This paper represents an analysis of small RNAs in Caenorhabditis elegans by deep sequencing. The authors carried out their study on the nonpolyadenylated and intermediate-sized (50- to 500-nt) transcriptome in a genome-wide fashion. The authors applied an enzymatic approach for selective removal of 5′ monophosphate RNAs (i.e., assumed RNA degradation fragments) in combination with 454 pyrosequencing. This analysis resulted in the identification of 473 novel intergenic and intronic novel intermediate-sized noncoding RNAs (is-ncRNAs) in C. elegans, of which more than 250 do not belong to any known functional class of is-ncRNAs. Their data also suggested the existence of a secondary 5′-terminal modification system in C. elegans that may be involved in the functional maturation of processed RNAs and RNA fragments.
Keywords: intermediate-sized noncoding RNAs
Abstract
Noncoding RNAs are increasingly being recognized as important players in eukaryote biology. However, despite major efforts in mapping the Caenorhabditis elegans transcriptome over the last couple of years, nonpolyadenylated and intermediate-size noncoding RNAs (is-ncRNAs) are still incompletely explored. We have combined an enzymatic approach with full-length RNA-Seq of is-ncRNAs in C. elegans. A total of 473 novel is-ncRNAs has been identified, of which a substantial fraction was associated with transcription factor binding sites and developmentally regulated expression patterns. Analysis of sequence and secondary structure permitted classification of more than 200 is-ncRNAs into several known RNA classes, while another 33 is-ncRNAs were identified as belonging to two previously uncharacterized groups of is-ncRNAs. Three of the unclassified is-ncRNAs contain the 5′ Alu domain common to SRP RNAs and specifically bound with the SRP9/14 heterodimer in vitro. One of these (inc394) showed 65% sequence identity with the human, neuron-specific BC200 RNA. Structure-based clustering analysis and in vitro binding experiments supported the notion that the nematode stem-bulge RNAs (sbRNAs) are homologs (or functional analogs) of the Y RNAs. Moreover, analysis of the differential libraries showed that some mature snoRNAs undergo secondary 5′ cap modification after processing of the primary transcript, thus suggesting the existence of a wider range of functional RNAs arising from processed and modified fragments of primary transcripts.
INTRODUCTION
In recent years, both the very small RNAs (e.g., miRNAs, siRNAs) and the long mRNA-like noncoding RNAs (e.g., lincRNAs) have been identified as potent regulators of animal gene expression (Carninci et al. 2005; Carthew and Sontheimer 2009; Guttman et al. 2009; Nagano and Fraser 2011). In the size range between these two (i.e., ∼50–500 nt), there is a group of heterogenous ncRNAs, including the previously well-studied tRNAs, snRNAs, snoRNAs, and numerous transcripts of unknown function, which may tentatively be lumped together as “intermediate-size ncRNAs” (is-ncRNAs) (Wang et al. 2011). An increasing body of experimental evidence suggests that is-ncRNAs have specific regulatory roles in various cellular processes. For example, the human brain-specific snoRNA HBII-52 is strongly associated with Prader-Wili syndrome through regulation of alternative pre-mRNA splicing (Kishore and Stamm 2006), and the mouse neuronal BC1 RNA (Tiedge et al. 1991) and its human analog BC200 RNA (Tiedge et al. 1993) specifically target the catalytic activity of eIF4A to repress mRNA translation (Lin et al. 2008).
Previous cloning and sequencing efforts in Caenorhabditis elegans have identified close to 300 ncRNAs, of which nearly 50 could not be assigned to previously known functional classes (Deng et al. 2006; Zemann et al. 2006). Subsequent tiling array analyses have identified around 6000 small (<500 nt) unannotated and probably noncoding intronic and intergenic loci expressed at various developmental stages (He et al. 2007; Wang et al. 2011). Similarly, machine-learning-based analyses of several data sets (RNA-Seq and tiling array data) assembled by the modENCODE project predicted the existence of more than 7000 novel ncRNA candidates expressed at a series of developmental stages (Gerstein et al. 2010; Lu et al. 2011). The vast majority of these putative is-ncRNAs are functionally unknown, suggesting that, despite its smaller genome size, the noncoding transcriptome is no less complex in C. elegans than in other higher eukaryotes (He et al. 2007).
Tiling array analyses cannot determine the orientation and length of the novel ncRNA candidates with the same accuracy as can RNA sequencing. On the other hand, the tiling array data suggested that the majority of the yet uncharacterized is-ncRNA are expressed at low levels and/or only during specific stages of C. elegans development (Wang et al. 2011) and would thus only be detectable by very large sequencing depths. We reasoned that cleavage fragments (or other processed fragments) of mature rRNAs and mRNAs would most likely have monophosphate 5′ termini and could thus largely be eliminated by treatment with Terminator 5′-phosphate-dependent exonuclease (TEX). As RNA polymerase II (pol II) products generally contain a 5′-terminal cap (Banerjee 1980), and RNA polymerase III (pol III) products carry 5′-terminal tri-phosphate (5′ PPP) groups (Shumyatsky et al. 1990), these would be protected from TEX digestion. On the other hand, TEX efficiency is affected by RNA secondary structure (Szittya et al. 2010), and insensitivity to TEX can, therefore, not be ascribed to the absence of 5′ monophospate termini with absolute certainty. Nonetheless, several studies have succeeded in using TEX to enrich sequencing libraries in primary transcripts, thereby discovering low-abundance RNA transcripts as well as specifically identifying transcriptional start sites occurring at “unorthodox” positions such as within operons or in opposite orientation within annotated genes (Albrecht et al. 2010; Irnov et al. 2010; Sharma et al. 2010).
We prepared two nonpolyadenylated transcriptome cDNA libraries, one from untreated RNA (control), and one in which the RNA was treated with TEX (TEX-treated). A total of 473 novel intergenic and intronic is-ncRNA candidates have been identified, of which more than 250 do not belong to any known functional class of is-ncRNAs. Further analysis of their conservation, secondary structures, developmental expression, potential TF binding sites, and binding protein partners provides a foundation for subsequent investigation of the functional roles of these novel is-ncRNAs.
RESULTS
Sequencing and data analysis
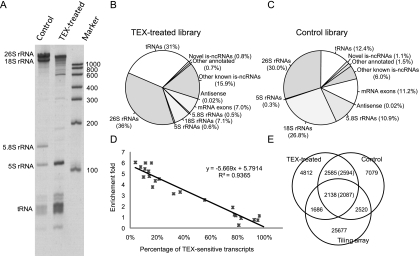
Preliminary analysis by PAGE gel electrophoresis suggested that the TEX treatment substantially depleted the amount of transcripts corresponding to mature 26S, 18S, and 5.8S rRNAs and correspondingly enriched the amount of RNA polymerase III-transcribed 5S rRNA (Fig. 1A). Quantitative measurement indicated that ∼87% of a total RNA sample was sensitive to TEX (e.g., consisting of 5′-terminal monophosphate RNAs).
FIGURE 1.
Sequencing and data analysis. (A) Effects of TEX treatment on total RNA. (B,C) Reads distribution in the two libraries. “Other annotated” mainly includes repeats, pseudogenes, transposons and ESTs; “Other known is-ncRNAs” include all annotated is-ncRNAs and previous literature (Deng et al. 2006; Zemann et al. 2006). (B) TEX-treated library. (C) Control library. (D) Correlation between fold enrichment in the TEX-treated library and percentage of TEX-insensitive fragments as calculated from the qRT-PCR results. (E) Overlap between 454 pyrosequencing contigs from the two libraries and tiling array fragments from Wang et al. (2011).
