The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. Telomerase holoenzyme proteins are required to fold telomerase RNA into its active conformation. In this study, the Stone laboratory employed a combination of single-molecule FRET and RNase protection mapping to demonstrate that the C-terminal domain of the Tetrahymena telomerase holoenzyme protein p65 is essential for its RNA folding activity. RNase probing experiments directly mapped the binding site of the p65 C-terminal domain within telomerase RNA and revealed an unanticipated region of p65-induced conformational change beyond the p65 binding site itself. These studies illuminate the structural principles of a hierarchical RNP assembly process.
Keywords: Tetrahymena telomerase, ribonucleoprotein biogenesis, p65, telomerase RNA, single-molecule FRET
Abstract
The unique cellular activity of the telomerase reverse transcriptase ribonucleoprotein (RNP) requires proper assembly of protein and RNA components into a functional complex. In the ciliate model organism Tetrahymena thermophila, the La-domain protein p65 is required for in vivo assembly of telomerase. Single-molecule and biochemical studies have shown that p65 promotes efficient RNA assembly with the telomerase reverse transcriptase (TERT) protein, in part by inducing a bend in the conserved stem IV region of telomerase RNA (TER). The domain architecture of p65 consists of an N-terminal domain, a La-RRM motif, and a C-terminal domain (CTD). Using single-molecule Förster resonance energy transfer (smFRET), we demonstrate the p65CTD is necessary for the RNA remodeling activity of the protein and is sufficient to induce a substantial conformational change in stem IV of TER. Moreover, nuclease protection assays directly map the site of p65CTD interaction to stem IV and reveal that, in addition to bending stem IV, p65 binding reorganizes nucleotides that comprise the low-affinity TERT binding site within stem–loop IV.
INTRODUCTION
Telomeres protect the ends of linear chromosomes from damage due to homologous recombination and other DNA repair mechanisms (Palm and de Lange 2008). They consist of a repetitive DNA sequence, (T2AG3)n in humans and (T2G4)n in Tetrahymena thermophila, which forms a scaffold for DNA-binding proteins that shelter the DNA. Intact telomeres are essential for cell viability; short telomeres are associated with cellular senescence, and telomere loss can lead to genomic instability and cell death (Gilson and Geli 2007).
Telomeres are progressively shortened during cell division due to the end replication problem; therefore, eukaryotic cells require the specialized enzyme telomerase to maintain telomere length over the course of multiple rounds of cell division. Telomerase dysfunction is associated with the disorders dyskeratosis congenita (DKC) and aplastic anemia (AA), which are marked by symptoms that disproportionately affect proliferative tissues (Vulliamy and Dokal 2008). Conversely, telomerase overexpression can also have negative consequences, helping to confer a high level of proliferative potential upon cells, as evidenced by the presence of telomerase activity in ∼90% of human cancer cell lines (Kim et al. 1994).
Telomerase is a ribonucleoprotein (RNP) enzyme, minimally composed of two elements: a protein subunit called telomerase reverse transcriptase (TERT) and the telomerase RNA (TER). TERT functions by repetitively reverse transcribing a short template region of TER into telomeric DNA sequence (Greider and Blackburn 1989). In addition to containing a template, all known TERs share a conserved structural organization including: a template boundary element (TBE), pseudoknot domain, and a stem terminal element (STE) (Lin et al. 2004; Blackburn and Collins 2010).
Due to the naturally low abundance of both TERT and TER, telomerase RNP assembly presents a serious challenge to the cell. In humans, many mutations associated with the telomerase dysfunction disorder DKC act at the level of telomerase assembly cofactors and not in TERT or TER (Vulliamy and Dokal 2008). Indeed, assembly cofactors play an essential role in telomerase assembly in many different model organisms. In vertebrates, the dyskerin/NHP2/NOP10 complex is required for telomerase biogenesis; in yeast, it is Sm proteins that facilitate telomerase assembly; and in ciliates La domain proteins are required for assembly (Mitchell et al. 1999; Seto et al. 1999; Aigner et al. 2003; Witkin and Collins 2004).
