Abstract
Background
Imipenem-resistant Pseudomonas aeruginosa (IRPA) is an emerging problem. The causal role of antibiotic selective pressure versus patient-to-patient transmission has not been assessed using a large cohort.
Methods
Patients who were admitted to the medical and surgical intensive care units (ICUs) at the University of Maryland Medical Center from 2001 through 2006 had multiple perianal culture samples collected. Using pulsed-field gel electrophoresis (PFGE), the number of patients who acquired IRPA as a result of patient-to-patient transmission was determined. We also analyzed a subset of patients who had a previous surveillance culture that grew an imipenem-susceptible P. aeruginosa (ISPA) and a subsequent culture that grew IRPA.
Results
Our cohort consisted of 7071 patients. Three hundred patients were colonized with IRPA. 151 patients had positive culture findings at ICU admission, and 149 patients acquired an IRPA. Among the patients who acquired IRPA, 46 (31%) had a PFGE pattern similar to that for another isolate, and 38 (26%) were found to be colonized with an ISPA on the basis of earlier culture results. Of the 38-patient subset, 28 (74%) had identical PFGE patterns.
Conclusions
Our data showed that, of those cases of IRPA acquisition, 46 (31%) were defined as cases of patient-to-patient transmission, and 28 (19%) were cases of acquisition by the patients’ endogenous flora.
Pseudomonas aeruginosa is a leading cause of nosocomial infection and is the most common gram-negative organism causing pneumonia in intensive care units (ICUs) in the United States [1]. Carbapenems are considered to be an optimal antimicrobial agent for the treatment of hard-to-manage pseudomonal infections. The prevalence of P. aeruginosa strains with resistance to carbapenems has significantly increased over time, thus limiting treatment options [1]. The emergence of this antimicrobial-resistant organism has infection-control practitioners, hospital epidemiologists, and clinicians struggling to control its spread. The understanding of the epidemiology and modes of transmission of imipenem-resistant P. aeruginosa (IRPA) in ICUs is needed to implement measures to control dissemination. Currently, studies that attempt to measure the fraction of acquisitions attributable to patient-to-patient transmission of antibiotic-resistant P. aeruginosa have been limited and underpowered. Studies that assess a particular class of antibiotics as the main cause for the emergence of IRPA have also been limited, and none have used a large cohort.
The objective of this study was to analyze the molecular epidemiology of patients who acquired an IRPA by use of perianal surveillance cultures over a 5-year period in a large ICU population. We set out to quantify the amount of patient-to-patient transmission versus endogenous acquisitions of IRPA among our large cohort of patients hospitalized in an ICU. Understanding the causal mechanisms of acquisition of IRPA will aid in providing the appropriate care to patients to prevent and control the dissemination of these drug-resistant organisms.
METHODS
Study design and patient population
This study was approved by the institutional review board of the University of Maryland, Baltimore. This study used a prospective cohort of adult patients who were admitted to the medical ICU (MICU) and surgical ICU (SICU) of the University of Maryland Medical Center (UMMC) between 1 September 2001 and 1 September 2006. Perirectal swab samples were obtained only while the patient was hospitalized in the ICU. UMMC is a tertiary-care facility in Baltimore, Maryland. The MICU is a 10-bed, private room unit providing care to patients who have acute or potentially life-threatening medical conditions, including hematologic and other malignancies. The SICU is a 19-bed private room unit that provides care to adult patients who undergo solid-organ transplantation and abdominal, genitourinary, orthopedic, and otolaryngologic surgical procedures.
During the study period, patients in the MICU and SICU had perianal culture samples obtained at ICU admission, once per week during hospitalization, and at ICU discharge. The culturing technique involved swabbing the perianal area in a circular motion. Samples for culture were obtained as part of an ongoing vancomycin-resistant enterococci active surveillance program in the MICU and SICU and were concurrently screened for IRPA.
We defined admission-positive patients as patients who had cultures of samples obtained at ICU admission that were positive for IRPA. We defined acquisition-positive patients as patients who had cultures of samples obtained at ICU admission that were negative for IRPA and had either a subsequent weekly culture or ICU discharge culture positive for IRPA.