We consequently size-fractioned (50–500 nt) total RNA from mixed-stage C. elegans, depleted the sample of ribosomal and polyadenylated RNAs, and treated one aliquot of the sample with TEX before cDNA preparation (Materials and Methods; Supplemental Fig. S1A). The 454 pyrosequencing yielded totals of 287,443 and 301,406 reads from the TEX-treated and untreated (control) libraries, respectively. After removal of the 5′ and 3′ adaptors as well as reads with abnormal adaptors or inserted sequences shorter than 30 nt, 261,150 and 259,818 reads from the two respective libraries could be mapped directly to the C. elegans genome (WS190) by BLAST (NCBI) and BLAT (Kent 2002) with a set of filtering criteria (Materials and Methods). The distribution of the TEX-treated sequence reads was markedly more skewed toward shorter lengths than the control reads (Wilcoxon test, P-value < 2.2 × 10−16); however, this difference in reads length was mainly confined to reads mapping to rRNAs and tRNAs (Supplemental Fig. S2). For most annotated genomic loci, the reads numbers were comparable in the TEX-treated and control libraries (Fig. 1B,C). The 5.8S rRNA was significantly depleted in the TEX-treated library (0.3% of all reads) compared to the control library (11% of all reads), whereas snRNAs and stem-bulge RNAs (sbRNAs), known to be primary RNA pol II transcripts bearing 5′-terminal cap or pol III transcripts with tri-phosphate 5′-terminal structures (Deng et al. 2006; Li et al. 2008), were strongly enriched in the TEX-treated library (Supplemental Fig. S3A).
Known is-ncRNAs (other than rRNAs and tRNAs) constituted 15.9% and 6% of the reads in the TEX-treated and control libraries, respectively. The percent reduction by TEX treatment was quantified by qRT-PCR of 24 known is-ncRNAs from different functional categories before and after TEX treatment (Supplemental Fig. S4). The data showed a strong negative correlation (R2 = 0.94) between the percentage of TEX-sensitive transcripts as calculated from the qRT-PCR results and the fold enrichment in the TEX-treated library (Fig. 1D). This correlation was subsequently utilized to estimate approximate percentages of TEX-sensitive and -insensitive transcripts at all loci.
Overlapping reads were clustered into contigs, and “wig” profiles for visualization in the Integrated Genome Browser (IGB) were generated (Nicol et al. 2009). The TEX-treated and control libraries produced 11,221 and 14,280 contigs, respectively (Supplemental Fig. S5). Comparison with previously published tiling microarray data (Wang et al. 2011) showed that 34% and 32% of these contigs, respectively, overlapped with transcribed fragments detected by the tiling arrays (Fig. 1E). However, only 24 contigs from TEX-treated and 16 from control libraries overlapped with the 7k ncRNA data (Lu et al. 2011) in the same strand.
The contigs were almost evenly distributed among the five C. elegans autosomes but were markedly more abundant on the X chromosome (Supplemental Fig. S1B). Of the TEX-treated and control library contigs, 9274 (83%) and 12,838 (90%), respectively, wholly or partially overlapped with mRNA exons in sense orientation, and only 57 and 42 contigs mapped to the antisense strand of mRNAs; thus, the majority of the contigs represented random degradation fragments or specific cellular cleavage products of mRNAs (Karginov et al. 2010). Known is-ncRNA loci were represented by a total of 849 contigs (812 and 811 contigs from the TEX-treated and control libraries, respectively) corresponding to ∼90% of such loci (Table 1).
TABLE 1.
Detection rates of known is-ncRNA loci for the TEX-treated and control libraries
As the 454 pyrosequencing enabled exact identification of the 5′ and 3′ termini of the mature is-ncRNA sequences, we could show that of the 849 annotated is-ncRNAs that we detected, 12 is-ncRNAs had previously been annotated with incorrect 5′ and 3′ termini, and nine other is-ncRNAs had been annotated with incorrect orientation (Supplemental Fig. S6; Supplemental Table S1).
Novel intergenic and intronic is-ncRNAs
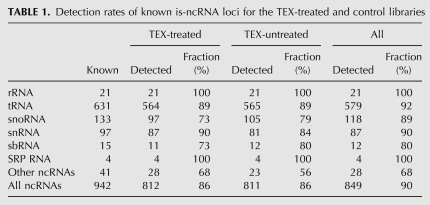
After removal of contigs overlapping coding genes, annotated repeats, pseudogenes, transposons and public EST data, there were 473 contigs (352 contigs from the TEX-treated library and 154 contigs from the control library) that mapped to single, unannotated intergenic or intronic loci, thus corresponding to 473 unique novel is-ncRNAs (Supplemental Table S2). Of these 473 putative is-ncRNAs, 408 were represented by a single read. This, and the higher number of loci identified by the TEX-treated library, suggests the advantage of enrichment by removal of degradation fragments for detection of the much lower expressed part of the noncoding transcriptome. For validation, 12 randomly selected is-ncRNAs (10 of which were represented by a single read) were subjected to RT-PCR, all amplifying a fragment of the expected size (Fig. 2A). In addition, among the is-ncRNAs with higher expression, six candidates were verified by Northern blot analysis (Fig. 2B), all with transcript lengths in accordance with the 454 pyrosequencing data.
FIGURE 2.
Validation of novel is-ncRNAs. (A) RT-PCR validation of novel is-ncRNAs. Of 12 randomly chosen is-ncRNAs, all were verified by RT-PCR. Each is-ncRNA is represented by three adjacent lanes, from left to right: (G) genomic DNA PCR, positive control); (+) DNase-treated RNA RT-PCR; (−) DNase-treated RNA PCR (no RT; negative control). (B) Six is-ncRNAs verified by Northern blot. Arrows indicate the size of the is-ncRNA as obtained from the sequencing data.
Conservation and expression analysis of the novel is-ncRNAs
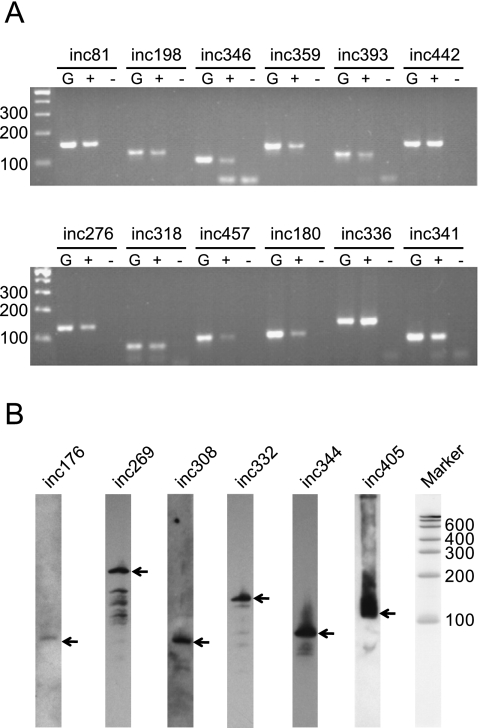
Of the 473 is-ncRNAs, 26% (124) were located in sense orientation to introns of protein-coding genes, while 7% (33) were located in antisense orientation to introns, the remaining 67% (316) being intergenic. Sixty-three percent (299) of the loci had average phastCons scores (Siepel et al. 2005) ≥0.2 when compared to other worm species (Fig. 3A). Nearly half of the functionally classifiable is-ncRNAs (74 out of 156) and 82 of the unclassifiable is-ncRNAs were well-conserved with phastCons scores ≥0.5. The 316 intergenic novel is-ncRNAs that were located within 2 kb upstream of or downstream from coding genes tended to be more conserved (P < 0.02 and P < 0.05, respectively) than more distant loci (Fig. 3B).
FIGURE 3.