In the well-studied ciliate model organism Tetrahymena thermophila, the La-domain protein p65 facilitates telomerase assembly in vivo and is required for telomere length maintenance (Witkin and Collins 2004). In vitro biochemical studies have shown that p65 improves the Kd of the interaction between TERT and TER (Prathapam et al. 2005), mapped the site of p65-TER interaction to stems I and IV of the RNA (O'Connor and Collins 2006), and demonstrated that the presence of p65 could rescue the catalytic activity of certain telomerase mutants (Berman et al. 2010). A recent study investigating the molecular mechanism of p65-induced telomerase RNP assembly employed single-molecule Förster resonance energy transfer (smFRET), an approach that exploits the distance-dependent energy transfer from a donor fluorophore to an acceptor fluorophore to directly measure conformational changes in biological macromolecules. This work revealed a hierarchal mechanism for telomerase RNP assembly, wherein p65 binds TER and induces a bend in TER stem IV. This positions separate TERT-binding elements in the RNA in the correct orientation for assembly, facilitating the TERT–TER interaction (Stone et al. 2007). Moreover, these studies demonstrated an essential role for the evolutionarily conserved GA bulge within TER stem IV during p65-directed telomerase RNP assembly, consistent with a structural characterization of this GA bulge by NMR spectroscopy that demonstrated conformational flexibility in this region of TER (Richards et al. 2006).
To further dissect the RNA remodeling activity of p65, we expressed various p65 domain constructs and analyzed their RNA binding properties using smFRET and RNase protection mapping. The results directly demonstrate that the novel C-terminal domain (CTD) of p65 is required for p65-mediated TER conformational rearrangement. RNase protection experiments precisely mapped the site of the p65CTD-TER interaction to stem IV. Interestingly, RNase probing experiments also revealed p65 binding to TER induces a substantial reorganization of nucleotides that comprise a low affinity TERT binding site within TER stem–loop IV (Lai et al. 2003). This result suggests the mechanism of p65-facilitated telomerase assembly involves precise RNA conformational changes beyond the previously described bending of TER stem IV.
RESULTS
The p65 C-terminal domain is essential for telomerase RNA remodeling activity
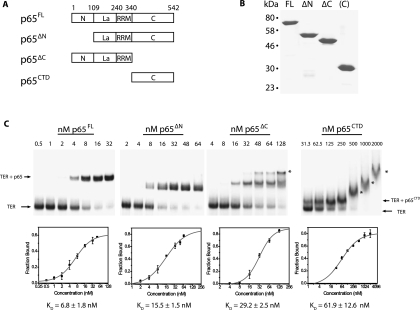
Previous studies of p65 have identified four domains: an N-terminal domain, a La domain, an RNA recognition motif (RRM) domain, and a C-terminal domain (Fig. 1A; Prathapam et al. 2005; O'Connor and Collins 2006). Neither the N-terminal domain nor the C-terminal domain has significant sequence homology with known protein motifs. La motifs are frequently found in conjunction with RRM motifs and often bind poly-U tracts at the 3′ end of RNA Pol III transcripts (Bayfield et al. 2010). Since Tetrahymena TER contains a 3′ U-tract and is transcribed by Pol III (Greider and Blackburn 1989), it is likely the p65 La-RRM domains bind to telomerase RNA in the vicinity of the 3′ terminus. A previous study suggested the uncharacterized C-terminal domain and not the La-RRM motifs were required to increase the affinity between p65-bound TER and the RNA binding domain of TERT (O'Connor and Collins 2006). However, no direct test of the role of p65CTD in promoting conformational rearrangements of TER has been reported.
FIGURE 1.
