Abstract
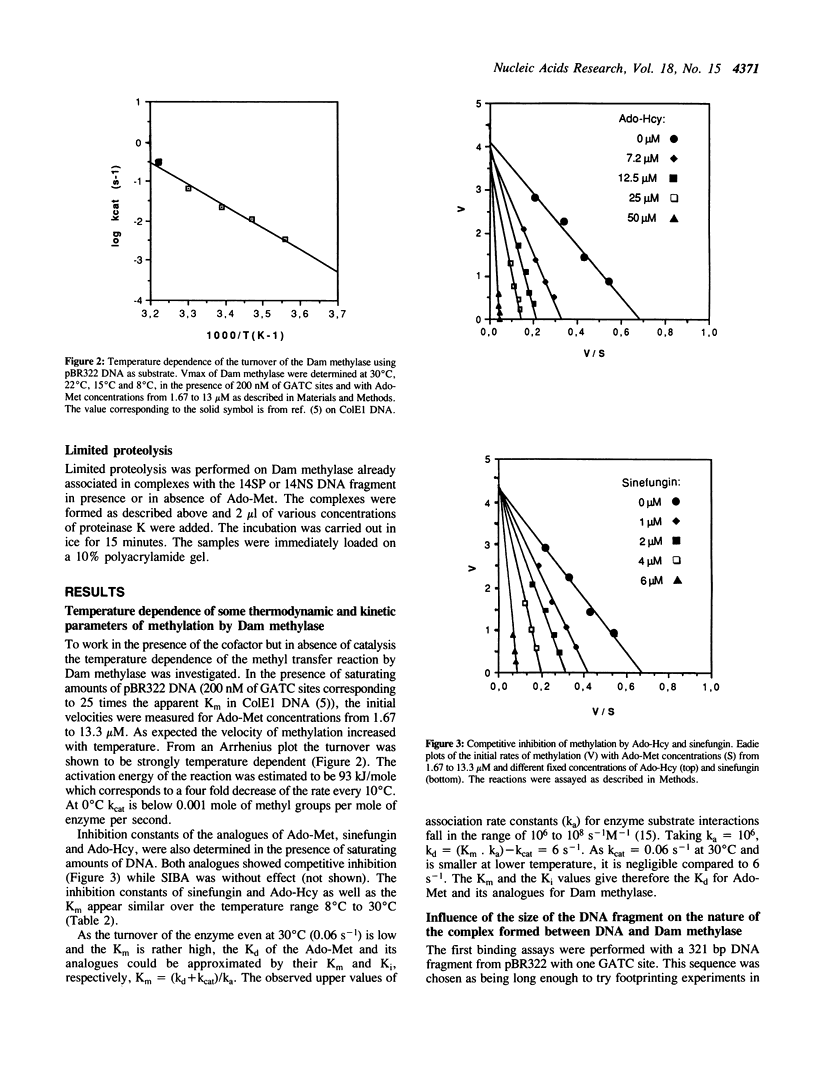
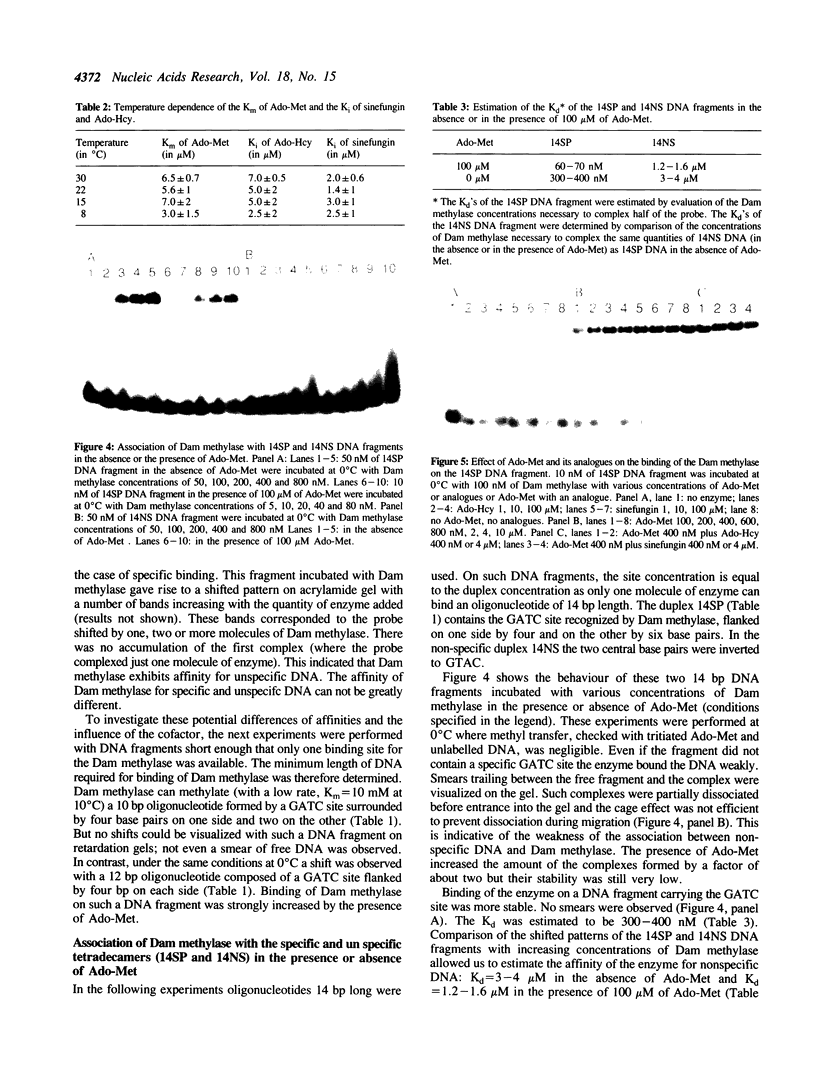
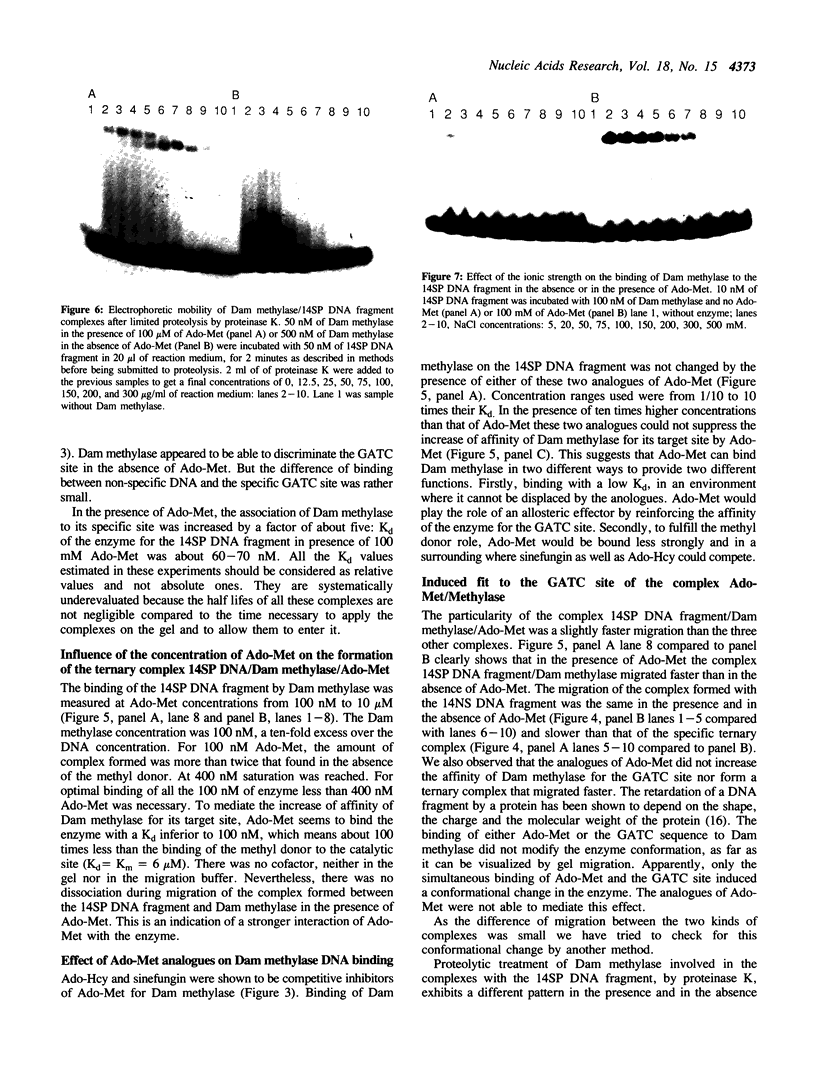
The turnover of DNA-adenine-methylase of E. coli strongly decreases when the temperature is lowered. This has allowed us to study the binding of Dam methylase on 14 bp DNA fragments at 0 degrees C by gel retardation in the presence of Ado-Met, but without methylation taking place. The enzyme can bind non-specific DNA with low affinity. Binding to the specific sequence occurs in the absence of S-adenosyl-methionine (Ado-Met), but is activated by the presence of the methyl donor. The two competitive inhibitors of Ado-Met, sinefungin and S-adenosyl-homocysteine, can neither activate this binding to DNA by themselves, nor inhibit this activation by Ado-Met. This suggests that Ado-Met could bind to Dam methylase in two different environments. In one of them, it could play the role of an allosteric effector which would reinforce the affinity of the enzyme for the GATC site. The analogues can not compete for such binding. In the other environment Ado-Met would be in the catalytic site and could be exchanged by its analogues. We have also visualized conformational changes in Dam methylase induced by the simultaneous binding of Ado-Met and the specific target sequence of the enzyme, by an anomaly of migration and partial resistance to proteolytic treatment of the ternary complex Ado-Met/Dam methylase/GATC.