Conservation and expression analysis. (A) Conservation of known is-ncRNAs, mRNA exons, and novel intergenic and intronic is-ncRNA contigs. (B) phastCons score distributions of contigs mapping to mRNA exons, introns, and intergenic regions. “Upstream” and “downstrean” denote contigs mapping within 2000 bp of coding genes, while “distant” denotes contigs mapping farther from coding genes. (C) qRT-PCR analysis of 12 is-ncRNAs across five developmental stages. For each is-ncRNA, the relative expression levels among various stages after normalizing to the amount of U6 snRNA (CeN4) in each sample are shown.
Among the 473 loci, there were 118 loci that overlapped transcribed regions detected by a recently published C. elegans stage-specific tiling array data (Wang et al. 2011). The tiling array expression profiles suggested that these 118 is-ncRNAs could be divided into ten clusters with predominant expression in one or several distinct developmental stages or conditions (Supplemental Fig. S7). These included is-ncRNAs with predominant expression in the four larval stages, in the mature adult, in the male, and in the Dauer stage. To further test whether the tendency toward developmentally regulated expression also applied to other novel loci (i.e., not overlapping the tiling array data), we selected 12 novel is-ncRNAs (all detected by a single read) whose loci are associated with the PHA-4 transcription factor binding sites (see below). The expression of these was analyzed by qRT-PCR across five larvae stages (L1, L2, L3, L4, and Dauer). All of the 12 loci showed markedly (more than twofold difference) increased or reduced expression in at least one stage (Fig. 3C), suggesting they were developmentally regulated.
Functional classification of the novel is-ncRNAs
Functional analysis suggested that nearly one-half (211) of the is-ncRNAs belonged to previously known functional classes of ncRNAs, the majority of these being snoRNAs. Seventeen C/D box snoRNA candidates were predicted by snoSeeker (Yang et al. 2006), snoReport (Hertel et al. 2008), and Rfam (Gardner et al. 2011), while 184 H/ACA box snoRNA candidates were predicted by snoGPS (Schattner et al. 2004). Other potential homologs of known ncRNAs were identified by alignment searches against 1371 ncRNA families in Rfam using the HMMER3 (http://hmmer.janelia.org/). Four snRNAs, one SRP RNA, three tRNA-like RNAs, and three potential miRNA precursors were also identified. The three potential miRNA precursors inc59, inc71, and inc162 (corresponding to mir-533, mir-84, and mir-574, respectively), were also predicted to be H/ACA box snoRNAs, suggesting that certain H/ACA box snoRNAs may have miRNA precursor-like features (Scott et al. 2009). Searches for two internal motifs previously identified in sbRNAs (Deng et al. 2006) yielded two potential sbRNA candidates (inc176 and inc332).
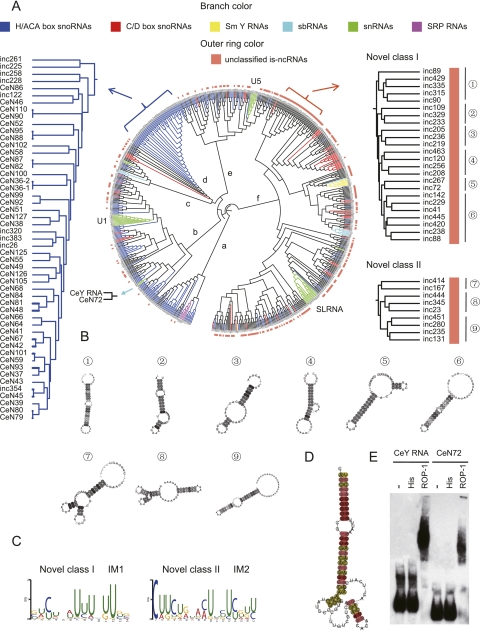
To further characterize the remaining 262 is-ncRNAs, we applied the LocARNA-based clustering approach (Will et al. 2007) to all 473 novel is-ncRNAs as well as to a set of 161 previously described ncRNAs (Deng et al. 2006). The method efficiently clustered annotated is-ncRNAs according to their respective classes, like snRNAs, snoRNAs, and SRP RNAs. The novel is-ncRNAs identified as snRNAs and SRP RNAs also clustered into their respective categories (Fig. 4A). We obtained several H/ACA box snoRNA clusters, of which the largest (represented by the “d” branch in Fig. 4A) contained 42 known H/ACA box snoRNAs and nine novel is-ncRNAs predicted as H/ACA box snoRNAs by snoGPS. The unclassifed is-ncRNAs were mostly found together with a variety of known ncRNA types in a large cluster (represented by branch “f”) and a smaller cluster (represented by branch “e”). Within the branch “f” cluster, the LocARNA software also identified two tightly knit clusters of, respectively, 24 and nine novel ncRNA contigs, potentially forming two novel ncRNA classes. The first of these (novel class I) includes 24 is-ncRNAs that range in size from 50 to 66 nt (Supplemental Document S2), and all share a common internal motif (IM1) and two structural loops (Fig. 4B,C). Seventeen of the 24 members of this class have recognizable counterparts in other worm species. The novel class II is-ncRNAs are between 62 and 69 nt in length and also share a common internal motif (IM2). Five of the novel class II is-ncRNAs are conserved in other worm species, and two sub-clusters of this potential class were predicted to form three structural loops.
FIGURE 4.
Structure-based analysis of the novel is-ncRNAs. (A) Clustering of the 473 novel is-ncRNAs and 161 known is-ncRNAs based on their predicted secondary structure. The branch color indicates the (predicted) functional classes of known is-ncRNA and novel is-ncRNAs, and the orange color of the outer ring indicates unclassified novel is-ncRNAs. The orange bracket indicates the two putatively novel class I and II contig clusters. (B) Predicted secondary structures of the novel class I and II transcripts. (C) Internal motifs (IM1 and IM2, respectively) of the novel class I and II contig clusters. (D) The consensus structure of CeY RNA and sbRNA (CeN72) was calculated by RNAalifold (Bernhart et al. 2008). (E) EMSA of biotin-labeled RNA probes corresponding to CeY RNA and sbRNA (CeN72) and His-ROP-1 fusion proteins. (—) Free probes. His alone was used as a negative control.
According to the LocARNA-based clustering, a previously identified sbRNA (CeN72) was tightly clustered with the only known CeY RNA in C. elegans. This suggests strong structure similarities (Fig. 4D) and is in accordance with a recent study (Boria et al. 2010) predicting that the nematode sbRNAs could be homologs of the vertebrate Y RNAs. We, therefore, carried out an electrophoretic mobility shift assay (EMSA) to test whether sbRNAs could potentially interact with the CeY RNA binding protein (ROP-1) (Van Horn et al. 1995). The results showed that not only sbRNA CeN72 but also two other novel sbRNA-like is-ncRNAs (inc176 and inc332) shifted position in the presence of ROP-1 (Fig. 4E; Supplemental Fig. S8).
An additional group of six novel is-ncRNAs contains the Sm protein binding site “AAU/AUUUUGGA” and a 3′-tail stem–loop structure common to snRNAs, SLRNAs, and Sm Y RNAs; however, these transcripts did not cluster in the LocARNA analysis. One transcript (inc464) is 75 nt long and has 52% sequence identity to the Ascaris lumbricoides Sm X RNA sequence (Supplemental Fig. S9C; Maroney et al. 1996). The remaining transcripts vary in length from 135 to 177 nt and are thus considerably longer than classified Sm Y RNAs (MacMorris et al. 2007). None of the six is-ncRNAs showed much sequence similarity to the Sm Y RNAs outside the Sm binding site (Supplemental Fig. S9A).