(A) Schematic diagram demonstrating the domain organization of p65. p65 has four domains: an N-terminal domain, a La motif, an RNA recognition motif (RRM) domain, and a C-terminal domain. Protein constructs were expressed for full-length p65 (p65FL), p65 lacking the N-terminal domain (p65ΔN), p65 lacking the C-terminal domain (p65ΔC), and only the C-terminal domain of p65 (p65CTD). (B) A Coomassie stained SDS-PAGE gel of the four purified p65 constructs. Numbers on the left indicate the position of molecular weight markers. (C) Affinities of p65 constructs for telomerase RNA as determined by gel shift assay. Radiolabeled TER was incubated with increasing concentrations of p65FL, p65ΔN, p65ΔC, and p65CTD and run on a native polyacrylamide gel, top panels. Gels were quantified to determine the fraction bound (bottom panels). Error bars represent the standard deviation of triplicate measurements. The line is a fit of the data used to determine the dissociation constant (Kd) of each p65 construct for its TER substrate. The data were fit to the equation F = [(Fmax)(cn)]/[(Kd)n(cn)], where F represents the fraction bound, c represents the concentration, Kd is the dissociation constant (the concentration at which 50% of the RNA is bound), Fmax represents the maximal value of F, and n represents the Hill coefficient. The following Hill coefficients were obtained: p65FL n = 1.3, p65ΔN n = 1.3, p65ΔC n = 2.1, p65CTD n = 0.9. Asterisks mark higher-order protein-RNA complexes formed at high protein concentrations.
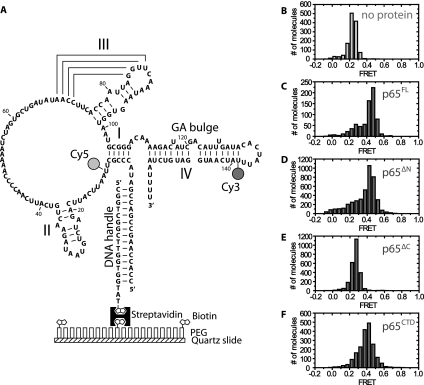
We set out to directly assay which domains were required for p65-mediated conformational rearrangement within TER. To this end, we expressed and purified full-length p65 (p65FL), a p65 N-terminal domain truncation (p65ΔN), a p65 C-terminal truncation (p65ΔC), and the CTD alone (p65CTD) (Fig. 1A,B). Gel shift assays demonstrated that all constructs effectively bound TER, although p65ΔN and p65ΔC had a slightly reduced affinity for TER, while the p65CTD had a further reduced affinity (Fig. 1C). We next employed the previously established smFRET assay of p65 activity (Stone et al. 2007), which measures the efficiency of energy transfer between a donor dye and an acceptor dye incorporated at specific sites within TER. A donor fluorophore was placed on U139 of the native TER sequence, and an acceptor dye was placed on U10, flanking the putative p65 binding site (Fig. 2A). To facilitate our single-molecule studies, a 5′ extension was incorporated into the RNA, which was then hybridized to a short biotinylated DNA oligonucleotide, immobilized onto a streptavidin-coated microscope slide, and imaged using prism-type total internal reflection microscopy (Axelrod et al. 1983). The dye-labeled RNA construct was generated using DNA-splinted RNA ligation of chemically synthesized RNA fragments (Akiyama and Stone 2009) and was shown previously to support wild-type levels of telomerase activity when reconstituted with TERT and p65 in vitro (Stone et al. 2007). We define FRET as IA/(IA + ID), where IA is the intensity of the acceptor dye, and ID is the intensity of the donor dye. In this assay, lower FRET values represent larger inter-dye distances indicative of a more extended stem I and stem IV RNA conformation, while higher FRET values correspond to smaller inter-dye distances and a more compact stem I and stem IV RNA structure.
FIGURE 2.