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bading H. Determination of the molecular weight of DNA-bound protein(s) responsible for gel electrophoretic mobility shift of linear DNA fragments examplified with purified viral myb protein. Nucleic Acids Res. 1988 Jun 24;16(12):5241–5248. doi: 10.1093/nar/16.12.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F., Marinus M. G. The great GATC: DNA methylation in E. coli. Trends Genet. 1989 May;5(5):139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- Bergerat A., Kriebardis A., Guschlbauer W. Preferential site-specific hemimethylation of GATC sites in pBR322 DNA by Dam methyltransferase from Escherichia coli. J Biol Chem. 1989 Mar 5;264(7):4064–4070. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Friedman S. Binding of the EcoRII methylase to azacytosine-containing DNA. Nucleic Acids Res. 1986 Jun 11;14(11):4543–4556. doi: 10.1093/nar/14.11.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz H. J., Belagaje R., Brown E. L., Fritz R. H., Jones R. A., Lees R. G., Khorana H. G. High-pressure liquid chromatography in polynucleotide synthesis. Biochemistry. 1978 Apr 4;17(7):1257–1267. doi: 10.1021/bi00600a020. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gersonde K., Sick H., Overkamp M., Smith K. M., Parish D. W. Bohr effect in monomeric insect haemoglobins controlled by O2 off-rate and modulated by haem-rotational disorder. Eur J Biochem. 1986 Jun 2;157(2):393–404. doi: 10.1111/j.1432-1033.1986.tb09681.x. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Bickle T. A., Yuan R. The role of S-adenosylmethionine in the cleavage of deoxyribonucleic acid by the restriction endonuclease from Escherichia coli K. J Biol Chem. 1975 Jun 10;250(11):4159–4164. [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Rubin R. A., Newman A., Modrich P. Structures and mechanisms of Eco RI DNA restriction and modification enzymes. Gene Amplif Anal. 1981;1:165–179. [PubMed] [Google Scholar]
- Kriebardis A., Guschlbauer W. dam methylase from E. coli. Circular dichroism investigations of the secondary structure and influence of S-adenosylmethionine. FEBS Lett. 1987 Mar 23;213(2):297–300. doi: 10.1016/0014-5793(87)81509-0. [DOI] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D., Michalak M., Daisley S. L., Glick R. A method for the purification of E. coli plasmid DNA by homogeneous lysis and polyethylene glycol precipitation. Mol Biol Rep. 1983 Aug;9(3):191–195. doi: 10.1007/BF00775367. [DOI] [PubMed] [Google Scholar]
- Som S., Friedman S. Direct photolabeling of the EcoRII methyltransferase with S-adenosyl-L-methionine. J Biol Chem. 1990 Mar 15;265(8):4278–4283. [PubMed] [Google Scholar]
- Steitz T. A., Harrison R., Weber I. T., Leahy M. Ligand-induced conformational changes in proteins. Ciba Found Symp. 1983;93:25–46. doi: 10.1002/9780470720752.ch3. [DOI] [PubMed] [Google Scholar]
- Wagner J., Danzin C., Mamont P. Reversed-phase ion-pair liquid chromatographic procedure for the simultaneous analysis of S-adenosylmethionine, its metabolites and the natural polyamines. J Chromatogr. 1982 Feb 12;227(2):349–368. doi: 10.1016/s0378-4347(00)80389-8. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]