Abstract
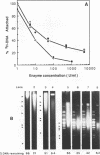
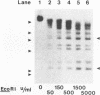
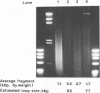
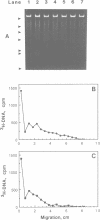
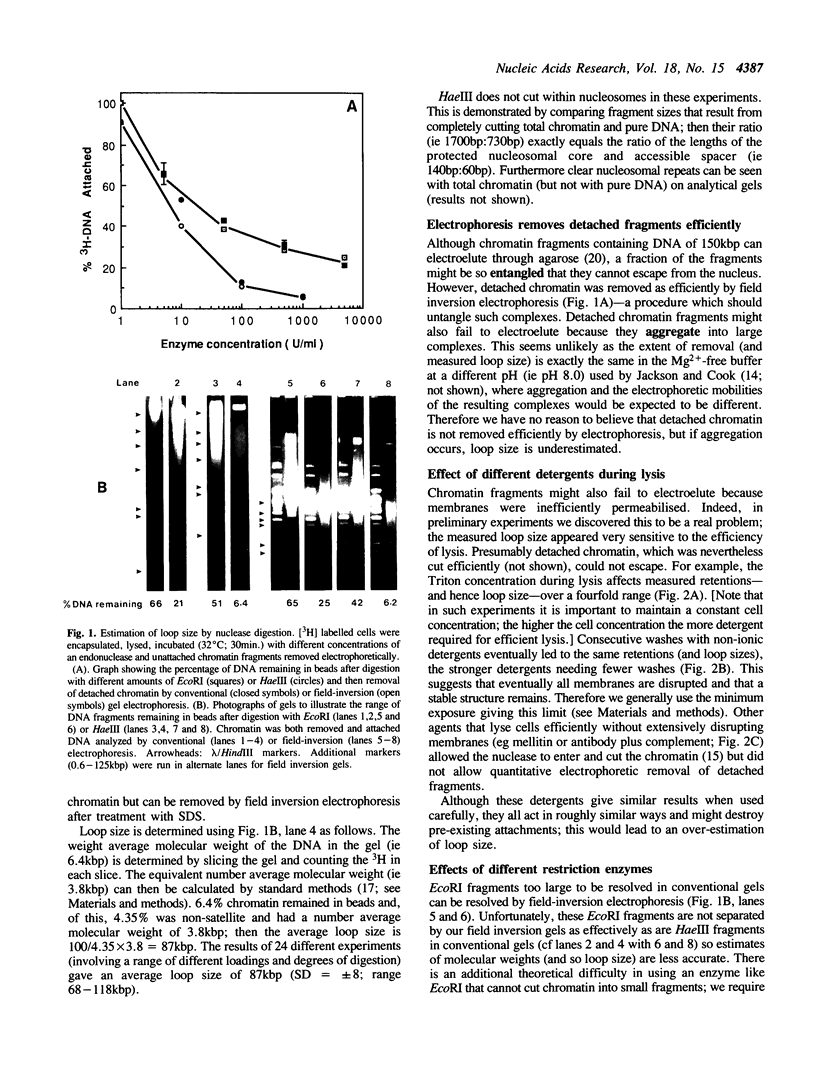
Although it is widely believed that eukaryotic DNA is looped by attachment to a nucleoskeleton, there is controversy about its composition and which sequences are attached to it. As most nuclear derivatives are isolated using unphysiological conditions, the criticism that attachments seen in vitro are generated artifactually has been difficult to rebut. Therefore we have re-investigated attachments of chromatin to the skeleton using physiological conditions. HeLa cells are encapsulated in agarose microbeads and lysed using Triton in a 'physiological' buffer. Then, most chromatin can be electroeluted after treatment with a restriction enzyme to leave some at the base of the loops still attached. Analysis of the size and amounts of these residual fragments indicates that the loops are 80-90kbp long. The residual fragments are stably attached, with about 1kbp of each fragment protected from nuclease attack. This is very much longer than a typical protein-binding site of 10-20bp.
