Abstract
Achieving drug-free tolerance or successfully using only small doses of immunosuppression is a major goal in organ transplantation. To investigate the potential mechanisms by which some kidney transplant recipients can achieve operational tolerance, we compared the expression profiles of microRNA in peripheral blood mononuclear cells of operationally tolerant patients with those of stable patients treated with conventional immunosuppression. B cells from operationally tolerant patients overexpressed miR-142-3p. The expression of miR-142-3p was stable over time and was not modulated by immunosuppression. In Raji B cells, overexpression of miR-142-3p modulated nearly 1000 genes related to the immune response of B cells, including potential miR-142-3p targets and molecules previously identified in the blood of operationally tolerant patients. Furthermore, our results suggested that a negative feedback loop involving TGF-β signaling and miR-142-3p expression in B cells may contribute to the maintenance of tolerance. In summary, miR-142-3p expression in peripheral blood mononuclear cells correlates with operational tolerance. Whether upregulation of miR-142-3p modulates inflammatory responses to promote tolerance or is a result of this tolerance state requires further study.
The use of minimal doses of immunosuppression or even achievement of drug-free tolerance is a major goal in organ transplantation.1,2 Although the kidney is less susceptible to successful immunosuppressive drug withdrawal than the liver, where around 20% of transplant patients can be successfully weaned off immunonsuppression,3,4 an increasing number of kidney transplant recipients who continue to display good graft function in the absence of immunosuppressive drugs have been described in the literature.5–8 We and others have looked at the gene expression profile in PBMCs of such “operationally tolerant” kidney transplant recipients.7–12 The blood is a popular choice for analysis because it provides a noninvasive means for potential biomarker discovery, which is important in the case of tolerance; performing biopsies in such patients can be challenging in terms of ethical considerations and patient adherence. In these different studies, several key pathways were highlighted, such as a pathway implicating the TGF-β gene;10 in addition, several other genes have been highlighted as “key leader genes,” such as BANK-1 (B-cell scaffold protein with ankyrin repeats 1),11 a modulator of B-cell hyperactivation through AKT upon CD40 activation.13 The implication of B-cell–related genes correlated with a high number of B cells14 and to a gene signature enriched in B-cell–related genes in the blood of operationally tolerant kidney recipients, which was subsequently confirmed in three different studies.7,8,12 The role of B cells has also been reported in experimental rodent models, in which transfer of B cells from tolerant rats prolonged graft survival when administered to untreated recipients.15
The mechanisms involved in the maintenance of this phenomenon remain elusive, and the identification of related biomarkers remains instrumental to achieving safe drug minimization or complete weaning in clinical practice.
miRNA are small, endogenous, noncoding RNAs that can regulate the expression of a variety of genes by directly destabilizing their target mRNA.16,17 Furthermore, the incomplete pattern of target recognition allows a single miRNA to target hundreds of mRNAs, and conversely, a single mRNA to be targeted by multiple miRNAs, thus affecting a broad range of gene networks.18 Numerous studies have reported on the differential expression of miRNA in physiologic disorders or diseases,19,20 and miRNA has been found to be modulated in biopsy specimens from kidney transplant recipients.21–23 In this study, we investigated whether miRNA is modulated in the blood of patients with operational tolerance compared with a cohort of patients with stable graft function under classic immunosuppression.
Results
miRNA Profiling in PBMC from Kidney Transplant Recipients
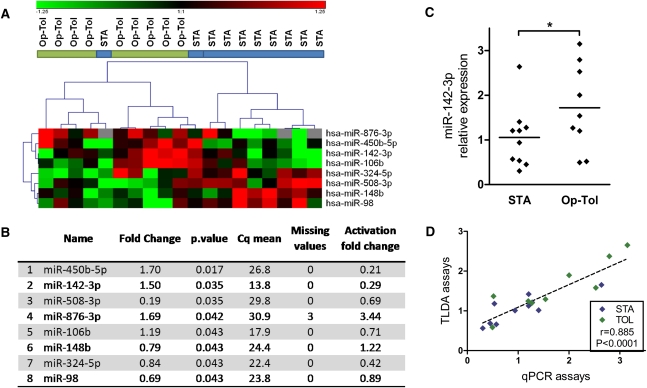
We first searched for a global miRNA profile in PBMCs from kidney transplant recipients using miRNA Taqman low-density arrays (TLDAs). The expression of 381 miRNAs was measured in PBMCs from 9 operationally tolerant patients and 10 STA recipients. A total of 266 miRNAs were expressed with a quantification cycle (Cq) inferior to 35 in at least half of samples from each group. We selected the eight top-ranked miRNAs on the basis of Mann–Whitney tests between the two groups of patients (Figure 1, A and B). According to the expression values of these eight miRNAs, the clear separation of the two groups of patients by clinical status was further observed using principal component analysis (Supplemental Figure 1). Among the eight differentially expressed miRNAs, four were overexpressed (miR-450b-5p, miR142-3p, miR-876-3p, and miR-106b) and four were underexpressed (miR508-3p, miR-148b, miR-324-5p, and miR-98) in PBMCs from operationally tolerant compared with STA patients.