A potential analog of the primate-specific BC200 RNA
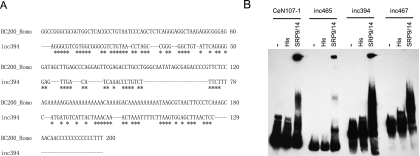
Comparison to Rfam (Gardner et al. 2011) showed that three of the is-ncRNAs (inc394, inc465, and inc467) had high sequence and structure similarity to the first 50 nucleotides of the C. elegans SRP RNAs, called the 5′Alu domain (Weichenrieder et al. 2000). The remaining sequence of these is-ncRNAs showed little similarity to the SRP RNAs (Supplemental Fig. S10A), suggesting that they are not partial fragments or pseudogenes of the SRP RNAs. Similar to the SRP RNAs, the 5′Alu domain of the three is-ncRNAs also contained a Box A element 10∼20 bp downstream from the transcription start site, indicative of transcription by the RNA polymerase III (Deng et al. 2006). Thus, these is-ncRNAs present a basic similarity to the neuron-specific human BC200 RNA, whose 5′-end region is homologous to the human Alu domain (Tiedge et al. 1993). Subsequently, a phylogenetic analysis (neighbor-joining tree) of these sequences by ClustalW2 (EBI) multiple alignments showed that inc394 had the strongest similarity to the BC200 RNA (Supplemental Fig. S10B). Further comparison indicated that inc394 had 65% sequence identity with the BC200 when sequence gaps were excluded (Fig. 5A).
FIGURE 5.
Analysis of three is-ncRNAs with 5′ Alu domains. (A) Sequence alignment of human BC200 RNA and inc394. (B) EMSA of biotin-labeled RNA probes corresponding to the SRP RNA (CeN107-1) and inc465, inc394, and inc467 with His-SRP9 and His-SRP14 fusion proteins. (—) Free probes. His alone was used as a negative control.
The protein heterodimer SRP9/14 binds strongly to the conserved core of the SRP RNA 5′Alu domain (Weichenrieder et al. 2000). Analysis by the RNAalifold program (Bernhart et al. 2008) suggested that all three is-ncRNAs, as well as the SRP RNAs, form similar structures, consisting of a U-turn connecting two helical stacks (Supplemental Fig. S10C; Weichenrieder et al. 2000). The BC200 RNA binds the heterodimer SRP9/14 in vitro (Bovia et al. 1997) and in vivo (Kremerskothen et al. 1998). To test whether the three novel is-ncRNAs interacted with SRP9/14, we carried out EMSA. All three is-ncRNAs showed specific shifts in the presence of the C. elegans SRP9/14 heterodimer (Fig. 5B). None of them shifted in the presence of the C. elegans ROP-1 protein (negative control) (Supplemental Fig. S11), suggesting that the binding to the SRP9/14 heterodimer was specific.
The novel is-ncRNA loci are associated with transcription factor binding sites
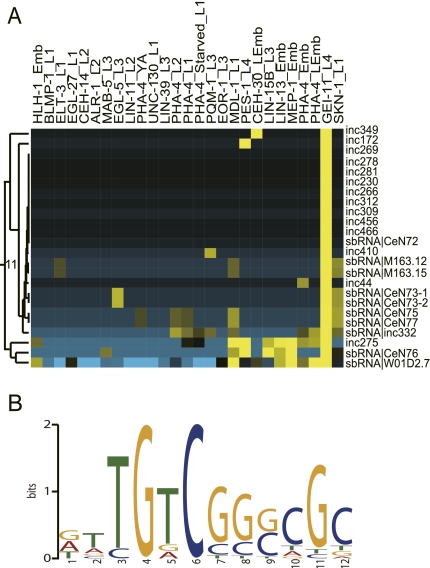
Given the variable spatial and temporal expression pattern of the novel ncRNA candidates, we next asked whether the genes for the novel is-ncRNAs were associated with the TF binding sites identified in ChIP-seq experiments (Gerstein et al. 2010; Niu et al. 2010). To this end, we searched the ChIP-seq data for possible TF binding sites in regions of ±150 bp surrounding the 5′-ends of the 473 novel is-ncRNAs as well as 15 known sbRNA loci. Ten of the sbRNA loci (Deng et al. 2006; Boria et al. 2010), as well as 193 of the novel is-ncRNAs loci, were associated with one or more binding sites for at least one of the 22 analyzed TFs. Comparing these to random genomic regions of the same size (see Lu et al. 2011) suggested an ∼2.8-fold enrichment of TF binding sites at the is-ncRNA loci (χ2 test, P-value < 2.2 × 10−16). The loci were associated with various TF binding sites, but most binding sites (>80 hits) were found for the TFs, MDL-1_L1 and PHA-4_L1 (Supplemental Fig. S12). The 203 loci with at least one TF binding site could be grouped into 11 clusters according to their TF binding peak signal scores. For half of the clusters, the cluster members had one TF in common. The is-ncRNAs loci of cluster 1, 2, 7, 8, 9, and 11 were mainly associated with binding sites for a single TF (BLMP-1, HLH-1, PQM-1, PHA-4, LIN-39, and GEI-11, respectively) (Supplemental Fig. S13). The is-ncRNAs were frequently expressed at the same developmental stage as the associated TFs. For example, binding sites for EGL-5_L3, LIN-39_L3, and PQM-1_L3 were found at the loci of inc302, inc422, and inc425, respectively, and these three is-ncRNAs are all expressed at the L3 stage.
The is-ncRNAs loci in cluster 11 were all associated with binding sites for transcription factor GEI-11, a SNAP190 ortholog (Fig. 6A). Previous studies have shown that SNAP190 is involved in specific binding of the SNAP complex to the proximal sequence element (PSE) located upstream of snRNAs (Wong et al. 1998). Our analyses showed that, in addition to a high fraction of snRNA and other is-ncRNA loci having an upstream PSE (or UM1) (Deng et al. 2006), GEI-11_L4 binding sites were found in the proximity of 59% (10/17) of the sbRNA loci, whose upstream promoter element (UM3) has been suggested to include a PSEB box (Boria et al. 2010). Sequence analysis of the 100 bp flanking the 5′ ends of the cluster 11 loci identified a motif with strong resemblance to the PSEB box present at all loci in this cluster (Fig. 6B), suggesting that this motif may be the GEI-11 binding site.
FIGURE 6.
Novel is-ncRNAs associated with TF binding sites. (A) Cluster 11 is associated with binding sites for GEI-11. This cluster includes 9 known sbRNAs and one novel is-ncRNA with sbRNA characteristics. (B) The GEI-11 potential binding site of cluster 11 is-ncRNA loci.
Analysis of exonic contigs
Several studies have suggested that RNA fragments overlapping coding exons and even exon-exon junctions contain 5′-terminal cap-like structures furnished by a secondary 5′-end capping mechanism (Fejes-Toth et al. 2009; Schoenberg and Maquat 2009), and analysis of RNA fragments overlapping coding and noncoding exons in C. elegans is consequently of considerable interest. A differential RNA-Seq approach employing TEX digestion might be ideally suited for such an analysis; however, owing to possible effects of RNA secondary structure upon TEX efficiency (Szittya et al. 2010), we have thus far only carried out a preliminary analysis of these data, the details of which are found in Supplemental Document S1.
Briefly, the analysis shows that the expression levels of coding exonic contigs with a high frequency of TEX-sensitive fragments are well-correlated with the expression level of the corresponding “host” mRNA (as would be expected for degradation fragments), whereas contigs consisting mainly of TEX-insensitive fragments are not (Supplemental Fig. S14). Similarly, the stage-specific expression of TEX-sensitive contigs resembled that of their “host” genes to a larger extent than did that of TEX-insensitive contigs. Analysis of a specific case (the largely TEX-insensitive contig 5839) spanning an exon-exon junction of the PUF family RNA-binding protein fbf-1 (H12I13.4) suggested that this contig represents a fragment with precise 5′ and 3′ termini which shows only weak expressional correlation to its host gene. No similar fragment was found at the paralog gene fbf-2 (F21H12.5) despite only minor sequence differences between the two paralogs (Supplemental Fig. S15F).