(A) Diagram of telomerase RNA construct used in the current study. Telomerase RNA was labeled at U139 with a donor fluorophore (Cy3) and at U10 with an acceptor fluorophore (Cy5) for smFRET studies. The RNA construct was deposited on a quartz slide for TIRF microscopy by means of a 5′ extension designed to hybridize with a biotinylated DNA handle. The biotinylated handle was immobilized on the quartz slide via a biotin-streptavidin linkage. (B) Histogram demonstrating the FRET distribution of dye-labeled RNA molecules in the absence of p65. The distribution is centered at 0.26 FRET. (C–F) Histograms of the FRET distribution of dye-labeled RNA molecules in the presence of 10 nM p65FL (C), 32 nM p65ΔN (D), 64 nM p65ΔC (E), or 750 nM p65CTD (F). FRET is defined as IA/(IA + ID), where IA is the intensity of the acceptor dye and ID is the intensity of the donor dye. The protein concentrations used were determined by EMSA to have a large fraction of p65-RNA complexes (see Fig. 1C).
As expected, smFRET measurements of the immobilized RNA yielded a dominant FRET distribution centered at 0.26 FRET (Fig. 2B), consistent with previously reported results on a similar construct (Stone et al. 2007). Also as anticipated, adding increasing amounts of full-length p65 (p65FL) to the immobilized RNA shifted a large fraction of the TER molecules to a 0.45 FRET state (Fig. 2C). Having confirmed that the smFRET assay provides an accurate measure of p65-mediated conformational rearrangement, we next set out to test which domains of p65 were necessary for its RNA remodeling activity. p65ΔN retained the ability to induce a conformational change in TER that was quantitatively very similar (FRET ∼ 0.43) to the FRET change observed with p65FL (cf. Fig. 2C,D). In contrast, p65ΔC failed to induce any detectable FRET change (Fig. 2E), demonstrating that the p65CTD is necessary for p65-mediated structural rearrangement of TER. Importantly, gel shift analysis determined the concentrations used in our smFRET assay were sufficient to support nearly quantitative binding of p65ΔC to TER (Fig. 1C).
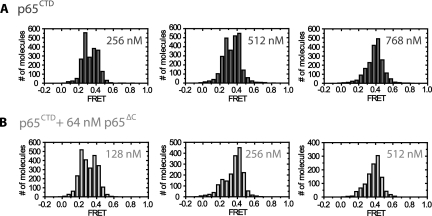
We next tested whether the p65CTD alone was sufficient to induce a TER conformational rearrangement. This construct induced a stable 0.40 FRET state upon protein binding (Fig. 2F; Supplemental Fig. S1), demonstrating that the C-terminal domain alone does induce an RNA structural change. The difference in FRET values between p65CTD and p65FL suggests the conformational states induced by the respective proteins are similar but not identical to one another. The origin of the difference between the p65FL- and p65CTD-induced FRET states may reflect the contributions of other p65 domains to the overall RNA fold. Alternatively, the 0.40 FRET state may represent a time-averaged measurement of rapid RNA conformational dynamics between the 0.26 and 0.45 FRET states. Importantly, the 0.40 FRET state does not appear to be an artifact of high protein concentrations since concentrations of p65ΔC as high as 250 nM induced no conformational change as observed by FRET (Supplemental Fig. S2). Interestingly, we observed that addition of p65CTD to p65ΔC-bound TER complexes shifted the FRET distribution to the 0.40 FRET state at lower concentrations than experiments conducted with p65CTD alone (Fig. 3A,B). This result demonstrates that p65CTD can be added in trans to the p65ΔC protein and suggests noncovalent interactions between the p65CTD and the remainder of p65 stabilize C-terminal domain binding.
FIGURE 3.
