klionsky 2005). Mitochondrial fragments containing Atg32 are then enwrapped by the expanding phagophore, ultimately being incorporated into an autophagosome. The detailed mechanism of this process is still under study.
In higher eukaryotes, autophagy also plays a critical role in degrading mitochondria. In fact, mitochondria were first detected inside an autophagosome in the 1950s (Eskelinen et al. 2011); however, a molecular understanding of this process is occurring only now. Studies suggest that the selective removal of mitochondria, especially damaged mitochondria, is part of an important homeostatic pathway for organelle quality control. Since mitochondria function is compromised in some PD models, a defect in mitochondria quality control may play a critical role in the pathogenesis of PD.
Mitophagy and PD
As mentioned above, several genes related to PD have been recently reported to participate in the removal of damaged mitochondria through autophagy. The PARK2 gene has been reported to be mutated in nearly 50% of autosomal recessive, and 10%–15% of sporadic early-onset PD. PARKIN, the gene product of PARK2 is a primarily cytosolic ubiquitin E3 ligase that contains a ubiquitin-like domain, two RING finger domains, and a conserved region between the RING domains (Schapira 2008). PARKIN has been previously reported to function in the cytosol, in the ER, on mitochondrial targets, and at the plasma membrane; however, no clear evidence had linked PARKIN function to the pathogenesis of PD. Recent studies from Richard Youle’s group and others, however, have provided a model for PARKIN’s role in eliminating impaired mitochondria (Narendra et al. 2008): PARKIN is specifically recruited to damaged mitochondria and promotes their autophagic degradation (Gasser 2009). At steady state, PARKIN is cytosolic. However, treatment of PARKIN-overexpessing cells with the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) leads to a rapid and significant relocation of PARKIN to mitochondria, followed by substantial mitochondria loss from the treated cells. The loss of mitochondria is dependent on the expression of PARKIN and the presence of autophagic proteins, demonstrating that degradation of mitochondria is through autophagy. Extensive mitochondria fragmentation is observed following CCCP treatment, in a PARKIN-independent manner. Microscopy studies show that PARKIN is selectively recruited to mitochondria fragments that have decreased or no membrane potential, suggesting a role for PARKIN in distinguishing between healthy and damaged mitochondria. Further observation shows that these PARKIN-marked mitochondrial fragments are LC3 (a mammalian homolog of yeast Atg8) positive, further demonstrating that clearance of damaged mitochondria occurs through autophagy. Overexpressed PARKIN is also recruited to mitochondria upon an increase in complex one-dependent ROS, which follows treatment with the herbicide paraquat, a toxin frequently used to induce a PD phenotype in some animal and cell culture models (Terzioglu and Galter 2008).
Mitochondrial Targets of PARKIN
The translocation of PARKIN to mitochondria is an indispensable step in PARKIN-dependent mitophagy. Therefore, the identification of mitochondrial targets of PARKIN is significant for elucidating the underlying mechanism of this cellular activity. Although the mitochondrial voltage-dependent anion channel 1 (VDAC1) was reported to be ubquitinated by PARKIN in HeLa cells, this ubquitination does not seem to be required for mitochondrial clustering or mitophagy (Geisler et al. 2010; Narendra et al. 2010a,b). The other putative mitochondrial targets of PARKIN include the mitochondrial fusion proteins MFN1 and MFN2 (Gegg et al. 2010; Ziviani et al. 2010). After translocation to mitochondria, PARKIN ubquitinates MFN1/2 causing their degradation, which facilitates mitochondrial fission; normal fission may be necessary for efficient mitophagy. However, if MFN1/2 are the only substrates of PARKIN, the latter might play a role in facilitating, but not activating mitophagy; it is thought that mitochondrial fission is required, but not sufficient to initiate mitophagy. Therefore, to determine the real role of PARKIN in mitophagy, some other specific substrates of PARKIN, if they exist, have to be identified. Along these lines, a recent study from David Chan's group suggests that PARKIN activates the ubiquitin-proteasome system, which results in the ubiquitination of a large number of mitochondrial proteins (Chan et al. 2011).
The Role of p62 in PARKIN-Dependent Mitophagy
p62 connects ubiquitinated proteins to LC3 for autophagic degradation (Pankiv et al. 2007). As accumulation of p62 is strikingly elevated when autophagy is blocked, it is widely used as an autophagy marker. The loss of mitochondrial membrane potential promotes the accumulation of p62 on clustered mitochondria in a PARKIN-dependent manner. Whether p62 is required for mitophagy, however, is controversial and further studies are needed to determine its role (Geisler et al. 2010; Narendra et al. 2010a).