Analysis of snoRNAs suggests secondary 5′-end modification
One particular case concerns contigs overlapping snoRNA loci. The snoRNA loci can be divided into three groups, the first group consisting of intronic snoRNA loci without any discernible upstream motif, and the second and third group being loci with either the UM1 or UM2 upstream motifs (Deng et al. 2006), respectively. The first group (45 intronic snoRNAs without discernible promoters) are assumed to be processed from their respective introns after (or during) splicing of their host gene mRNA (Deng et al. 2006) and would consequently have 5′-end monophosphates. These snoRNAs were almost exclusively composed of TEX-sensitive fragments (Supplemental Fig. S3B). The second group (UM1 snoRNAs) is assumed to be transcribed by RNA polymerase II (Deng et al. 2006) and produced mostly TEX-insensitive fragments (median values 80%–90%) (Supplemental Fig. S3B), suggesting that these transcripts are furnished with a 5′-end cap similar to other pol II transcripts. The differences in TEX-sensitivity between these two groups of snoRNAs are, thus, more easily explained by differences in their 5′-end structure than by differences in secondary structure.
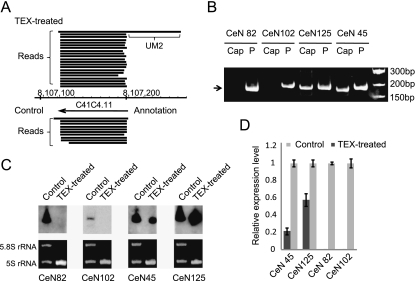
The contigs of the third group (48 UM2-snoRNAs) were for the most part (40/48) not significantly enriched in either library (one-tailed Fisher's exact test, P > 0.01), indicating that these loci each gave rise to 75%–90% TEX-sensitive fragments and 10%–25% TEX-insensitive fragments (Supplemental Fig. S3B). UM2 snoRNAs are probably processed from longer primary transcripts encompassing the UM2 sequence (Li et al. 2008), which is likely to generate mature snoRNAs with 5′ monophosphate termini. The sequencing data indicated that “UM2-snoRNA” primary transcripts could be detected for 16 of the 48 UM2 snoRNAs (Fig. 7A). To investigate the 5′-end status of the UM2 snoRNAs, we carried out 5′ cap- and 5′ monophosphate-dependent rapid amplification of cDNA ends (RACE) of two randomly chosen UM2 snoRNAs (CeN125 and CeN45) and two randomly chosen intronic snoRNAs (CeN82 and CeN102). The two intronic snoRNAs were only amplified by 5′ monophosphate-dependent RACE (“P”) but not by 5′ cap-dependent RACE (“Cap”), while the two UM2 snoRNAs were amplified by both types of RACE (Fig. 7B), showing that the mature UM2 snoRNAs populations consist of RNAs with both 5′ capped and 5′ monophosphate termini. Subsequently, Northern blot analysis and qRT-PCR results indicated that the two UM2 snoRNAs (CeN45 and CeN125) were composed of, respectively, 20% and 50% of TEX-insensitive transcripts, while the two intronic snoRNAs consisted almost exclusively of TEX-sensitive transcripts (Fig. 7C,D). The two UM2 snoRNAs and the two intronic snoRNAs are all H/ACA box snoRNAs and were all included in the same LocARNA cluster (Fig. 4A), indicating that they have similar secondary structures. Taken together, these data strongly suggest that processed snoRNAs in C. elegans may undergo varying degrees of secondary 5′-end modification.
FIGURE 7.
Analysis of UM2 snoRNA 5′ termini. (A) An example of a primary UM2 snoRNA transcript (C41C4.11) in the TEX-treated library. (B) 5′ cap (Cap) and 5′ monophosphate-dependent RACE (P) of mature snoRNAs. The two intronic, processed snoRNAs (CeN82 and CeN102) were only amplified by 5′ monophosphate-dependent RACE but not by 5′ cap-dependent RACE, while the two UM2 snoRNAs (CeN125 and CeN45) were amplified by both types of RACE. (C) Northern blot analysis of UM2 snoRNAs (CeN45 and CeN125) and intronic processed snoRNAs (CeN82 and CeN102) before and after TEX-treatment. 5.8S rRNA and 5S rRNA were used as loading controls. (D) qRT-PCR analysis of the relative expression levels of the four snoRNAs in the TEX-treated and control samples. The average expression levels were determined after normalizing to 5S rRNA in each sample.
DISCUSSION
It has become increasingly clear in recent years that eukaryotic transcriptomes are remarkably complex, containing numerous noncoding RNAs (ncRNAs) that have previously not been characterized. Our study identified 473 contigs probably representing unannotated, full-length intergenic and intronic is-ncRNA loci, of which a substantial fraction were associated with transcription factor binding sites and/or showed developmentally regulated expression in a recent tiling array study (Wang et al. 2011). More than 65% of the loci had not been detected in earlier tiling array (He et al. 2007; Wang et al. 2011) or modENCODE (Gerstein et al. 2010; Lu et al. 2011) studies, and the majority (81%) of these were only detected in the TEX-treated library, suggesting that our differential RNA-sequencing approach is well-suited for identifying is-ncRNAs with limited expression ranges or levels.
Approximately 33% of the novel is-ncRNA loci had been detected by previously published tiling array studies (He et al. 2007; Wang et al. 2011), while only 2% overlapped with the 7k modENCODE data (Lu et al. 2011). With respect to the tiling array data, the lack of overlap may, at least in part, owe to the following two reasons: (1) While the tiling array study analyzed RNAs from several developmental stages, the RNA samples used in this study were derived from mix-stage worms and may thus have missed stage-specific, lowly expressed transcripts. The fact that most of these contigs were represented by only one or a few sequence reads and frequently overlapped stage-specific tiling array fragments supports the notion that the expression of many of these fragments is restricted to one or a few developmental stages or conditions through the C. elegans life cycle; and (2) The tiling array data may have a somewhat higher false positive rate due to cross-hybridization (Agarwal et al. 2010; van Bakel et al. 2010), as indicated by its 80% validation rate (Wang et al. 2011) compared to the 100% validation rate obtained for the contigs in this study. The poor overlap with the 7k modENCODE data (Lu et al. 2011) is more difficult to account for. It is possible that our dedicated aim at nonpolyadenylated, intermediate-size ncRNAs, with enrichment of transcripts with non-5′ monophosphate terminals, has picked up a specific segment of the transcriptome that, to a large extent, had been missed by the modENCODE approaches, which concentrated on either polyadenylated RNAs or total RNA of all size fractions (Gerstein et al. 2010). Alternatively, the application of computational prediction to the experimental data may have filtered signals in the intermediate-size ncRNA segment too stringently, thus missing a substantial fraction of these data.
The presence of an even wider repertoire of transcripts carrying the Sm protein binding site is interesting. The nematode Sm Y RNAs (as defined by MacMorris et al. [2007]) includes twelve transcripts with similar size (77–88 nt) and sequence features. In addition to these, the A. lumbricoides Sm X RNA (Maroney et al. 1996) and the C. elegans CeN115 (Deng et al. 2006) also contain Sm protein binding sites but display little sequence similarity to the Sm Y RNAs. The finding in this study of six additional transcripts that do not conform to the Sm Y RNAs in neither length nor sequence characteristics suggests that the repertoire of transcripts employed by the nematode splicing apparatus may be more variable than hitherto assumed. The fact that the SmY-10 apparently does not coprecipitate with (most) other Sm Y RNAs (MacMorris et al. 2007) could likewise suggest that the Sm Y RNAs are not a functionally homogenous group, or, alternatively, that the Sm Y RNAs are just one cluster within an extended spectrum of splicing-related transcripts occurring within the nematodes.