Data used to calculate the Charlson Comorbidity Index, Chronic Disease Score (CDS), and length of stay were obtained from the UMMC Central Data Repository. For each study patient, we collected demographic data including sex, age, ICU admission date, ICU discharge date, CDS, and Charlson Comorbidity Index. The CDS is a measure of patient comorbidity that uses patient medications as indicators for comorbid conditions [2]. The CDS includes 17 different comorbid conditions, such as diabetes, respiratory illness, cancer, and hyper-tension. Each condition contributes 1–5 points to the total score, and the potential range of values for each patient's CDS is 0–35. The CDS was calculated using the medications ordered for each patient within the first 24 h after ICU admission. Discharge International Classification of Diseases, Ninth Revision, codes were used to calculate the Charlson Comorbidity Index for each study participant [3]. The potential range of values for this index is 0–37. The length of stay in the ICU was calculated by subtracting the ICU discharge date from the ICU admission date.
Microbiological methods
Perianal surveillance cultures were plated to MacConkey (Becton-Dickinson) agar plates and MacConkey with 8 μg/mL imipenem. Non–lactose-fermenting, oxidase-positive isolates were identified as P. aeruginosa if they did not produce an acid reaction in triple-sugar iron agar (Becton-Dickinson), grew at 42°C, and produced blue-green pigment on Pseudomonas P agar (Remel). Nonfermenters not matching these characteristics were identified using API 20 NE (BioMérieux) test strips. Antimicrobial susceptibility testing was done by disk diffusion according to Clinical Laboratory Standards Institute guidelines [4].
Molecular methods and definition of patient-to-patient transmission
Pulsed-field gel electrophoresis (PFGE) was performed as described elsewhere [5]. All isolates were digested with SpeI or XbaI, and the resulting fragments were separated by electrophoresis in 1% agarose gels with the CHEF DR II (Bio-Rad Laboratories) for 24 h with switch times ranging from 2 s and 40 s in Tris-Borate ethylenediaminetetraacetic acid (TBE) buffer containing 50 μM thiourea [6]. Photographic images of the gels were saved digitally with the Geldoc EQ (Bio-Rad Laboratories) and saved as TIFF files for gel analysis with GelCompar II Software (Applied Maths). The band patterns were compared by means of the Dice coefficient using the unweighted pair-group method to determine band similarity using the criteria established by Tenover et al [7] to define the pulsed-field type clusters. PFGE types were defined as isolates that are 100% identical, and genetic relatedness was determined by isolates that had ≥80% similarity.
For patients who acquired an IRPA, we explored the following criteria for whether the strain was acquired as a result of patient-to-patient transmission: (1) the isolates were defined as similar on the basis of the PFGE type, and (2) they were defined as epidemiologically related on the basis of any overlap in hospital length of stay. Hospital length of stay overlap was defined as any amount of time in which patients were in the hospital at the same time, regardless of what unit they were in. Endogenous flora was defined as present in patients with an imipenem-susceptible P. aeruginosa and a subsequent IRPA with identical PFGE patterns.
Statistical analysis
Statistical comparisons were performed by using the Wilcoxon rank sum test. Statistical significance was defined as P < .05. Statistical analysis was performed with SAS, version 9.1 (SAS).
RESULTS
The 5-year cohort of patients who were admitted to the MICU and SICU at University of Maryland Medical Center included 7071 patients who had ICU admission and discharge culture samples obtained, resulting in >17,656 perirectal swabs. Compliance with obtaining perianal surveillance culture samples (at ICU admission and discharge) was >90%.
Three hundred patients had an IRPA; 151 of these patients had an IRPA at admission to the ICU and were defined as admission-positive patients. There were 149 patients who had a culture obtained at ICU admission that was negative for IRPA and a subsequent weekly or ICU discharge culture that was positive for IRPA; such patients were defined as having acquired an IRPA. One hundred and ten patients (37%) who were colonized with an IRPA strain had a clinical infection due to an IRPA. Demographic characteristics for these 300 patients were as follows: mean age, 57.5 years; male sex, 56.6%; mean Charlson score, 2.2; and mean chronic disease score, 7.9.
The molecular typing experiments discriminated the 300 IRPA isolates into 243 PFGE groups. Three isolates were unable to be digested by SpeI and were digested with XbaI; these isolates produced 2 different PFGE patterns. There were 2 patients who were colonized with 2 different P. aeruginosa strains, which showed 2 different PFGE patterns. These 7 isolates were included in the PFGE types determined.