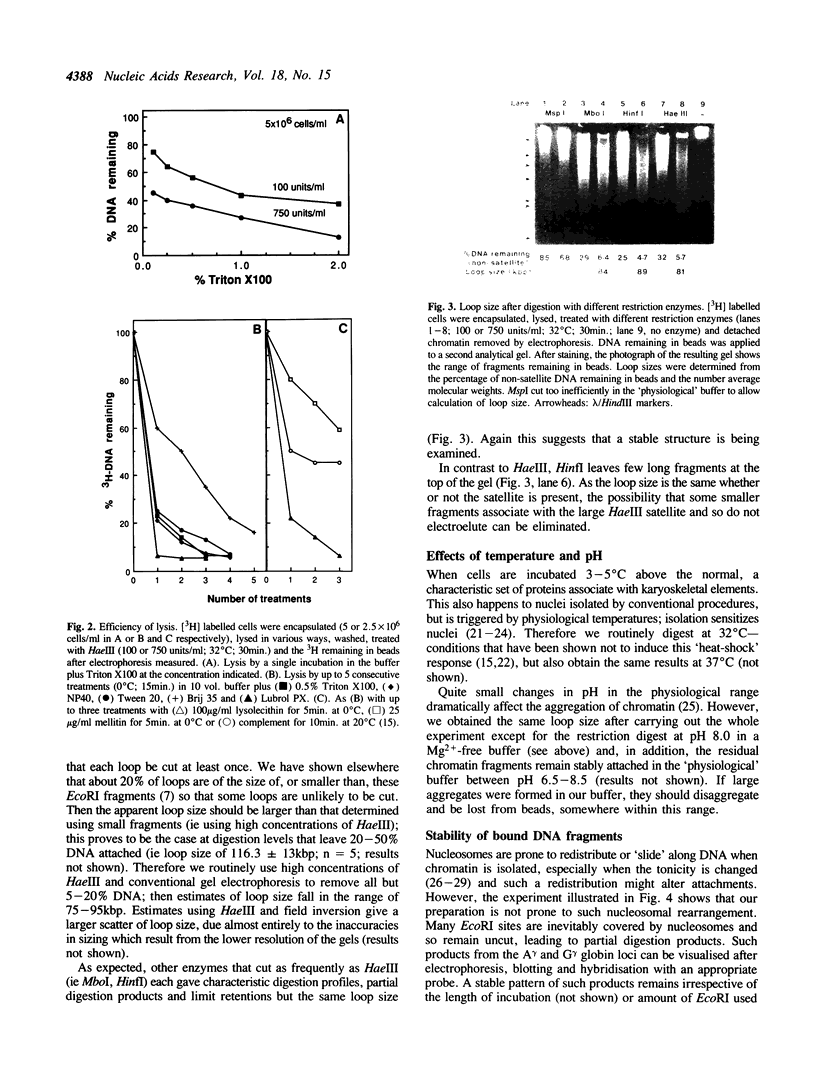
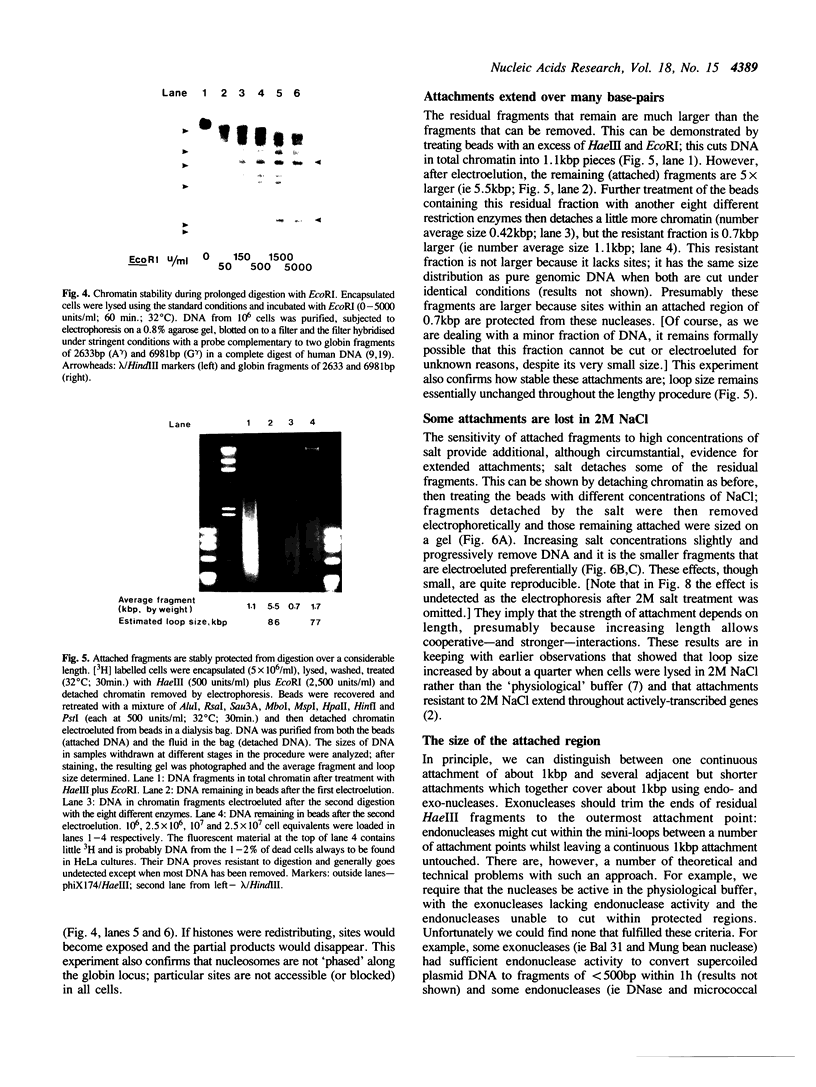
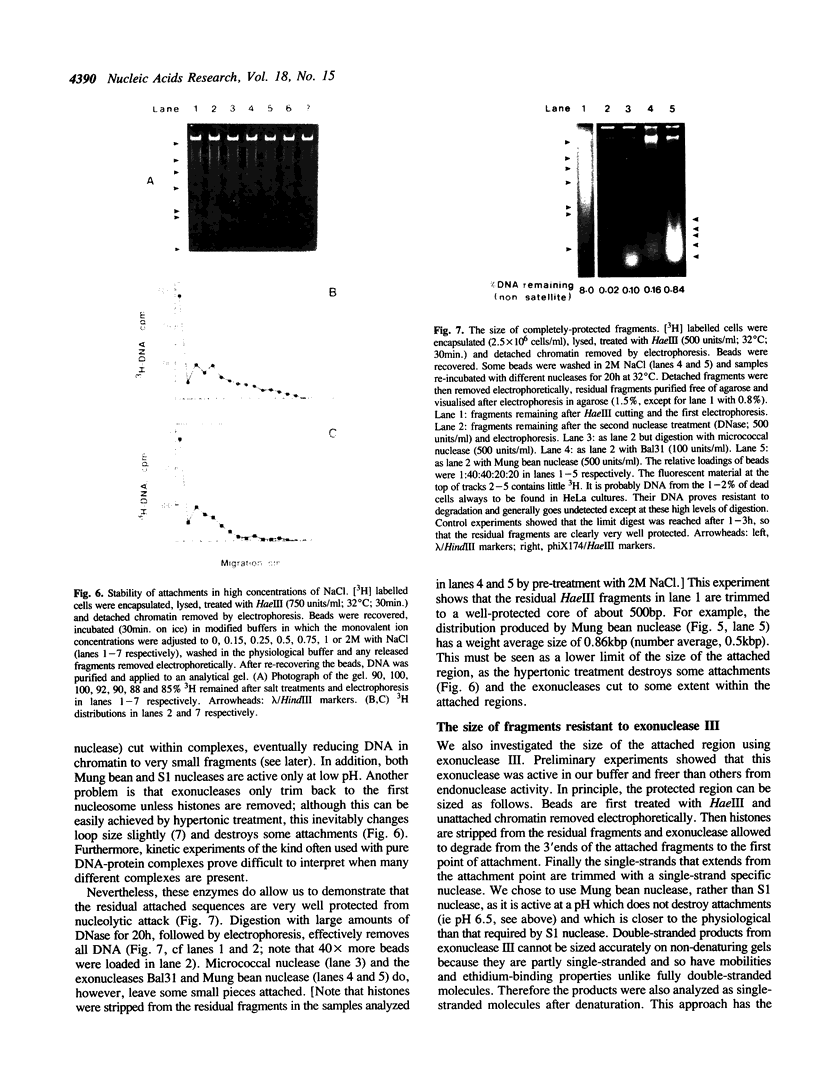
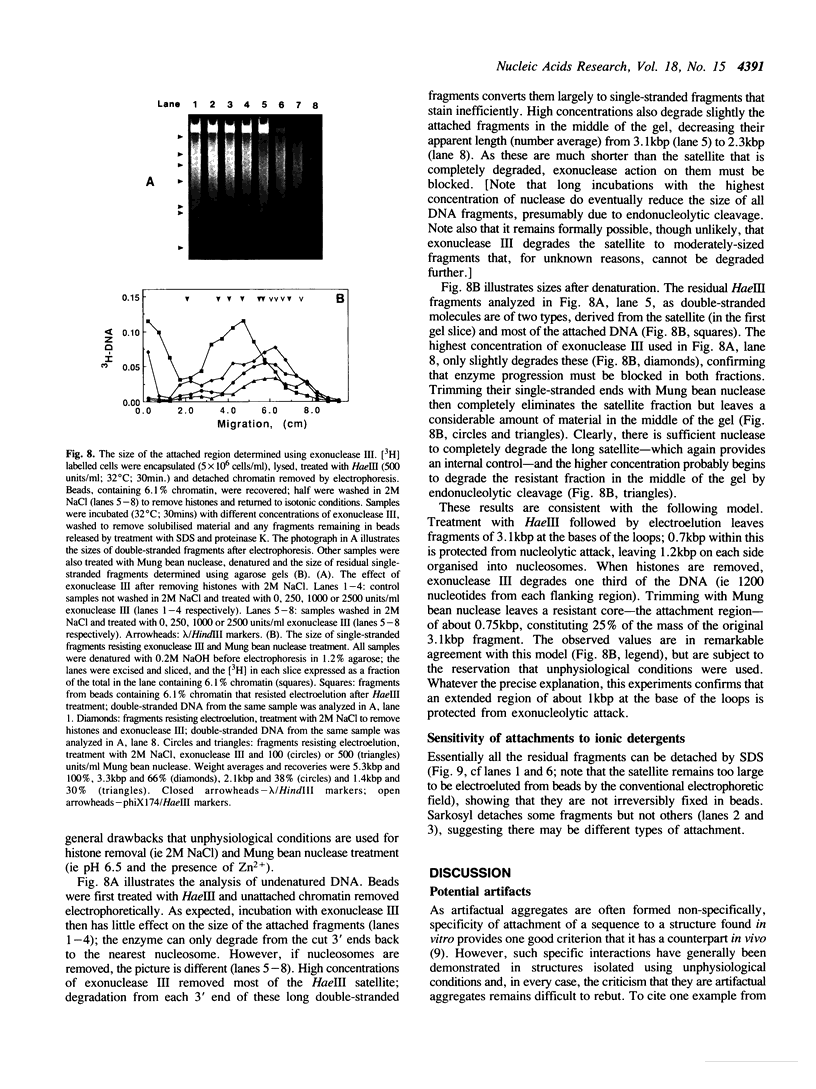
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Beard P. Mobility of histones on the chromosome of simian virus 40. Cell. 1978 Nov;15(3):955–967. doi: 10.1016/0092-8674(78)90279-9. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berrios S., Fisher P. A. Thermal stabilization of putative karyoskeletal protein-enriched fractions from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):4573–4575. doi: 10.1128/mcb.8.10.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar J. W., Ward D. C. Highly recurring sequence elements identified in eukaryotic DNAs by computer analysis are often homologous to regulatory sequences or protein binding sites. Nucleic Acids Res. 1987 Feb 25;15(4):1835–1851. doi: 10.1093/nar/15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., McKenna G., Sharp P. A. Cleavage of mouse DNA by a restriction enzyme as a clue to the arrangement of genes. Cold Spring Harb Symp Quant Biol. 1974;38:383–395. doi: 10.1101/sqb.1974.038.01.041. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Spectrofluorometric measurement of the binding of ethidium to superhelical DNA from cell nuclei. Eur J Biochem. 1978 Mar 15;84(2):465–477. doi: 10.1111/j.1432-1033.1978.tb12188.x. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Cook P. R. The nucleoskeleton: artefact, passive framework or active site? J Cell Sci. 1988 May;90(Pt 1):1–6. doi: 10.1242/jcs.90.1.1. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Hancock D. C. Studies on the interaction of the human c-myc protein with cell nuclei: p62c-myc as a member of a discrete subset of nuclear proteins. Cell. 1985 Nov;43(1):253–261. doi: 10.1016/0092-8674(85)90030-3. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Grosse F., Nasheuer H. P., Scholtissek S., Schomburg U. Lactate dehydrogenase and glyceraldehyde-phosphate dehydrogenase are single-stranded DNA-binding proteins that affect the DNA-polymerase-alpha-primase complex. Eur J Biochem. 1986 Nov 3;160(3):459–467. doi: 10.1111/j.1432-1033.1986.tb10062.x. [DOI] [PubMed] [Google Scholar]
- Guo X. W., Cole R. D. Chromatin aggregation changes substantially as pH varies within the physiological range. J Biol Chem. 1989 Jul 15;264(20):11653–11657. [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. A general method for preparing chromatin containing intact DNA. EMBO J. 1985 Apr;4(4):913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. Transcription occurs at a nucleoskeleton. EMBO J. 1985 Apr;4(4):919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Dickinson P., Cook P. R. The size of chromatin loops in HeLa cells. EMBO J. 1990 Feb;9(2):567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., McCready S. J., Cook P. R. Replication and transcription depend on attachment of DNA to the nuclear cage. J Cell Sci Suppl. 