Three of the novel is-ncRNAs share a structure similar to the 5′ Alu domain of the SRP RNAs. Beyond the 5′ Alu domain, they all showed little sequence similarity to the C. elegans SRP RNAs, but all three bound strongly to the heterodimer SRP9/14 in vitro. The neuron-specific BC200 RNA also contains an Alu domain that specifically binds SRP9/14 in vivo and plays a role in translational regulation of dendritic proteins in neurons (Kremerskothen et al. 1998; Khanam et al. 2007). Although BC200 homologs have only been found in anthropoid primates (Tiedge et al. 1993), the rodent BC1 RNA shares common regulatory functions and neuron-specific expression patterns with the BC200 RNA and is regarded as its functional analog (Tiedge et al. 1991). The inc394 RNA resembles BC200 in that they both contain a 5′ Alu domain and central A-rich region. The 3′-terminal region of inc394 is shorter than that of BC200, but several shorter motifs are common to both RNAs. It is thus tempting to speculate that the inc394 RNA may operate as a functional analog of BC200 RNA in C. elegans.
Analysis of average phastCons scores of the novel intronic and intergenic is-ncRNAs showed a dual conservation distribution, with 37% of the is-ncRNAs being specific to C. elegans, and the rest having more or less conserved counterparts in other worm species. However, lack of conservation may not necessarily imply lack of function (Pang et al. 2006). Homologous ncRNAs may share secondary structures rather than primary sequence, as numerous ncRNAs with similar secondary structures are functional and, thereby, well-conserved over evolutionary timescales (Washietl et al. 2005). To explore the structural common features of the unclassified novel is-ncRNAs, a structure-based clustering approach was carried out and revealed two potential ncRNA families with similar secondary structure and common internal motifs for further investigation. Consistent with a recent study (Boria et al. 2010), we also found that a previously identified sbRNA (CeN72) was tightly clustered with the CeY RNA, and EMSA demonstrated that CeN72, as well as two newly characterized sbRNAs, are potential ROP-1 binding partners. A previous analysis of ROP-1 binding RNAs in C. elegans embryos only identified the CeY RNA (Van Horn et al. 1995); however, since sbRNAs are predominantly expressed after L3 (Deng et al. 2006), ROP-1 may bind with sbRNAs in later stages of worm development. Moreover, the ROP-1 protein appears to undergo proteolytic processing between the L2 and L3 larval stages that shift it size by ∼3.5 KDa (Labbe et al. 2000), but it is not known whether or how this influences its RNA binding properties.
Nearly 40% of the 473 loci were associated with binding sites for one or more of 22 known transcription factors. These associations were commonly limited to one or a few specific TFs, suggesting that groups of the novel ncRNAs would be under common regulatory regimes, possibly accounting for their apparently well-regulated expression patterns. In addition, some TFs (e.g., GEI-11) were associated with both polymerase II and polymerase III transcribed ncRNAs, consistent with a recent study showing that transcription factors associated with Pol II are often associated with Pol III promoters (Raha et al. 2010). GEI-11 was previously known as a homolog of SNAP190 that specially directs the SNAP complex to bind to the proximal sequence element (PSE) (Wong et al. 1998). Our data suggest that GEI-11 could regulate both the majority of the snRNAs (Niu et al. 2010) (transcribed by Pol II or Pol III) and sbRNAs (transcribed by Pol III) by targeting a common DNA motif with strong resemblance to the PSEB box in C. elegans. In contrast, a recent study found that the SNAP complex recognizes the PSEA box sequence in Drosophila melanogaster (Kim et al. 2010), suggesting some variation in the SNAP complex binding preferences in different organisms.
Preliminary analysis of the TEX enzyme activity suggests that the TEX treatment could efficiently distinguish between types of RNAs known to possess (e.g., 5.8S rRNA) and not possess (e.g., 5S rRNA) 5′ monophosphate termini (Fig. 1A). We also observed that different is-ncRNAs species showed considerable variation in their relative enrichment in the two libraries (Supplemental Fig. S3), indicative of varying degrees of 5′-terminal modification for the different is-ncRNA classes. This accords with observations in Arabidopsis showing that a majority of mRNAs possessed various amounts/fractions of 5′ monophosphate and 5′ capped transcripts (Jiao et al. 2008). On the other hand, we cannot exclude that interactions between concentration and secondary structure may influence the efficiency by which TEX degrades individual RNA species (Szittya et al. 2010). For example, although most tRNA 5′ ends are generated by RNase P cleavage, resulting in a 5′-terminal monophosphate (Frank and Pace 1998), the tRNAs were markedly enriched after TEX treatment (Fig. 1A).
UM2 snoRNA loci give rise to primary transcripts that resemble the dicistronic primary tRNA-snoRNA transcripts found in yeast and plants, which are cleaved by RNase Z, thereby generating the mature snoRNAs with 5′ monophosphate termini (Kruszka et al. 2003; Guffanti et al. 2006). The processing mode of UM2 snoRNA primary transcripts is not known but is likely to generate mature snoRNAs with 5′ monophosphate termini (since both endonuclease and exonuclease activity creates this type of 5′ terminus). However, while the intronic processed snoRNA contigs consisted nearly exclusively of TEX-sensitive fragments, most mature UM2 snoRNA contigs were composed of both TEX-sensitive and TEX-insensitive fragments, and the 5′ end-dependent RACE analyses suggested that the differential TEX-sensitivity reflected differences in 5′-end structure. It is unlikely that the processing of UM2 snoRNAs should create two types of 5′ termini, and the most parsimonious explanation is, therefore, that the TEX-insensitive type is caused by some form of secondary 5′-terminal modification. Recent research has indicated that post-transcriptional RNA cleavage event produces substantial numbers of RNA fragments containing 5′-terminal cap-like structures which apparently are produced by a secondary 5′-end capping mechanism (Fejes-Toth et al. 2009; Mercer et al. 2010). Moreover, a cytoplasmic complex with transcription-independent 5′-end capping activity has been demonstrated in other organisms (Otsuka et al. 2009; Schoenberg and Maquat 2009). A study aiming at detecting polyadenylated small RNAs with methylguanosine-cap structures in the rice blast fungus also found numerous transcripts of different size mapping to rRNA, tRNA, snRNA, and other loci (Gowda et al. 2010) that might represent ncRNA transcripts which subsequently underwent secondary 5′-end modification. The apparent secondary 5′-terminal cap modification of snoRNAs is not likely to be a random process as it is somehow limited to UM2 snoRNAs, and the differing degree of TEX-insensitivity among different mature UM2 snoRNAs could also be indicative of a regulated process.
In summary, our data show that an enzymatic approach may be useful in revealing the low abundance fraction of transcriptome. Our data also suggest that RNA fragments may undergo 5′-terminal modification after post-transcriptional cleavage or processing. Thus, despite its organismal simplicity, C. elegans possesses a nearly full complement of protein-coding genes (Hillier et al. 2005), as well as substantial ncRNAs repertoires from intronic and intergenic regions, which indicates the existence of a complicated network of functional is-ncRNAs and protein-coding genes.