Comparison of RNA folding activity between p65CTD and p65CTD coincubated with p65ΔC. The smFRET assay was performed on TER incubated with the indicated concentrations of p65CTD in the absence (A) or presence (B) of 64 nM p65ΔC. Incubating TER with p65ΔC increased the RNA folding activity of p65CTD, as evidenced by the reduced concentrations of p65CTD required to reach the higher FRET state.
p65CTD binds stem IV and reorganizes bases within stem–loop IV
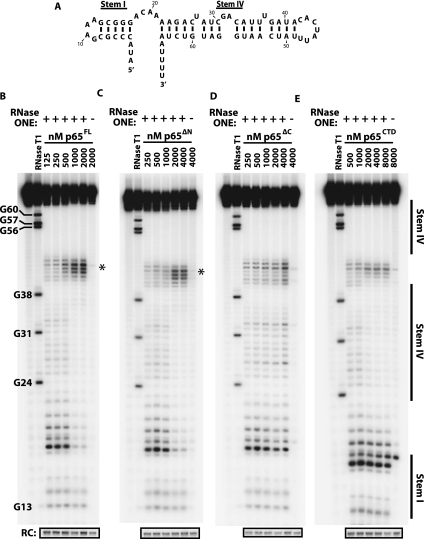
To further dissect the mechanism of p65CTD-induced RNA folding, we next sought to directly identify the p65CTD binding site using RNase protection experiments. We observed that full-length TER was prone to bind multiple p65 proteins at the high protein concentrations required for our footprinting analysis (Supplemental Fig. S3). For this reason, we employed a previously described RNA construct containing only the putative p65 binding site, stem I and stem IV of TER (Fig. 4A; O'Connor and Collins 2006). The stem I-IV TER construct binds p65FL with comparable affinity to the full-length TER and permitted titrations with higher concentrations of the p65 constructs prior to forming higher order protein complexes that could lead to misleading footprinting results. Furthermore, the shorter RNA construct facilitated the unambiguous resolution and quantification of all of the relevant nucleotides on a single gel. We digested the stem I-IV TER construct with RNase ONE, a nominally single-stranded RNA nuclease that lacks sequence specificity. RNase ONE cleavage of the stem I-IV construct produced cleavage products corresponding to predicted unpaired nucleotides. Surprisingly, we also observed cleavage products corresponding to the top strand of stem IV, but no cleavage was observed for the bottom strand. We hypothesize that the bulged residues present in the top strand of stem IV may permit RNase ONE to cleave in this region.
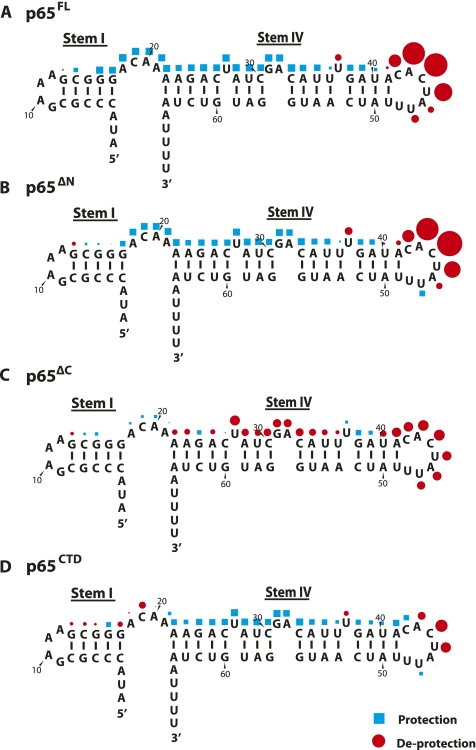
FIGURE 4.
(A) Shorter RNA construct used in RNA footprinting studies containing the putative RNA binding site of p65, stems I and IV of Tetrahymena telomerase RNA. A GAAA tetraloop has been introduced in stem I in place of the remainder of the RNA. (B–E) RNase ONE digestion of stem I–IV construct RNA in increasing concentrations of p65FL (B), p65ΔN (C), p65ΔC (D), and p65CTD (E). RNase T1 digestion was used to generate a ladder to identify nucleotide position. p65FL, p65ΔN, and p65CTD showed strong protection against cleavage in stem IV, suggesting this is the binding site of the C-terminal domain of p65. These constructs also demonstrated hypersensitivity in stem–loop IV (asterisks) upon protein binding, suggesting that p65 remodels this region of the RNA. (RC) recovery control.