Figure 1.
Differential miRNA in PBMCs from operationally tolerant compared with STA kidney transplant recipients. (A) The eight top-ranked miRNAs according to Mann–Whitney tests are represented in this heat map, in which blue bars represent STA patients (n=10) and green bars represent operationally tolerant (n=10) patients. The heat map represents normalized and color-coded relative expression values (2−ΔΔCq), in which red values indicate overexpression and green values indicate underexpression. (B) The eight miRNA are classified according to their uncorrected P values, which were used only for ranking miRNA rather than for an absolute identification of differential miRNA. FCs of miRNA after PHA/IL-2 activation of PBMCs from healthy volunteers are displayed in last column. (C) Relative expression values of miR-142-3p from TLDA assays are displayed in a scatter plot. (D) Correlation between TLDA assays and individual Taqman qPCR assays for miR-142-3p (9 operationally tolerant and 10 STA patients).
miRNA Profiling in PBMCs from Healthy Volunteers after Polyclonal Activation
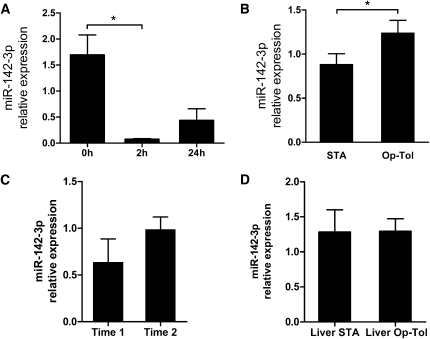
To further analyze the basal expression and modulation of these eight specific miRNAs, we performed miRNA profiling in PBMCs from healthy volunteers before and 24 hours after phytohemagglutinin A (PHA) and IL-2 stimulation (Supplemental Figure 2). Fifty-two miRNAs displayed a fold change (FC) superior to 2 (i.e., log2FC>1) and 59 others, an FC inferior to 2 (i.e., log2FC>-1), indicating a strong modulation of expression after PHA/IL-2 stimulation (Supplemental Table 5 and Supplemental Figure 2). Among the eight miRNAs differentially expressed between operationally tolerant and STA recipients, miR-450b-5p, miR-142-3p, and miR-324-5p were downregulated (FC=0.21, 0.29, and 0.42, respectively; Figure 1B) and miR-876-3p was upregulated (FC=3.4; Figure 1B) after PHA/IL-2 stimulation. We confirmed the downregulation of miR-142-3p as early as 2 hours and at 24 hours after PHA/IL-2 stimulation, using individual Taqman microRNA assays (Figure 2A).
Figure 2.
miR-142-3p expression in PBMCs. (A) miR-142-3p was downregulated in PBMCs from healthy volunteers after PHA/IL-2 stimulation as early as 2 hours and remained low for up to 24 hours. Mean± SEM of miR-142-3p relative expression (2−ΔΔCq relative to RNU6) in total PBMCs from three healthy volunteers stimulated with PHA (2 μg/ml) and IL-2 (150 U/ml) are represented. (B) miR-142-3p was significantly overexpressed in PBMCs from operationally tolerant (Op-Tol) kidney transplant recipients compared with STA patients. qPCR measurements were performed using individual Taqman qPCR assays with independent PBMC samples from TLDA assays (10 STA and 6 operationally tolerant patients). Means ± SEM of miR-142-3p relative expression (2−ΔΔCq relative to miR-374b) are represented. (C) miR-142-3p expression did not differ in PBMCs from liver transplant recipients before (n=11) and after (n=16) immunosuppression weaning. Means ± SEM of miR-142-3p relative expression (2−ΔΔCq relative to miR-374b) are represented. (D) miR-142-3p expression was stable between the two time points (5.5, 11, and 13 months between blood collections) for three operationally tolerant patients. Means ± SEM of miR-142-3p relative expression (2−ΔΔCq relative to miR-374b) are represented. *P<0.05.
Because miR-142-3p (1) appeared as the highest differentially expressed miRNA between operationally tolerant and STA recipients, (2) was underexpressed after PHA/IL-2 stimulation (Figure 2A), (3) is specific to the hematopoietic lineage,24–26 and (4) plays a role in lymphocyte functions,27 we focused on the potential role of this miRNA in our study. This miRNA is overexpressed in operationally tolerant compared with STA patients according to TLDA assays (Figure 1C).
Validation of miR-142-3p Overexpression and Stability over Time in PBMCs from Operationally Tolerant Recipients
Individual miR-142-3p Taqman quantitative PCR (qPCR) assays displays a good correlation (r=0.885; P<0.0001) (Figure 1D). Furthermore, we validated the overexpression of miR-142-3p in operationally tolerant recipients on 16 independent PBMC samples (6 operationally tolerant and 10 STA patients) (P=0.02; FC=1.58) (Figure 2B). The post-transplantation time was significantly higher in operationally tolerant than in STA patients (P=0.02; Table 1); however, we did not found any correlation between this time and miR-142-3p expression value (r=0.0342; P=0.845; Supplemental Figure 3). No significant difference was observed between the subgroups of patients for any of the clinical variables tested (age, sex, creatine level in blood, proteinuria, number of HLA mismatches). Finally, the expression of mir-142-3p was stable over time, as tested in PBMCs collected from three operationally tolerant recipients at two different time points (5.5, 11, and 13 months) (Figure 2C).