PINK1
PARKIN interacts with another PD-related protein, PTEN-induced kinase 1 (PINK1), a mitochondrial membrane-anchored kinase. In Drosophila melanogaster, the phenotype resulting from the loss of PINK1 is rescued on overexpression of PARKIN; however, loss of PARKIN is not rescued by the overexpression of PINK1 (Clark et al. 2006; Yang et al. 2006), suggesting that PINK1 acts upstream of PARKIN. Subsequent studies show that PINK1 plays a role in the recruitment of PARKIN (Geisler et al. 2010; Matsuda et al. 2010; Narendra et al. 2010b; Vives-Bauza et al. 2010). Expression of PINK1 on individual mitochondria is regulated by voltage-dependent proteolysis; thus, low levels of PINK1 are maintained on healthy, polarized mitochondria. In steady-state cells, PINK1 is imported into the mitochondrial inner membrane in a membrane potential-dependent manner. When imported into the inner membrane, the mitochondrial inner membrane rhomboid protease presenilin-associated rhomboidlike protein (PARL) mediates the cleavage of PINK1 (Jin et al. 2010). Upon mitochondria depolarization, PINK1 import into the inner membrane is impaired, leading to a rapid PINK1 accumulation on the outer membrane of damaged mitochondria. PINK1 accumulation on mitochondria is both necessary and sufficient for PARKIN recruitment to mitochondria. How recruited PARKIN on damaged mitochondria can promote their degradation is still under extensive investigation. An intriguing possibility is that PARKIN may mediate the ubiquitination of certain substrates on mitochondria, and the ubiquitinated substrates may serve as a recognition target for p62/SQSTM1, a ubiquitin-binding protein that interacts with LC3 and is proposed to play a role in cargo recruitment to the phagophore (Gegg et al. 2010; Geisler et al. 2010). Importantly, several follow-up studies show that disease-associated PARK2 and PARK6 mutations result in defective mitophagy, thereby implicating mitophagy defects in the development of PD (Lee et al. 2010; Matsuda et al. 2010; Narendra et al. 2010b).
Functions of Different Isoforms of PINK1
PINK1 has at least two isoforms: a full-length form and an N-terminally truncated form (Beilina et al. 2005; Silvestri et al. 2005). PINK1 cleavage is mediated by the mitochondrial protease rhomboid-7/PARL in flies and mammalian cells (Whitworth et al. 2008; Deas et al. 2010; Jin et al. 2010). However, which is the functional isoform of PINK1 remains unclear. Early work indicated the cleaved PINK1 might be the functional form, as the expression of a cytoplasmic, cleaved PINK1 is sufficient to protect neurons from mitochondrial stress by MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) (Haque et al. 2008). In contrast, recent work suggests that full-length PINK1 is the only functional form. Full-length PINK1 is rapidly degraded in normal conditions, but accumulates in dysfunctional mitochondria to activate mitophagy when mitochondria lose their membrane potential (Matsuda et al. 2010; Narendra et al. 2010a,b). These latter results imply that truncated PINK1 is an intermediate product destined for degradation. As different isoforms of PINK1 are related to different cellular locations and functions and might respond to different stresses, further studies are still needed to elucidate this issue.
Protective Function of PINK1 in Different Animal Models
Although the significant role of PINK1 in neuron protection is clear, an apparent difference of displayed phenotypes is observed between fly and mouse models when PINK1 is depleted. In Pink1 mutant Drosophila, the obvious phenotypes, including loss of dopaminergic neurons, reduced life span, mitochondrial impairment, and mobility abnormalities, are strikingly similar to the PD pathology in humans (Clark et al. 2006; Park et al. 2006). However, dopaminergic neurons remain normal in the park6−/− mouse, which implies an even more complicated mechanism of PINK1 function in PD (Kitada et al. 2007).
Other PD-Related Genes and Autophagy
Although 95% of PD cases are sporadic, identification of genes responsible for monogenic forms has improved our knowledge of this neurodegenerative disease. In addition to SNCA (encoding α-synuclein), PARK2, and PARK6, two other monogenic PD-related genes, encoding the leucine-rich repeat kinase 2 (LRRK2) and DJ-1, also play a role in autophagy or mitochondrial dynamics.
Mutation of the gene encoding LRRK2 is responsible for an autosomal dominant form of PD. LRRK2 is mainly localized in membrane microdomains, multivesicular bodies, and autophagic vesicles. Mutation or depletion of LRRK2 results in autophagy impairment and the accumulation of the autophagy marker proteins LC3 and p62 (Alegre-Abarrategui et al. 2009; Tong et al. 2010). In contrast, DJ-1 was identified as mediating autosomal recessive PD. Recent studies also made a link between autophagy and DJ-1, as depletion of DJ-1 in both human neuroblastoma cells and Drosophila results in mitochondrial dysfunction and impaired autophagy (Hao et al. 2010; Thomas et al. 2010).