1984;1:59–79. doi: 10.1242/jcs.1984.supplement_1.5. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Yuan J., Cook P. R. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988 Jul;90(Pt 3):365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T. D., Hancock D. C., Evan G. I. Characterization of a heat shock-induced insoluble complex in the nuclei of cells. J Cell Sci. 1987 Aug;88(Pt 1):65–72. doi: 10.1242/jcs.88.1.65. [DOI] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. The salt dependence of chicken and yeast chromatin structure. Effects on internucleosomal organization and relation to active chromatin. J Biol Chem. 1986 Jul 25;261(21):9904–9914. [PubMed] [Google Scholar]
- Lothstein L., Arenstorf H. P., Chung S. Y., Walker B. W., Wooley J. C., LeStourgeon W. M. General organization of protein in HeLa 40S nuclear ribonucleoprotein particles. J Cell Biol. 1985 May;100(5):1570–1581. doi: 10.1083/jcb.100.5.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M. P., Laemmli U. K. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979 Aug;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- McConnell M., Whalen A. M., Smith D. E., Fisher P. A. Heat shock-induced changes in the structural stability of proteinaceous karyoskeletal elements in vitro and morphological effects in situ. J Cell Biol. 1987 Sep;105(3):1087–1098. doi: 10.1083/jcb.105.