Table 1.
Summary of clinical data for the 35 kidney recipients used for measurement of miR-142-3p expression
| Patient Group | Recipient Age (yr) | Recipient Gender (Female/All) (n/n) | Time between Graft and Analysis (mo) | Blood Creatine Level (μΜ) | Proteinuria (g/24 hr) | Nonliving Donor/Living Donor (n/n) | Time between Immunosuppression Withdrawal and Analysis (yr) | HLA Mismatches (n: n) |
|---|---|---|---|---|---|---|---|---|
| STA | ||||||||
| patients (n) | 20 | 9/20 | 20 | 20 | 17 | 0/20 | 20 | |
| 0: 0 | ||||||||
| mean ± SD | 53.8±11.9 | 138.7±84.5 | 121.1±32.5 | 0.23±0.17 | 1: 0 | |||
| 2: 4 | ||||||||
| min | 20 | 36 | 53.0 | 0.07 | 3: 8 | |||
| max | 75 | 281 | 174.0 | 0.58 | ≥4: 8 | |||
| Operationally tolerant | ||||||||
| patients (n) | 15 | 5/15 | 15 | 15 | 11 | 3/15 | 15 | |
| 0: 3 | ||||||||
| mean ± SD | 50.6±17.0 | 208.3±74.0 | 110.1±22.1 | 0.25±0.34 | 7.4 | 1: 2 | ||
| 3.3 | 2: 1 | |||||||
| min | 20 | 105 | 68.0 | 0.00 | 1.8 | 3: 5 | ||
| max | 80 | 333 | 144.0 | 0.93 | 14.9 | ≥4: 4 | ||
| P value | 0.37 | 0.32 | 0.02 | 0.68 | 0.14 | 0.57 |
Mean, minimum (min), and maximum (max) values apply only to recipient age, recipient gender, time between graft and analysis, creatine level, and proteinuria.
Overexpression of miR-142-3p in the Blood of Operationally Tolerant Recipients Is Not Due to the Absence of Immunosuppression
Because operationally tolerant patients display stable graft function but no longer receive immunosuppression and because healthy volunteers also displayed an increased expression of miR-142-3p compared with STA patients (P=0.0038; FC=1.54) (data not shown), we hypothesized that immunosuppression treatment may modulate the blood expression of miR-142-3p. We thus measured its expression in PBMCs from tolerant liver transplant recipients before and after the patients entered an immunosuppressive drug weaning protocol (n=27), a population not available in the context of kidney transplantation. No difference was found in the PBMC expression of miR-142-3p with or without immunosuppression regimen in these liver recipients (Figure 2D).
miR-142-3p Is Overexpressed in B Cells from Operationally Tolerant Patients
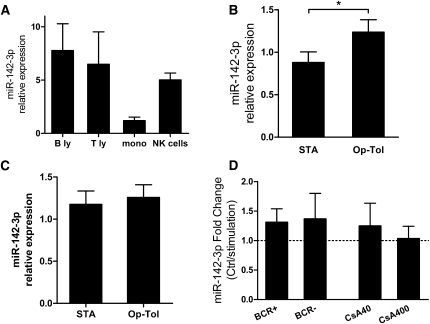
We then analyzed the expression of miR-142-3p in purified blood leukocyte subpopulations (T and B lymphocytes, monocytes, and NK cells) from healthy volunteers (n=3). Purification was typically greater than 95%. Figure 3A shows that miR-142-3p is expressed in all blood cell subsets, confirming previous reports.24–26 Because miR-142-3p was highly expressed in T and B lymphocytes from healthy volunteers, we next analyzed its expression in these two populations in transplant recipients. No difference was observed in the expression of miR-142-3p in the T lymphocyte subset (Figure 3C). In contrast, the expression of miR-142-3p was significantly higher in total B cells purified from operationally tolerant compared with STA recipients (P=0.01; FC= 2.84) (Figure 3B), whereas no difference was observed compared with healthy volunteers (data not shown). Therefore, purified B cells from healthy volunteers were cultured over 24 hours with physiologic and high doses of cyclosporine A (40 and 400 ng/µl). As shown in Figure 3D, no difference was observed for either dose of cyclosporine A used. These results are concordant with the absence of modulation of miR-142-3p in blood from liver-tolerant recipients before and after withdrawal of the treatment and suggest that miR-142-3p is not affected by immunosuppressive drugs in total PBMC or purified B cells.