MATERIALS AND METHODS
Total RNA preparation
Total RNA was isolated from mixed-stage worms by the Trizol (Invitrogen) protocol according to the manufacturer's instructions. Subsequently, total RNA preparations were subjected to RNase-free DNase I (Ambion) digestion for 30 min at 37°C, and the RNA was purified with acid-phenol:chloroform (pH 4.5; Ambion) followed by three volumes ethanol precipitation, and resuspended in nuclease-free water (AMRESCO). The RNA quality was checked by loading the samples on a 1% formaldehyde-agarose gel and quantified via absorbance spectroscopy.
Quantification of the TEX effect
Two identical samples of 10 μg of total RNA each were either treated with 4 μL of Terminator 5′-phosphate-dependent exonuclease (TEX; Epicentre), or with 4 μL of water (control), in TEX reaction buffer at 30°C for 2 h, followed by purification with acid-phenol:chloroform (pH 4.5; Ambion), and 3 volumes ethanol precipitation. The results were visualized by PAGE gel.
is-ncRNA-specific library construction
Two-hundred micrograms of DNase I-treated total RNA (see Total RNA Preparation) was fractionated on denaturing 6% polyacrylamide gels (7 M urea, 1× TBE buffer). RNA in the size ranging from 50 to 500 nt was excised from the gel and electroeluted in TBE buffer (0.5×) using the D-Tube Dialyzer Maxi MWCO 3.5 kDa (Merck). After polyadenylated RNAs and rRNAs had been removed using an adapted MicrobExpress kit (Ambion), the RNA sample was split into two aliquots, one of which was treated with TEX (Epicentre), and the other left untreated. Subsequently, the two RNA samples were dephosphorylated with FastAP (Fermentas), followed by ligation to the 3′ adaptor oligonucleotide (UUUUGACCACGGTACCCAG; underlined bases are RNA) by T4 RNA ligase (Promega). The 3′ end ligated RNAs were reverse-transcribed by SMARTer PCR cDNA Synthesis kit (Clontech) using an oligo complementary to the 3′ RT adaptor (CTGGGTACCGTGGTCAAA). The two cDNA libraries were amplified using the Advantage 2 PCR kit (Clontech), followed by purification of the PCR products using PureLink PCR Purification Kit (Invitrogen). The two libraries were then sequenced on a Roche/454 GS-FLX system.
Computational analysis
The 5′ and 3′ adaptors were removed by Crossmatch (Ewing et al. 1998), and the resulting reads were mapped to the C. elegans genome (WS190) by NCBI BLAST (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). For each read, only the top hit locus (i.e., the hit with the lowest E-value) was kept, and only if the percent coverage of the alignment (alignment length/read length) was ≥95%. (In the case of several hits having the same lowest E-value and the same sequence identity, these were all kept). In order to identify reads spanning exon-exon junctions, the unmapped reads were subsequently mapped by BLAT (Kent 2002) with identity ≥95% and filtered by the pslCDnaFilter program (http://hgdownload.cse.ucsc.edu/admin/exe/). Genome annotation, sequence, and conservation data were downloaded from WormBase (version WS190) (Harris et al. 2010) and the UCSC genome browser (version ce6) (Fujita et al. 2011). Gene expression profiles from various developmental stages (RNA-seq), H3K4me3 histone modification data, and binding data for 22 transcription factors were downloaded from the modENCODE consortium and related papers (Hillier et al. 2009; Niu et al. 2010; Liu et al. 2011) and lifted over to WS190. The Gene Ontology term enrichment was analyzed using DAVID (Huang et al. 2009). GO terms with significantly (P-value < 0.01) different enrichment between the group I and group II contig host genes were identified using the one-tailed Fisher's exact test and the Benjamini and Hochberg false discovery rate for multiple testing corrections. Conservation analysis was carried out using phastCons data (Siepel et al. 2005). Potential H/ACA and C/D box snoRNAs were identified by combining all predictions by snoGPS (Schattner et al. 2004), snoSeeker (Yang et al. 2006), and snoReport (Hertel et al. 2008). Other potential homologs of known ncRNA were predicted by a search with the HMMER3 software (http://hmmer.janelia.org/) against the Rfam database (Gardner et al. 2011). LocARNA-based clustering (Will et al. 2007) was applied to identify ncRNA classes with similar secondary structures, and the tree was generated by iTOL (Letunic and Bork 2011). The sequence motif detection was performed with MEME (Bailey et al. 2009) and WebLogo (Crooks et al. 2004).
RT-PCR
Two micrograms of DNase I-treated total RNA was polyadenylated by poly(A) polymerase (NEB). The RNA was then reverse-transcribed using oligo (dT) reverse primers in a 20-μL reaction. One microliter of the RT product was used for PCR with gene-specific primers for the transcripts. The primers used in these experiments are listed in Supplemental Document S3.
Quantitative RT-PCR
DNase I-treated total RNA (2.5 μg) were treated with 2 μL of TEX (Epicentre) at 30°C for 2 h and the reaction was terminated by adding 1 μL of 100 mM EDTA (TEX-treated). An identical aliquot was treated likewise, with the exception that TEX was substituted by an equal volume of nuclease-free water (AMRESCO) (untreated control). Subsequently, qRT-PCR analysis of 24 known is-ncRNAs was performed on Rotor-Gene 6000 (Corbett Life Science) with its application software (version 1.7), using the TransScript II Green One-step qRT-PCR SuperMix (TransGen) according to the manufacturer's instructions and 0.5 μL of TEX-treated or untreated total RNA as template. The average expression level in the TEX-treated sample relative to the control sample was determined after normalizing to the amount of 5S rRNA present in each sample by the comparative CT method (Schmittgen and Livak 2008). Quantitative RT-PCR analysis of the expression patterns of 12 novel is-ncRNAs across five larvae stages was performed likewise, and a U6 snRNA (CeN4) was used as a normalizer to calculate the relative expression level of each is-ncRNAs. The primers used in these experiments are listed in Supplemental Document S3.
5′ Cap- or monophosphate-dependent RACE
Of two identical 2 μg of DNase I-treated total RNA aliquots, one was directly ligated to the 5′ adaptor oligonucleotide by 4 μL of T4 RNA ligase (Promega) at 37°C for 2 h, whereas the other was first treated with 2 μL of calf intestine alkaline phosphatase (Promega) and then with 2 μL of tobacco acid pyrophosphatase (Epicentre), followed by ligation to the 5′ adaptor oligonucleotide as given above. Subsequently, 5′ RACE was performed using TransScript II One-step RT-PCR SuperMix (TransGen) with gene-specific downstream primers and the 5′ adaptor upstream primer (Supplemental Document S3).
Northern blot analysis
RNA probes were synthesized and labeled by in vitro transcription of plasmids with T7 RNA polymerase (Fermentas) and Dig-11-UTP (Roche). Total RNA samples (20 μg) were heated at 65°C for 10 min in 2× RNA loading buffer (Fermentas) and resolved by 6% denaturing (8 M urea) polyacrylamide electrophoresis. Subsequently, the RNAs were transferred to Hybond-N+ membranes (Amersham Biosciences) by an ECL semi-dry transfer unit (Amersham Biosciences). Northern blotting was performed per standard and manufacturer's protocols. Blots were hybridized in ULTRAhyb (Ambion) at 60°C overnight, then treated with Blocking and Washing Buffer (Roche) and detected by CDP-star (Roche). The primers used in these experiments are listed in Supplemental Document S3.