These findings imply that mitochondria and autophagy might play a significant role or even be the convergence points for different monogenic PD-related mutations that give rise to similar symptoms. An alternative possibility is that these PD-related gene products might function together. However, to date, only PINK1 and PARKIN have been shown to genetically and physically interact, especially in modulating neuron protection, mitochondrial function, and mitophagy. Thus, even though many studies have begun to uncover the connections among PINK1, PARKIN, and mitochondria, several controversies remain to be resolved.
BASIC SCIENCE RESEARCH AND CLINICAL TREATMENT
Based on the significant roles of mitochondria and autophagy in PD, maintaining and stabilizing mitochondrial function or promoting the degradation of damaged mitochondria might benefit the protection of dopaminergic neurons. Data on the possible connection between defects in mitophagy and PD suggest that modulation of autophagy might be one avenue for treating some types of this disease. However, autophagy is described as a double-edged sword, because both reduced and excessive autophagy can be detrimental; therefore, simply upregulating autophagy is not a practical course of action, and the application of autophagy-inducing drugs must be undertaken with extreme caution.
CONCLUDING REMARKS
The turnover of proteins has been the focus of attention across neurodegenerative diseases, given that many, if not all, of these diseases show characteristic protein aggregation as part of their cellular pathology. There have been tremendous advances in our understanding of the causes of PD. Novel genes causing familial PD have been discovered, and have been shown to be involved in the autophagy pathway, one of the major proteolytic systems that maintain cellular protein homeostasis. Because autophagy is part of the cell’s homeostatic machinery, maintaining a proper level of autophagy is important for minimizing abnormal protein aggregates and for facilitating organelle turnover. Discovery of therapeutic agents that boost autophagic activity or that directly maintain mitochondrial homeostasis, could potentially reduce neuronal loss and slow down disease progression. A better understanding of the regulatory mechanism of autophagy in the pathogenesis of PD will enable the identification of possible methods for clinical intervention.
ACKNOWLEDGMENTS
This work was supported by NIH Grant GM53396 to D.J.K.
Footnotes
Editor: Serge Przedborski
Additional Perspectives on Parkinson’s Disease available at www.perspectivesinmedicine.org
REFERENCES
- Alegre-Abarrategui J, Wade-Martins R 2009. Parkinson disease, LRRK2 and the endocytic-autophagic pathway. Autophagy 5: 1208–1210 [DOI] [PubMed] [Google Scholar]
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R 2009. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 18: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U, Cuervo AM 2007. Chaperone-mediated autophagy in aging and neurodegeneration: Lessons from α-synuclein. Exp Gerontol 42: 120–128 [DOI] [PubMed] [Google Scholar]
- Banerjee R, Beal MF, Thomas B 2010. Autophagy in neurodegenerative disorders: Pathogenic roles and therapeutic implications. Trends Neurosci 33: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR 2005. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci 102: 5703–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Wallimann TW 2000. Protective effect of the energy precursor creatine against toxicity of glutamate and β-amyloid in rat hippocampal neurons. J Neurochem 74: 1968–1978 [DOI] [PubMed] [Google Scholar]
- Celsi F, Pizzo P, Brini M, Leo S, Fotino C, Pinton P, Rizzuto R 2009. Mitochondria, calcium and cell death: A deadly triad in neurodegeneration. Biochim Biophys Acta 1787: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC 2011. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 20: 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, Zharkovsky A, Kaasik A 2011. Mutant A53T α-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem 286: 10814–10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M 2006. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Clayton DF, George JM 1998. The synucleins: A family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci 21: 249–254 [DOI] [PubMed] [Google Scholar]
- Clayton DF, George JM 1999. Synucleins in synaptic plasticity and neurodegenerative disorders. J Neurosci Res 58: 120–129 [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D 2004. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, et al. 2010. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E-L, Reggiori F, Baba M, Kovacs AL, Seglen PO 2011. Seeing is believing. The impact of electron microscopy on autophagy research. Autophagy 7: 935–956 [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Jenkins BG, Dedeoglu A, Kuemmerle S, Kubilus JK, Kaddurah-Daouk R, Hersch SM, Beal MF 2000. Neuroprotective effects of creatine in a transgenic mouse model of Huntington’s disease. J Neurosci 20: 4389–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T 2009. Mendelian forms of Parkinson’s disease. Biochim Biophys Acta 1792: 587–596 [DOI] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW 2010. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 19: 4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Hao LY, Giasson BI, Bonini NM 2010. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci 107: 9747–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS 2008. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci 105: 1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Lewis P, Revesz T, Lees A, Paisan-Ruiz C 2009. The genetics of Parkinson’s syndromes: A critical review. Curr Opin Genet Dev 19: 254–265 [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ 2007. Autophagy and human disease. Cell Cycle 6: 1837–1849 [DOI] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, et al. 2011. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ 2001. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem 276: 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ 2010. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ 2009. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble JA, Gilmore AP 2007. Apoptosis commitment—Translating survival signals into decisions on mitochondria. Cell Res 17: 976–984 [DOI] [PubMed] [Google Scholar]
- Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, Martella G, Bonsi P, Zhang C, Pothos EN, Shen J 2007. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci 104: 11441–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ 2005. The molecular machinery of autophagy: Unanswered questions. J Cell Sci 118: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS 1992. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 119: 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Dunn WA Jr, Levine B, van der Klei I, Seglen PO 2007. How shall I eat thee? Autophagy 3: 413–416 [DOI] [PubMed] [Google Scholar]
- Kunz JB, Schwarz H, Mayer A 2004. Determination of four sequential stages during microautophagy in vitro. J Biol Chem 279: 9987–9996 [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Brand MD 2004. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I). J Biol Chem 279: 39414–39420 [DOI] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP 2010. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol 189: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littarru GP, Tiano L 2007. Bioenergetic and antioxidant properties of coenzyme Q10: Recent developments. Mol Biotechnol 37: 31–37 [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci 107: 7811–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeski AE, Dice JF 2004. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol 36: 2435–2444 [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ 2011. Two MAPK signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 193: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Tallóczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, et al. 2008. Dopamine-modified α-synuclein blocks chaperone-mediated autophagy. J Clin Invest 118: 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S 2006. Mitochondria: More than just a powerhouse. Curr Biol 16: R551–R560 [DOI] [PubMed] [Google Scholar]
- Naderi J, Somayajulu-Nitu M, Mukerji A, Sharda P, Sikorska M, Borowy-Borowski H, Antonsson B, Pandey S 2006. Water-soluble formulation of Coenzyme Q10 inhibits Bax-induced destabilization of mitochondria in mammalian cells. Apoptosis 11: 1359–1369 [DOI] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem 280: 41785–41788 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ 2010a. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6: 1090–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ 2010b. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Papucci L, Schiavone N, Witort E, Donnini M, Lapucci A, Tempestini A, Formigli L, Zecchi-Orlandini S, Orlandini G, Carella G, et al. 2003. Coenzyme q10 prevents apoptosis by inhibiting mitochondrial depolarization independently of its free radical scavenging property. J Biol Chem 278: 28220–28228 [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim J-M, et al. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. 1997. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276: 2045–2047 [DOI] [PubMed] [Google Scholar]
- Schapira AH 1993. Mitochondrial complex I deficiency in Parkinson’s disease. Adv Neurol 60: 288–291 [PubMed] [Google Scholar]
- Schapira AH 2008. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol 7: 97–109 [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD 1989. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet 1: 1269. [DOI] [PubMed] [Google Scholar]
- Schuler F, Casida JE 2001. Functional coupling of PSST and ND1 subunits in NADH:ubiquinone oxidoreductase established by photoaffinity labeling. Biochim Biophys Acta 1506: 79–87 [DOI] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, Casari G 2005. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet 14: 3477–3492 [DOI] [PubMed] [Google Scholar]
- Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E 2009. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in α-synuclein models of Parkinson’s and Lewy body diseases. J Neurosci 29: 13578–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM-Y, Trojanowski JQ, Jakes R, Goedert M 1997. α-Synuclein in Lewy bodies. Nature 388: 839–840 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M 1998. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci 95: 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA 2001. Expression of A53T mutant but not wild-type α-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci 21: 9549–9560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto HH 2008. Development of mitochondria-targeted aromatic-cationic peptides for neurodegenerative diseases. Ann NY Acad Sci 1147: 112–121 [DOI] [PubMed] [Google Scholar]
- Terzioglu M, Galter D 2008. Parkinson’s disease: Genetic versus toxin-induced rodent models. FEBS J 275: 1384–1391 [DOI] [PubMed] [Google Scholar]
- Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR 2010. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet 20: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ III, Shen J 2010. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of α-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci 107: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci 107: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L 2008. Wild type α-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem 283: 23542–23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet 39: 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ 2011. Mitochondria removal by autophagy. Autophagy 7: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA 2008. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech 1: 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow AR, Chen C-W, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, et al. 2010. α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J Cell Biol 190: 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xilouri M, Vogiatzi T, Vekrellis K, Park D, Stefanis L 2009. Abberant α-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One 4: e5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang J-W, Yang L, Beal MF, Vogel H, Lu B 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci 103: 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z 2009. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323: 124–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ 2005. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 16: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T 2011. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem 286: 13704–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Zhao GM, Wu D, Soong Y, Birk AV, Schiller PW, Szeto HH 2004. Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J Biol Chem 279: 34682–34690 [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ 2010. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci 107: 5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]