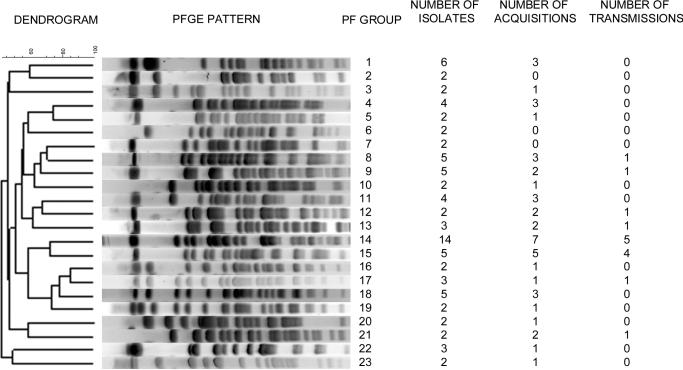
There were 23 different PFGE groups that contained isolates from >1 unique patient and, therefore, could be considered to meet the definition for patient-to-patient transmission (figure 1). Forty six (31%) of 149 patients with IRPA acquisitions acquired an IRPA strain that was classified by PFGE to have a genetic profile similar to that of another patients’ isolate. Of these patients, 16 (11%) of 149 had isolates with similar PFGE patterns and overlapping lengths of stay in the hospital.
Figure 1.
Pulsed-field (PF) gel electrophoresis (PFGE) dendrogram of 23 of the 248 PFGE groups of imipenem-resistant Pseudomonas aeruginosa. These 24 groups are the only PFGE types that have >1 isolate within the group. The data consist of the number of isolates, acquisitions, and transmissions within each group.
During the 5-year study period, there were 149 patients who acquired an IRPA, and 38 (26%) of these patients had an imipenem-susceptible P. aeruginosa isolate obtained from an ICU admission surveillance culture. Twenty-seven of these patients had drug-resistant isolates that were identical by PFGE to the susceptible isolate. One patient had a drug-resistant isolate that had a 1-band difference from the susceptible isolate, and therefore, we grouped the isolate in the identical category. Ten patients had a drug-resistant isolate that was unrelated by PFGE to their imipenem-susceptible P. aeruginosa isolate.
Seventy-four percent of patients (28 of 38 patients) who had imipenem-susceptible and imipenem-resistant isolates with closely related or identical PFGE patterns could be further divided into 4 groups defined by changes in the pattern of the isolate susceptibility to antibiotics other than imipenem. Table 1 shows that these 4 groups consist of isolates that changed with respect to (1) imipenem susceptibility only; (2) carbapenem (imipenem and meropenem) susceptibility; (3) carbapenems and other β-lactam antibiotic susceptibility; and (4) susceptibility to all β-lactams, fluoroquinolones, and gentamicin. The majority of the isolates had susceptibility patterns that changed only with respect to imipenem (39% of isolates) and/or meropenem (43% of isolates). Of the isolates with unrelated PFGE patterns, 1 IRPA acquisition was determined to be the results of patient-to-patient transmission by similar PFGE pattern and hospital stay overlap with that of another patient colonized with IRPA.
Table 1.
Susceptibility Data for Pseudomonas aeruginosa Isolates that Are Identical by Pulsed-Field Gel Electrophoresis
| Patient | Imipenem | Meropenem | Ceftazidime | Cefepime | Pipercillin | Piperacillin Tazobacam | Ciprofloxacin | Levofloxacin | Amikacin | Gentamicin |
|---|---|---|---|---|---|---|---|---|---|---|
| 1S | S | S | S | S | S | S | I | R | S | S |
| 1R | R | S | S | S | S | S | S | S | R | R |
| 2S | S | S | S | S | S | S | S | S | S | S |
| 2R | R | S | S | S | S | S | S | S | S | S |
| 3S | S | R | S | R | S | S | R | R | S | R |
| 3R | R | R | R | R | R | R | R | R | S | R |
| 4S | S | S | R | S | R | R | S | S | S | S |
| 4R | R | S | S | S | S | S | S | S | S | S |
| 5S | S | S | S | S | R | S | R | R | S | S |
| 5R | R | S | S | S | S | S | R | R | I | R |