Electrophoretic mobility shift assay
SRP RNA (CeN107-1), inc394, inc465, inc467, CeY RNA, CeN72, inc176, and inc332 were transcribed in vitro from plasmids or PCR templates containing a T7 promoter, and the resulting transcripts were 3′ end biotinylated with cytidine (bis) phosphate nucleotide using the 3′ End Biotinylation Kit (Pierce). The ROP-1, SRP9, and SRP14 mRNAs were amplified by RT-PCR from C. elegans total RNAs, using oligodT as the reverse transcription primer, and cloned into the pEASY-E1 vector (TransGen), and expressed in Escherichia coli (BL21[DE3]). 6xHis-tagged proteins or the 6xHis tag (negative control) were purified using Ni-NTA Agarose beads (Qiagen), and ∼2 μg of protein, together with the above biotinylated probes, were used for EMSA (LightShift Chemiluminescent RNA EMSA Kit; Pierce) according to the manufacturer's instructions. The primers used in these experiments are listed in Supplemental Document S3.
ACCESSION NUMBER
Raw 454 pyrosequencing data can be accessed from SRA by SRP007195.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Jian Yang for early discussion on 454 pyrosequencing and Xiaopeng Zhu for instructions in sequencing data and statistical analysis. The C. elegans strain used in this work was provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health—National Center for Research Resources. This work was supported by the Chinese Academy of Science Knowledge Innovation Project (KSCX2-EW-R-01-02), Chinese Academy of Science Strategic Project of Leading Science and Technology (XDA01020402), Innovation Program of Beijing Institute of Life Science (2010-Biols-CAS-0302), and National Key Basic Research and Development Program 973 (2009CB825401).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030965.111.
REFERENCES
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, Sasidharan R, Reinke V, Waterston RH, Gerstein M 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383 doi: 10.1186/1471-2164-11-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK 1980. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 44: 175–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF 2008. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9: 474 doi: 10.1186/1471-2105-9-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boria I, Gruber AR, Tanzer A, Bernhart SH, Lorenz R, Mueller MM, Hofacker IL, Stadler PF 2010. Nematode sbRNAs: Homologs of vertebrate Y RNAs. J Mol Evol 70: 346–358 [DOI] [PubMed] [Google Scholar]
- Bovia F, Wolff N, Ryser S, Strub K 1997. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res 25: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. 2006. Organization of the Caenorhabditis elegans small noncoding transcriptome: Genomic features, biogenesis, and expression. Genome Res 16: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon G, Kapranov P, Foissac S, Willingham A, Duttagupta R, Dumais E, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Pace NR 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem 67: 153–180 [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res 39: D876–D882 doi: 10.1093/nar/gkq963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. 2011. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–D145 doi: 10.1093/nar/gkq1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Nunes CC, Sailsbery J, Xue M, Chen F, Nelson CA, Brown DE, Oh Y, Meng S, Mitchell T, et al. 2010. Genome-wide characterization of methylguanosine-capped and polyadenylated small RNAs in the rice blast fungus Magnaporthe oryzae. Nucleic Acids Res 38: 7558–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti E, Ferrari R, Preti M, Forloni M, Harismendy O, Lefebvre O, Dieci G 2006. A minimal promoter for TFIIIC-dependent in vitro transcription of snoRNA and tRNA genes by RNA polymerase III. J Biol Chem 281: 23945–23957 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. 2010. WormBase: A comprehensive resource for nematode research. Nucleic Acids Res 38: D463–D467 doi: 10.1093/nar/gkp952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang J, Liu T, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, et al. 2007. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res 17: 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Hofacker IL, Stadler PF 2008. SnoReport: Computational identification of snoRNAs with unknown targets. Bioinformatics 24: 158–164 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Coulson A, Murray JI, Bao Z, Sulston JE, Waterston RH 2005. Genomics in C. elegans: So many genes, such a little worm. Genome Res 15: 1651–1660 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38: 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Riechmann JL, Meyerowitz EM 2008. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ 2010. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell 38: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanam T, Rozhdestvensky TS, Bundman M, Galiveti CR, Handel S, Sukonina V, Jordan U, Brosius J, Skryabin BV 2007. Two primate-specific small non-protein-coding RNAs in transgenic mice: Neuronal expression, subcellular localization and binding partners. Nucleic Acids Res 35: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Kang YS, Lai HT, Barakat NH, Magante D, Stumph WE 2010. Identification of SNAPc subunit domains that interact with specific nucleotide positions in the U1 and U6 gene promoters. Mol Cell Biol 30: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S 2006. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232 [DOI] [PubMed] [Google Scholar]
- Kremerskothen J, Zopf D, Walter P, Cheng JG, Nettermann M, Niewerth U, Maraia RJ, Brosius J 1998. Heterodimer SRP9/14 is an integral part of the neural BC200 RNP in primate brain. Neurosci Lett 245: 123–126 [DOI] [PubMed] [Google Scholar]
- Kruszka K, Barneche F, Guyot R, Ailhas J, Meneau I, Schiffer S, Marchfelder A, Echeverria M 2003. Plant dicistronic tRNA-snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J 22: 621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe JC, Burgess J, Rokeach LA, Hekimi S 2000. ROP-1, an RNA quality-control pathway component, affects Caenorhabditis elegans dauer formation. Proc Natl Acad Sci 97: 13233–13238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P 2011. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39: W475–W478 doi: 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, He H, Wang Y, Zheng H, Skogerbo G, Chen R 2008. In vivo analysis of Caenorhabditis elegans noncoding RNA promoter motifs. BMC Mol Biol 9: 71 doi: 10.1186/1471-2199-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Pestova TV, Hellen CU, Tiedge H 2008. Translational control by a small RNA: Dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol 28: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, Yip KY, Wang G, Shou C, Hillier LW, Khurana E, Agarwal A, Auerbach R, Rozowsky J, Cheng C, et al. 2011. Prediction and characterization of noncoding RNAs in C. elegans by integrating conservation, secondary structure, and high-throughput sequencing and array data. Genome Res 21: 276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris M, Kumar M, Lasda E, Larsen A, Kraemer B, Blumenthal T 2007. A novel family of C. elegans snRNPs contains proteins associated with trans-splicing. RNA 13: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu YT, Jankowska M, Nilsen TW 1996. Direct analysis of nematode cis- and trans-spliceosomes: A functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA 2: 735–745 [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. 2010. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res 20: 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P 2011. No-nonsense functions for long noncoding RNAs. Cell 145: 178–181 [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE 2009. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. 2010. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS 2006. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet 22: 1–5 [DOI] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Decatur WA, Davis CA, Ares M Jr, Fournier MJ, Lowe TM 2004. Genome-wide searching for pseudouridylation guide snoRNAs: Analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res 32: 4281–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE 2009. Re-capping the message. Trends Biochem Sci 34: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ 2009. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 5: e1000507 doi: 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Shumyatsky GP, Tillib SV, Kramerov DA 1990. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res 18: 6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Pantaleo V, Toth G, Rusholme Pilcher RL, Moulton V, Burgyan J, Dalmay T 2010. Structural and functional analysis of viral siRNAs. PLoS Pathog 6: e1000838 doi: 10.1371/journal.ppat.1000838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Fremeau RT Jr, Weinstock PH, Arancio O, Brosius J 1991. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci 88: 2093–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci 13: 2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Eisenberg D, O'Brien CA, Wolin SL 1995. Caenorhabditis elegans embryos contain only one major species of Ro RNP. RNA 1: 293–303 [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen J, Wei G, He H, Zhu X, Xiao T, Yuan J, Dong B, He S, Skogerbo G, et al. 2011. The Caenorhabditis elegans intermediate-size transcriptome shows high degree of stage-specific expression. Nucleic Acids Res 39: 5203–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173 [DOI] [PubMed] [Google Scholar]
- Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R 2007. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol 3: e65 doi: 10.1371/journal.pcbi.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MW, Henry RW, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N 1998. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol 18: 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH 2006. snoSeeker: An advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res 34: 5112–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J 2006. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res 34: 2676–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]