Figure 3.
miR-142-3p expression in purified blood cell populations. (A) miR-142-3p expression in isolated subpopulations from PBMCs of three healthy volunteers (except NK cells; n=2). B ly, B lymphocytes; mono, mononuclear cells; T ly, T lymphocytes. (B) miR-142-3p expression exhibited increased expression in total B lymphocytes isolated from kidney operationally tolerant (Op-Tol) recipients (n=5) compared with STA patients (n=12) (P=0.0098; FC= 2.84). (C) miR-142-3p expression was similar in total T lymphocytes isolated from kidney operationally tolerant recipients (n=5) compared with STA patients (n=8). (D) miR-142-3p expression was not significantly modulated in purified B lymphocytes from healthy volunteers after B-cell–independent and B-cell–dependent stimulation and after cyclosporine A incubation during 24 hours of culture (n=4). Means ± SEM of miR-142-3p relative expression (2−ΔΔCq relative to RNU6) are represented. BCR, B-cell receptor. *P<0.05.
miR-142-3p Transfection in the Raji B-Cell Line
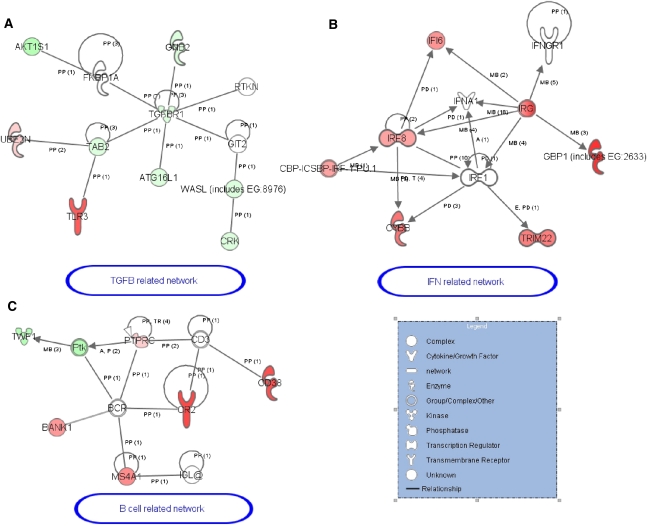
Because miRNA regulate mRNA levels,16,17 we overexpressed mir-142-3p in the Raji B-cell line using synthetic mimics and performed gene expression profiling using microarrays 24 hours after transfection. A total of 22,332 spots were filtered and the overexpression of miR-142-3p was found to induce the up- and downregulation of 492 and 489 transcripts, respectively. To provide a more comprehensive biologic interpretation of our finding, GOminer software was used to identify the over-represented gene ontology (GO) categories based on the differential gene lists compared with all other expressed genes in the microarray.28 Among the 492 overexpressed genes, the GO categories “immune response” (GO:0006955) and “B-cell activation” (GO:0042113) were identified with a total of 25 genes. Among the 489 underexpressed genes, GO categories related to cell communication (GO:0007154), “vesicle-mediated transport” (GO:0016192), and “small GTPase [guanosine triphosphatase] mediated signal transduction” (GO:0007264) were identified. We then identified 66 potential miR-142-3p targets downregulated subsequent to the miR-142-3p overexpression among the 242 genes predicted to be miR-142-3p targets (among at least 4 of 11 established miRNA target prediction databases computed by miRecords software).29 Mixing these 66 potential target genes and the 25 upregulated immune-related genes and using Ingenuity pathways analysis software, a gene network related to “Inflammation response, antimicrobial response and cell development” with 49 of these genes was found (Supplemental Figure 4). Within this gene network, we identified a subgroup of TGF-β–related genes in which the TGF-β receptor 1 gene (TGFBR1), a possible target of miR-142-3p, plays a central role (Figure 4A), together with a subgroup of IFN-γ–related genes (Figure 4B), and a subgroup of genes related to B cells (Figure 4C).
Figure 4.
Three subgroups of genes with modulated function after miR-142-3p overexpression in Raji cells. (A–C) These three subgroups of genes were extracted from the gene network created using IPA software (Supplemental Figure 3). Genes in red are upregulated and genes in green are downregulated in Raji transfected by miR-142-3p compared with control mimic. PP, protein–protein binding; PD, protein–DNA binding; MB, group/complex membership; TR, translocation.
The analysis of the detailed B-cell–related gene network after miR-142-3p transfection highlighted the upregulation of genes previously identified in PBMCs from operationally tolerant recipients, such as the key molecules MS4A1 (CD20), CR2, CD38,10 and BANK-1.11,12
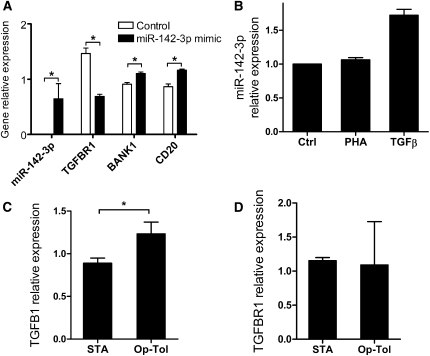
We next validated the array expression patterns of a few select targets and miRNA expression. The upregulation of miR-142-3p, MS4A1, and BANK1 in Raji cells transfected with miR-142-3p mimics was confirmed using individual Taqman assays (Figure 5A).