| 6S | S | S | S | S | S | S | S | S | S | S |
| 6R | R | S | S | S | S | S | S | S | S | S |
| 7S | S | S | S | S | S | S | S | S | S | S |
| 7R | R | S | S | S | S | S | S | S | S | S |
| 8S | S | S | R | S | R | R | I | R | S | S |
| 8R | R | S | R | I | R | R | R | R | S | S |
| 9S | S | S | S | S | S | S | S | S | S | S |
| 9R | R | S | S | S | S | S | S | S | S | S |
| 10S | S | R | S | S | S | S | R | R | S | S |
| 10R | R | R | S | S | S | S | R | R | S | S |
| 11S | S | R | S | S | S | S | R | R | S | R |
| 11R | R | R | S | S | S | S | R | R | S | R |
| 12S | S | S | S | S | S | S | S | S | S | S |
| 12R | I | R | S | S | S | S | S | S | S | S |
| 13S | S | S | S | S | S | S | S | S | S | S |
| 13R | R | R | S | S | S | S | S | S | S | S |
| 14S | S | S | S | S | S | S | R | R | S | S |
| 14R | I | R | S | S | S | S | R | R | S | S |
| 15S | S | S | R | R | R | R | R | R | S | S |
| 15R | R | R | R | R | R | R | I | R | S | S |
| 16S | S | S | S | S | R | R | R | R | I | R |
| 16R | R | R | S | S | R | R | R | R | I | R |
| 17S | S | S | R | I | R | R | I | R | S | I |
| 17R | R | R | R | I | R | R | R | R | S | R |
| 18S | S | S | R | I | R | R | S | I | S | S |
| 18R | R | R | R | R | R | R | S | S | S | S |
| 19S | S | S | S | S | S | S | R | R | S | R |
| 19R | R | R | S | S | S | S | R | R | S | R |
| 20S | S | S | S | S | S | S | R | R | S | R |
| 20R | R | R | S | S | S | S | R | R | S | R |
| 21S | S | S | R | I | R | R | S | R | S | S |
| 21R | R | R | R | R | R | R | S | I | S | S |
| 22S | S | S | R | I | R | R | S | S | S | S |
| 22R | R | I | R | I | R | R | S | S | S | S |
| 23S | S | S | S | S | S | S | S | S | S | S |
| 23R | R | R | S | S | S | S | S | S | S | S |
| 24S | S | S | S | S | S | S | I | R | S | R |
| 24R | R | S | R | R | R | R | R | R | S | R |
| 25S | S | S | S | S | S | S | S | S | S | S |
| 25R | R | R | R | R | R | R | S | S | S | S |
| 26S | S | S | S | S | R | S | S | S | S | S |
| 26R | R | R | I | S | R | R | S | S | S | S |
| 27S | S | S | S | S | S | S | S | S | S | S |
| 27R | R | R | R | R | R | R | S | S | S | S |
| 28S | S | S | S | S | S | S | S | S | S | S |
| 28R | R | R | R | R | R | R | R | R | S | R |
NOTE. I, Intermediate; R resistant; S susceptible.
We analyzed the length of ICU stay among patients who acquired an IRPA. The mean ICU length of stay was 21 days (median length of stay, 12.9 days; 25th–75th percentile, 6.0–26.5 days). When the IRPA acquisitions were classified into subsets of patient-to-patient transmission and endogeneous acquisition, the mean ICU length of stay was 21 days (median length of stay, 11.0 days; 25th–75th percentile, 4.8–22.8 days) and 27 days (median length of stay, 18.3 days; 25th–75th percentile, 8.9–37.6 days), respectively (P = .27).
DISCUSSION
In this study, we analyzed the molecular epidemiology of patients who acquired an IRPA strain by use of perianal surveillance cultures obtained over a 5-year period in a large ICU population. We found that, of the 149 patients who acquired colonization with IRPA, 46 (31%) had isolates with PFGE patterns that were similar to those of isolates obtained from other patients; 16 (11%) of 149 patients also had hospital stay overlap, and 28 (19%) acquired colonization by endogenous flora.