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S. J., Akrigg A., Cook P. R. Electron-microscopy of intact nuclear DNA from human cells. J Cell Sci. 1979 Oct;39:53–62. doi: 10.1242/jcs.39.1.53. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mullinger A. M., Johnson R. T. The organization of supercoiled DNA from human chromosomes. J Cell Sci. 1979 Aug;38:369–389. doi: 10.1242/jcs.38.1.369. [DOI] [PubMed] [Google Scholar]
- Nelson W. G., Pienta K. J., Barrack E. R., Coffey D. S. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen R., van Venrooij W., Ramaekers F. The nuclear matrix: structure and composition. J Cell Sci. 1988 May;90(Pt 1):11–36. doi: 10.1242/jcs.90.1.11. [DOI] [PubMed] [Google Scholar]
- Walker P. R., Sikorska M. Modulation of the sensitivity of chromatin to exogenous nucleases: implications for the apparent increased sensitivity of transcriptionally active genes. Biochemistry. 1986 Jul 1;25(13):3839–3845. doi: 10.1021/bi00361a015. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M. Steroid receptors: elements for modulation of eukaryotic transcription. Annu Rev Biochem. 1976;45:721–746. doi: 10.1146/annurev.bi.45.070176.003445. [DOI] [PubMed] [Google Scholar]