Figure 5.
miR-142-3p expression in Raji cells. (A) Confirmation of the overexpression of miR-142-3p, BANK1, and CD20 and the underexpression of TGFBR1 in Raji cells transfected with miR-142-3p mimic compared with a control mimic (n=4). (B) miR-142-3p expression was increased by TGF-β in Raji cells, whereas PHA had marginal effects in Raji cells, compared with control (PBS), after 24 hours of culture. Means ± SEM of miR-142-3p FC (control/stimulation) are represented. (C) TGF-β expression was significantly increased (P=0.04) in purified B cells from operationally tolerant (Op-Tol) (n=4) and STA (n=6) patients, whereas TGFBR1 was not. (D) Means ± SEM of gene relative expression (2−ΔΔCq relative to RNU6 for miR-142-3p or relative to HPRT1 for other genes) are represented. *P<0.05.
Implication of miR-142-3p in TGF-β Pathway
Using individual qPCR, we also confirmed the significant downregulation of TGFBR1 gene transcripts subsequent to miR-142-3p transfection in the Raji cell line compared with control (P=0.03; FC=0.47; Figure 5A). We then noted that the addition of TGF-β to the Raji culture media (5 ng/ml over 24 hours) induced an increase in miR-142-3p expression, whereas PHA stimulation did not (Figure 5B). Similarly, purified B cells from healthy volunteers were activated with independent and dependent B-cell receptor signaling for 24 hours (Figure 3D). As observed after PHA stimulation in Raji cells (Figure 5B), there is only nonsignificant and marginal regulation of miR-142-3p. This finding reinforces the fact that miR-142-3p needs specific stimulation, such as TGF-β, for its modulation (Figure 5B).
We then measured TGF-β1 and TGFBR1 transcript levels in purified B cells from operationally tolerant and STA patients. We found that TGF-β1 expression is increased in B cells from operationally tolerant patients (FC=1.4 compared with STA; P=0.04) (Figure 5C), whereas the level of TGFBR1 did not significantly differ between operationally tolerant and STA patients (Figure 5D).
Discussion
The achievement of long-term drug-free tolerance in solid organ transplantation is thought to be possible on the basis of observations in both liver4 and kidney3,5 transplantation. We and others have reported on an increased number of peripheral B cells and have identified modifications in blood gene expression in operationally tolerant kidney transplant recipients that involve TGF-β signaling and B-cell–related pathways.7,8,10,12,14 However, a clear mechanism or biologic process of peripheral regulation has yet to be identified in these patients. miRNAs are small RNA molecules with important roles in immune modulation, homeostasis, the development of immune diseases and the regulation of physiologic processes.19 Deregulated miRNA expression has been shown to be involved in several human immune-related diseases, such as multiple sclerosis,30,31 cancer,32,33 and rheumatoid arthritis,34 but their function and regulation processes are still far from being totally understood. In renal transplantation, two studies reported on the identification of miRNA profile signatures in biopsy samples from kidney transplant patients with acute rejection episodes,21,23 suggesting that miRNA expression profiling may be used to monitor allograft status. To our knowledge, and particularly in operationally tolerant kidney transplant recipients, no miRNA expression analyses have been performed in the blood.
In this study, we report on the modulation of expression of eight miRNAs in PBMCs from kidney graft recipients with drug-free operational tolerance compared with patients with stable graft function under immunosuppression. Our choice to compare operationally tolerant patients with patients who have stable graft function under immunosuppression was based on the fact that the latter population is the most likely to benefit from immunosuppression minimization, whereas patients with chronic rejection or healthy volunteers would not. Unsupervised hierarchical clustering analysis based on the expression of these eight miRNAs only led to the clustering of operationally tolerant patients together (Figure 1A). Among these eight miRNA, miR-142-3p was highly expressed in PBMCs from operationally tolerant patients (Figure 1C and Figure 2B). This miRNA has been described as a hematopoietic-restricted lineage miRNA.24–27 We found that miR-142-3p was decreased after PHA/IL-2 activation, further favoring a regulatory loop, as already described for other miRNAs, such as miR-125b, miR-16b, or miR-148b.35–37 miR-142-3p expression has been reported as playing a role in CD4+CD25+Treg function.27 Although miR-142-3p was highly expressed in T lymphocytes, we did not observe any differential expression in T lymphocytes between operationally tolerant and STA recipients (Figure 3C). In contrast, we found a significant overexpression of miR-142-3p in the B-lymphocyte subset of operationally tolerant compared with STA patients (P=0.0098; FC=2.84) (Figure 3B) and also compared with patients with signs of chronic antibody-mediated rejection (data not shown). We also found that its expression was not modulated by immunosuppressive treatment in tolerant liver transplant recipients (Figure 2D) or when purified B cells from human volunteers were cultured with cyclosporine A in vitro (Figure 3D), indicating that its overexpression in operationally tolerant kidney transplant recipients was not just a consequence of the absence of immunosuppression.