Our study is unique in that we are able to study the acquisition of antibiotic-susceptible and/or antibiotic-resistant organisms over a period of several years in a unique population of >7000 patients. Previous studies have generally studied drug-susceptible P. aeruginosa and have disagreed on the amount of patient-to-patient transmission that occurs when patients acquire a P. aeruginosa strain while in the ICU. These studies also included far fewer patients in their cohorts [8–13]. Three major studies have found that cross-transmission played an important role in the acquisition of P. aeruginosa, with cross-transmission accounting for >30% of cases of P. aeruginosa acquisition. Olson et al [12] found that cross-transmission accounted for 12 (32%) of 37 cases in which patients acquired a susceptible P. aeruginosa strain. Pena et al [14] studied the acquisition of carbapenem-resistant P. aeruginosa using 2 serial incidence surveys that monitored colonization during two 1-month periods that were 1 year apart. The data showed that 9 (30%) of the patients in this study were found to have similar genotypes, but it was unclear whether these patients had hospital overlap. Bertrand et al [9] found that 107 (51%) of 208 patients who acquired a P. aeruginosa strain acquired that strain as a result of cross-transmission. Other studies, however, have indicated that cross-transmission does not play a major role in the acquisition of P. aeruginosa. Bonten et al [11] concluded from their study of 44 cases of acquisition that transmission of P. aeruginosa was polyclonal; they found that endogenous acquisition played a major role and that patient-to- patient transmission was not significant (occurring in only 8% of cases). Two other studies agreed that endogenous sources played an important role and that clonal dissemination was not a significant factor in the acquisition of antibiotic-resistant P. aeruginosa in an ICU population [10, 13]. Reuter et al [8] concluded that faucets play an important role in the transmission of P. aeruginosa. Their study analyzed a cohort of 45 patients colonized or infected with P. aeruginosa in a surgical ICU and found that 33% of these patients had genotypes that were identical to those of isolates grown from the faucet of the patient's room.
The literature on the role of patient-to-patient transmission of drug-susceptible P. aeruginosa is confusing, and there is even less data pertaining to IRPA. Therefore, we looked at a large cohort of patients over a 5-year period to determine the role of patient-to-patient transmission and antibiotic selective pressure on endogenous flora of IRPA colonization in the ICU population. In our study, we defined cases of patient-to-patient transmission on the basis of isolates with similar PFGE patterns and an overlap in hospital stay. We chose overlap in hospital stay, because other investigators have used a definition that extends beyond ICU stay, and we believe that this is an important variable in capturing all possible cases of patient-to-patient transmission [15–17]. However, our data are puzzling, because 46 (31%) of 149 patients who acquired this organism had isolates with genotypes similar to those of isolates obtained from other patients, but when we combined this data with hospital overlap data, there were only 16 (11%) of 149 patients who fulfilled both criteria. When we used this same definition of patient-to-patient transmission for other antimicrobial-resistant gram-negative organisms, we did not see this discrepancy between hospital overlap and PFGE results [18, 19]. This finding could suggest that P. aeruginosa has a reservoir that is undiscovered or that P. aeruginosa can survive for many months in the environment. On the other hand, it could be possible that, in some of these patients, imipenem-susceptible strains were acquired through patient-to-patient transmission and then became resistant to imipenem as a result of selective antibiotic pressure.
Within this cohort, there is a population of patients for whom we did not determine the mode of IRPA acquisition. We believe that there are several possible explanations. Imipenem-susceptible P. aeruginosa could have been transmitted by patient-to-patient spread and not determined in our study. Environmental contamination could have been a mode of transmission, because Pseudomonas species are known to be an environmental pathogen found in water sources. Environmental sources were not evaluated in our prospective cohort study. Obtaining perianal cultures daily for each patient would have been laborious and time-consuming but could have increased the chance of obtaining more patients with drug-susceptible strains that became resistant to imipenem while the patient was hospitalized in the ICU or could have pinpointed more patients with culture results positive for an IRPA, possibly adding to the patient-to-patient transmission group. The use of swab samples from an infection-control intervention study for vancomycin-resistant enterococci could have decreased the recovery of IRPA within this cohort of patients. However, we performed a validation study of the swab samples after multiple cultures and freeze/thaw cycles and showed that we were able to recover all but 1 (5%) of the IRPA isolates [20].
In conclusion, our study suggests that patient-to-patient transmission plays a role in the acquisition of IRPA colonization in the ICU at a tertiary-care hospital in a nonoutbreak setting. Our data also suggest that there is not a single mechanism of acquisition, and although patient-to-patient transmission is important, a patient's endogenous flora is also important. This suggests that a multifaceted approach is needed to control the emergence of IRPA.
Acknowledgments
We thank Colleen Reilly and Jingkun Zhu for their database support.
Financial support: the National Institutes of Health (grant K12 RR023250–03 to J.K.J. and grant R01 AI60859–01A1 and A.D.H.), the University of Maryland General Clinical Research Center (grant M01 RR 16500), and General Clinical Research Centers Program, National Center for Research Resources, National Institutes of Health.
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 25–28 October 2008 (abstract C2-202 and K-3493).