Interestingly, these data are in accordance with an increased number of peripheral B cells14 and with the specific B-cell–enriched gene profile that we and others previously reported in the blood of these patients.7,8,10,12 Although the increased expression of miR-142-3p in PBMCs from operationally tolerant kidney recipients is probably in part due to the increased number of blood B cells in these patients, because miR-142-3p expression in PBMCs is correlated with B cell number (data not shown), we report that purified B cells from operationally tolerant also expressed more miR-142-3p. This miRNA has already been reported to be expressed specifically in hematopoietic tissues and particularly in B cells, but a precise role in the B-cell compartment has yet to be attributed.24–27,38
miR-142-3p expression has also been associated with tubular atrophy and interstitial fibrosis of renal transplants.22 Moreover, miR-142-3p was overexpressed in B lymphocytes from the blood of operationally tolerant patients, and this analysis performed in peripheral blood does not exclude a different expression profile within the graft itself. In addition, we did not find any correlation between miR-142-3p expression and post-transplantation time and did find miR-142-3p expression to be stable over time (Figure 2C); these results indicate that miR-142-3p is independent of post-transplantation time.
Recently, Guo and colleagues demonstrated that miRNA decreased protein production mostly by lowering mRNA levels.17 Thus, we used gene microarrays to measure the effect of miR-142-3p overexpression in B cells, looking at the effect of experimental miR-142-3p transfection in a stable Raji B-cell line. Whereas the overexpression of this single miRNA repressed many genes, we observed the paradoxical biologic effect of miR-142-3p, which also induced gene upregulation, as already described for other miRNAs.39 A large number of genes related to B cells, IFN-γ, and TGF-β signaling were upregulated after overexpression of miR-142-3p in the Raji B-cell line. Of note, in previous studies we had already observed some of these genes, such as MS4A1 (CD20); this gene was part of the 49-gene signature that correctly classified kidney operationally tolerant patients10 and was part of the best-classifier genes in the blood and urine of operationally tolerant patients in the study by Newell and colleagues.7 Similarly, miR-142-3p overexpression led to the upregulation of BANK1 transcripts, one of the key leader genes upregulated in the blood of kidney operationally tolerant recipients,11 independent of immunosuppressive treatment.12 BANK1 is an inhibitory adaptor protein highly expressed in peripheral B cells and is a modulator of hyperactive B-cell responses by inhibiting AKT activation upon CD40 signaling.13
Finally, the TGF-β signaling pathway was also affected after miR-142-3p overexpression in the Raji cell line and in B cells from operationally tolerant patients. Interestingly, miR-106b, one of the eight differentially overexpressed miRNA in PBMCs from operationally tolerant kidney transplant recipients, also affects downstream effector molecules of TGF-β signaling.40,41 In a previous report, we showed that among the specific and unique blood signature of 49 genes associated with tolerance, 27% of the genes modulated in blood from operationally tolerant patients could be regulated by the TGF-β even though TGF-β was not significantly increased in total PBMCs from operationally tolerant patients (increased from 30% only).10 TGF-β is involved in various animal models of tolerance,42–44 plays a role in immune regulation and homeostasis of Treg cells,45 and is known for its intrinsic suppressive properties.46
We also report here that TGF-β stimulation of Raji cells induced an increased expression of miR-142-3p and that TGF-β1 expression is increased in B cells from operationally tolerant patients, whereas TGFBR1 is not; this finding suggests a negative feedback loop between this cytokine and miR-142-3p in B cells. Such a process of regulation, already described for different miRNAs,40 suggests that miRNAs act as key gene switches and as fine-tuning molecules, depending on the compartment and the specific biologic context.20 Therefore, our data suggest that mir-142-3p is also implicated in the TGF-β pathway. However, because the modulation of one miRNA may affect multiple mRNAs that are also regulated by several other miRNAs, a direct link between these two molecules cannot be predicted at this stage.
Our findings show that overexpression of miR-142-3p in B cells correlates to the state of operational tolerance in kidney transplant recipients. They also point toward a possible negative feedback loop between TGF-β and miR-142-3p in B cells. The mechanism driving and maintaining spontaneous tolerance in which TGF-β could be implicated remains unclear. Further investigations are now needed to find out whether this overexpression of miR-142-3p in B cells contributes to controlling inflammatory responses and tolerance maintenance or is only a consequence of this tolerance state.
Concise Methods
Patients
A total of 86 individuals were enrolled in this study: 15 operationally tolerant patients, 34 STA patients, 10 healthy volunteers, 11 liver recipients with stable graft function, and 16 drug-free liver recipients from the Nantes hospital in France and the Barcelona hospital in Spain. The two local ethics committees approved all aspects of the study, and all patients gave informed consent. The clinical information is described in the Supplemental Materials and Methods, clinical data are summarized in Table 1, and detailed clinical data are provided in Supplemental Table 6.
miRNA Profiling
miRNA profiling was performed using TLDA microRNA Cards pool A set, version 2.0 (Applied Biosystems, Foster City, CA), in accordance with the manufacturer’s recommendations. Normalization was performed by subtracting the mean Cq of the measured miRNA.47 After normalization, miRNA were ranked using P values from nonparametric Mann–Whitney tests, which do not require normal assumptions or asymptotic conditions. These uncorrected P values for multiple testing were used only for ranking miRNA and are not an absolute identification of differential miRNA.
miRNA Individual Assays
Individual miRNA expression was measured with Taqman miRNA assays (Applied Biosystems) using probes for miR-142-3p (assay ID: 000464), RNU6 (assay ID: 001973), and miR-374b (assay ID: 001319), starting with 10 ng of total RNA, on an ABI Prism 7900 HT.
Gene Expression Microarray Analysis
Gene expression was measured using whole human genome 4×44K Agilent microarrays, following the manufacturer’s two-color protocol (Agilent Technologies Inc., Palo Alto, CA). A total of 22,332 spots were filtered. GOminer software and Ingenuity Pathway Analysis 6.5 software (Ingenuity Systems Inc.) were used to assess biologic significance of genes selected with Mann–Whitney tests. Raw microarray data were deposited in the Gene Expression Ominbus (GEO) database (accession number GSE28456).
Statistical Analyses
The nonparametric Mann–Whitney test, Kruskal Wallis test, or paired Wilcoxon test was used for group comparisons using Graph PadPrism software, version 4. Differences were defined as statistically significant with P<0.05 and highly significant with P<0.01.
Additional details can be found in Supplemental Materials and Methods.
Disclosures
None.
Acknowledgments
We thank all the patients who participated in this study and the physicians who helped us recruit patients: J.F. Subra, F. Villemain, C. Legendre, E. Thervet, F.J. Bemelman, G. Roussey, G. Orlando, A. Garnier, H. Jambon, H. Le Monies De Sagazan, L. Braun, C. Noël, E. Pillebout, M.C. Moal, C. Cantarell, A. Hoitsma, M. Ranbant, A. Testa. We thank the transcriptome core facility of Nantes for technical assistance with gene expression microarrays. We also thank Yohann Foucher for critical review of the manuscript.
The Institut de Transplantation Urologie belongs to the Fondation Centaure, which supports a French research network in transplantation. R.D. was supported by the Fondation Centaure and by a grant from the Fondation pour la Recherche Médicale.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2011060543/-/DCSupplemental.
References
- 1.Danger R, Giral M, Soulillou JP, Brouard S: Rationale and criteria of eligibility for calcineurin inhibitor interruption following kidney transplantation. Curr Opin Organ Transplant 13: 609–613, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Ashton-Chess J, Giral M, Brouard S, Soulillou JP: Spontaneous operational tolerance after immunosuppressive drug withdrawal in clinical renal allotransplantation. Transplantation 84: 1215–1219, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Orlando G, Soker S, Wood K: Operational tolerance after liver transplantation. J Hepatol 50: 1247–1257, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Martínez-Llordella M, Puig-Pey I, Orlando G, Ramoni M, Tisone G, Rimola A, Lerut J, Latinne D, Margarit C, Bilbao I, Brouard S, Hernández-Fuentes M, Soulillou JP, Sánchez-Fueyo A: Multiparameter immune profiling of operational tolerance in liver transplantation. Am J Transplant 7: 309–319, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Roussey-Kesler G, Giral M, Moreau A, Subra JF, Legendre C, Noël C, Pillebout E, Brouard S, Soulillou JP: Clinical operational tolerance after kidney transplantation. Am J Transplant 6: 736–746, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, Shaffer J, Preffer FI, Ding R, Sharma V, Fishman JA, Dey B, Ko DS, Hertl M, Goes NB, Wong W, Williams WW, Jr, Colvin RB, Sykes M, Sachs DH: HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med 358: 353–361, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, Hernandez-Fuentes MP, Turka LA, Seyfert-Margolis VL. Immune Tolerance Network ST507 Study Group: Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 120: 1836–1847, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, Chapman S, Craciun L, Sergeant R, Brouard S, Rovis F, Jimenez E, Ballow A, Giral M, Rebollo-Mesa I, Le Moine A, Braudeau C, Hilton R, Gerstmayer B, Bourcier K, Sharif A, Krajewska M, Lord GM, Roberts I, Goldman M, Wood KJ, Newell K, Seyfert-Margolis V, Warrens AN, Janssen U, Volk HD, Soulillou JP, Hernandez-Fuentes MP, Lechler RI: Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest 120: 1848–1861, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braud C, Baeten D, Giral M, Pallier A, Ashton-Chess J, Braudeau C, Chevalier C, Lebars A, Léger J, Moreau A, Pechkova E, Nicolini C, Soulillou JP, Brouard S: Immunosuppressive drug-free operational immune tolerance in human kidney transplant recipients: Part I. Blood gene expression statistical analysis. J Cell Biochem 103: 1681–1692, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Brouard S, Mansfield E, Braud C, Li L, Giral M, Hsieh SC, Baeten D, Zhang M, Ashton-Chess J, Braudeau C, Hsieh F, Dupont A, Pallier A, Moreau A, Louis S, Ruiz C, Salvatierra O, Soulillou JP, Sarwal M: Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci USA 104: 15448–15453, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivozhelezov V, Braud C, Giacomelli L, Pechkova E, Giral M, Soulillou JP, Brouard S, Nicolini C: Immunosuppressive drug-free operational immune tolerance in human kidney transplants recipients. Part II. Non-statistical gene microarray analysis. J Cell Biochem 103: 1693–1706, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Pallier A, Hillion S, Danger R, Giral M, Racapé M, Degauque N, Dugast E, Ashton-Chess J, Pettré S, Lozano JJ, Bataille R, Devys A, Cesbron-Gautier A, Braudeau C, Larrose C, Soulillou JP, Brouard S: Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype. Kidney Int 78: 503–513, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, Kurosaki T: BANK negatively regulates Akt activation and subsequent B cell responses. Immunity 24: 259–268, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Louis S, Braudeau C, Giral M, Dupont A, Moizant F, Robillard N, Moreau A, Soulillou JP, Brouard S: Contrasting CD25hiCD4+T cells/FOXP3 patterns in chronic rejection and operational drug-free tolerance. Transplantation 81: 398–407, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Le Texier L, Thebault P, Lavault A, Usal C, Merieau E, Quillard T, Charreau B, Soulillou JP, Cuturi MC, Brouard S, Chiffoleau E: Long-term allograft tolerance is characterized by the accumulation of B cells exhibiting an inhibited profile. Am J Transplant 11: 429–438, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP: The impact of microRNAs on protein output. Nature 455: 64–71, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H, Ingolia NT, Weissman JS, Bartel DP: Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter ME: Targeting of mRNAs by multiple miRNAs: The next step. Oncogene 29: 2161–2164, 2010 [DOI] [PubMed] [Google Scholar]