References
- 1.Gaynes R, Edwards JR. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 2.Von Korff M, Wagner EH, Saunders K. A chronic disease score from automated pharmacy data. J Clin Epidemiol. 1992;45:197–203. doi: 10.1016/0895-4356(92)90016-g. [DOI] [PubMed] [Google Scholar]
- 3.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol. 1992;45:613–9. doi: 10.1016/0895-4356(92)90133-8. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute . Methods for dilution antimicrobial susceptibility test for bacteria that grow areobically; approved standard, 7th ed. CLSI document M7-A7. Clinical and Laboratory Standards Institute; Wayne, PA: 2006. [Google Scholar]
- 5.Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:3707–2. doi: 10.1128/JCM.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romling U, Tummler B. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J Clin Microbiol. 2000;38:464–5. doi: 10.1128/jcm.38.1.464-465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reuter S, Sigge A, Wiedeck H, Trautmann M. Analysis of transmission pathways of Pseudomonas aeruginosa between patients and tap water outlets. Crit Care Med. 2002;30:2222–8. doi: 10.1097/00003246-200210000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand X, Thouverez M, Talon D, et al. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med. 2001;27:1263–8. doi: 10.1007/s001340100979. [DOI] [PubMed] [Google Scholar]
- 10.Berthelot P, Grattard F, Mahul P, et al. Prospective study of nosocomial colonization and infection due to Pseudomonas aeruginosa in mechanically ventilated patients. Intensive Care Med. 2001;27:503–12. doi: 10.1007/s001340100870. [DOI] [PubMed] [Google Scholar]
- 11.Bonten MJ, Bergmans DC, Speijer H, Stobberingh EE. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units: implications for infection control. Am J Respir Crit Care Med. 1999;160:1212–9. doi: 10.1164/ajrccm.160.4.9809031. [DOI] [PubMed] [Google Scholar]
- 12.Olson B, Weinstein RA, Nathan C, Chamberlin W, Kabins SA. Epidemiology of endemic Pseudomonas aeruginosa: why infection control efforts have failed. J Infect Dis. 1984;150:808–16. doi: 10.1093/infdis/150.6.808. [DOI] [PubMed] [Google Scholar]
- 13.Juan C, Gutierrez O, Oliver A, Ayestaran JI, Borrell N, Perez JL. Contribution of clonal dissemination and selection of mutants during therapy to Pseudomonas aeruginosa antimicrobial resistance in an intensive care unit setting. Clin Microbiol Infect. 2005;11:887–92. doi: 10.1111/j.1469-0691.2005.01251.x. [DOI] [PubMed] [Google Scholar]
- 14.Pena C, Guzman A, Suarez C, et al. Effects of carbapenem exposure on the risk for digestive tract carriage of intensive care unit–endemic carbapenem-resistant Pseudomonas aeruginosa strains in critically ill patients. Antimicrob Agents Chemother. 2007;51:1967–71. doi: 10.1128/AAC.01483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beyersmann J, Gastmeier P, Grundmann H, et al. Transmission-associated nosocomial infections: prolongation of intensive care unit stay and risk factor analysis using multistate models. Am J Infect Control. 2008;36:98–103. doi: 10.1016/j.ajic.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Grundmann H, Barwolff S, Tami A, et al. How many infections are caused by patient-to-patient transmission in intensive care units? Crit Care Med. 2005;33:946–51. doi: 10.1097/01.ccm.0000163223.26234.56. [DOI] [PubMed] [Google Scholar]
- 17.Vonberg RP, Wolter A, Chaberny IF, et al. Epidemiology of multi-drug-resistant gram-negative bacteria: data from an university hospital over a 36-month period. Int J Hyg Environ Health. 2008;211:251–7. doi: 10.1016/j.ijheh.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Harris AD, Kotetishvili M, Shurland S, et al. How important is patient-to-patient transmission in extended-spectrum β-lactamase Escherichia coli acquisition. Am J Infect Control. 2007;35:97–101. doi: 10.1016/j.ajic.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Harris AD, Perencevich EN, Johnson JK, et al. Patient-to-patient transmission is important in extended-spectrum β-lactamase-producing Klebsiella pneumoniae acquisition. Clin Infect Dis. 2007;45:1347–50. doi: 10.1086/522657. [DOI] [PubMed] [Google Scholar]
- 20.Green HP, Johnson JA, Furuno JP, et al. Impact of freezing on the future utility of archived surveillance culture specimens. Infect Control Hosp Epidemiol. 2007;28:886–8. doi: 10.1086/518843. [DOI] [PubMed] [Google Scholar]