- 19.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D: Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 10: 111–122, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Schott J, Stoecklin G: Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA 1: 432–456, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M: MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA 106:5330–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scian MJ, Maluf DG, Archer KJ, Suh JL, Massey D, Fassnacht RC, Whitehill B, Sharma A, King A, Gehr T, Cotterell A, Posner MP, Mas V: Gene expression changes are associated with loss of kidney graft function and interstitial fibrosis and tubular atrophy: Diagnosis versus prediction. Transplantation 91: 657–665, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H: Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol 19: 81–85, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Chen CZ, Li L, Lodish HF, Bartel DP: MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T: A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkerova M, Belickova M, Bruchova H: Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol 81: 304–310, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, Zhang GM, Feng ZH: miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep 10: 180–185, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN: GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol 4: R28–R28, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T: miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res 37[Database issue]: D105–D110, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P, Inza I, Lozano JA, Castillo-Triviño T, Asensio A, Olaskoaga J, López de Munain A: Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE 4: e6309, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E: MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132: 3342–3352, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Farazi TA, Spitzer JI, Morozov P, Tuschl T: miRNAs in human cancer. J Pathol 223: 102–115, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD: The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood 117: 1121–1129, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK: Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther 10: R101, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM: Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X: MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol 185: 7244–7251, 2010 [DOI] [PubMed] [Google Scholar]
- 37.De Santis G, Ferracin M, Biondani A, Caniatti L, Rosaria Tola M, Castellazzi M, Zagatti B, Battistini L, Borsellino G, Fainardi E, Gavioli R, Negrini M, Furlan R, Granieri E: Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol 226: 165–171, 2010 [DOI] [PubMed] [Google Scholar]
- 38.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS: Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113: 3754–3764, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikantan S, Marasa BS, Becker KG, Gorospe M, Abdelmohsen K: Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle 10: 751–759, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A: E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H, Liu Y, Ma C, Huang L, Zhang L, Qin C: miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-β type II receptor. Brain Res 1357: 166–174, 2010 [DOI] [PubMed] [Google Scholar]
- 42.Chiffoleau E, Bériou G, Dutartre P, Usal C, Soulillou JP, Cuturi MC: Role for thymic and splenic regulatory CD4+ T cells induced by donor dendritic cells in allograft tolerance by LF15-0195 treatment. J Immunol 168: 5058–5069, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Gagne K, Brouard S, Guillet M, Cuturi MC, Soulillou JP: TGF-beta1 and donor dendritic cells are common key components in donor-specific blood transfusion and anti-class II heart graft enhancement, whereas tolerance induction also required inflammatory cytokines down-regulation. Eur J Immunol 31: 3111–3120, 2001 [DOI] [PubMed] [Google Scholar]
- 44.Josien R, Douillard P, Guillot C, Müschen M, Anegon I, Chetritt J, Menoret S, Vignes C, Soulillou JP, Cuturi MC: A critical role for transforming growth factor-beta in donor transfusion-induced allograft tolerance. J Clin Invest 102: 1920–1926, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bommireddy R, Doetschman T: TGFbeta1 and Treg cells: Alliance for tolerance. Trends Mol Med 13: 492–501, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan YY, Flavell RA: ‘Yin-Yang’ functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol Rev 220: 199–213, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J: A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10: R64, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]