Curiously, addition of p65 to our binding reaction, even at concentrations insufficient to drive formation of the p65-TER complex, had a stimulatory effect on RNase ONE cleavage (Supplemental Fig. S4). As a result, cleavage efficiencies were substantially reduced in the absence of p65 which precluded us from using this condition as a baseline for protection mapping. Therefore, for each individual experiment, a protein concentration that showed little to no binding in our gel shift analyses (Supplemental Fig. S5) was used as the reference for quantifying our RNase protection patterns. Importantly, this approach yielded an unambiguous protection trend upon p65 titration across the range of protein concentrations used in this study.
The stem I-IV TER molecules were 5′-end-labeled with 32P, incubated with RNase ONE in the presence of increasing concentrations of each protein construct (p65FL, p65ΔN, p65ΔC, or p65CTD) under single-hit conditions, and then resolved on denaturing PAGE sequencing gels. In the case of p65FL, a clear footprint emerges at higher protein concentrations, demonstrating that p65 binds across the GA bulge on stem IV, consistent with this evolutionarily conserved RNA structural feature being an essential determinant of p65 activity (Fig. 4B; Ye and Romero 2002; Stone et al. 2007). The quantified results reveal that the footprint extends from the GA bulge to the 4-nt linker joining stems I and IV (Fig. 5A, blue squares). No protection was observed in stem I and the lack of RNA cleavage in the 3′ poly-U tract precluded our ability to measure any protein protection in this region of the RNA. Strikingly, we observe a strong hypersensitivity to cleavage in stem–loop IV (Fig. 4B, asterisk; Fig. 5A, red circles), suggesting that, in addition to repositioning stem–loop IV for TERT–TER interaction, p65 also remodels base-pairing contacts in this region, exposing stem–loop IV to single-stranded RNA digestion. In contrast to this result, a previous RNase ONE probing study with p65FL and the native full-length TER sequence showed protection in the same stem–loop IV residues (Berman et al. 2010). We, therefore, repeated our experiments using full-length TER and found the hypersensitivity to RNase ONE cleavage persisted under our binding and cleavage conditions (Supplemental Fig. S6). The source of the discrepancy between the two studies is unclear but may be due to differences in the binding and cleavage conditions.
FIGURE 5.
Quantification of RNase ONE protection results displayed in Figure 4 for p65FL (A), p65ΔN (B), p65ΔC (C), and p65CTD (D). The semi-automated footprinting analysis (SAFA) software package was used to quantify cleavage at individual nucleotides in the RNA (Das et al. 2005). Cleavage was compared at each nucleotide for the highest and the lowest protein concentration to determine the percent protection (blue squares) or deprotection (red circles) upon protein addition. The relative size of the symbols in the figure was scaled to the amount of protection or deprotection observed at that nucleotide. A comparison of the four constructs suggests that the C-terminal domain binds stem IV of telomerase RNA. In addition, the binding of p65 to stem IV sensitizes stem–loop IV nucleotides to RNase ONE cleavage.
Experiments with stem I-IV bound by p65ΔN yielded an identical protection pattern to full-length p65 (Figs. 4C, 5B). In contrast, p65ΔC showed modest hypersensitivity throughout stem IV rather than protection, as well as a reduction in the degree of hypersensitivity in stem–loop IV (Figs. 4D, 5C). Last, protection mapping experiments performed in the presence of p65CTD yielded protection and hypersensitivity patterns that, while slightly less pronounced, are qualitatively very similar to the patterns observed for p65FL (Figs. 4E, 5D). Taken together, these results demonstrate the p65CTD is responsible for the protection of stem IV across the GA bulge, consistent with our smFRET experiments. Furthermore, the nuclease mapping experiments reveal a heretofore unappreciated activity of p65: the remodeling of bases in stem–loop IV which comprise a known low-affinity TERT binding site.
DISCUSSION
Model of p65 function
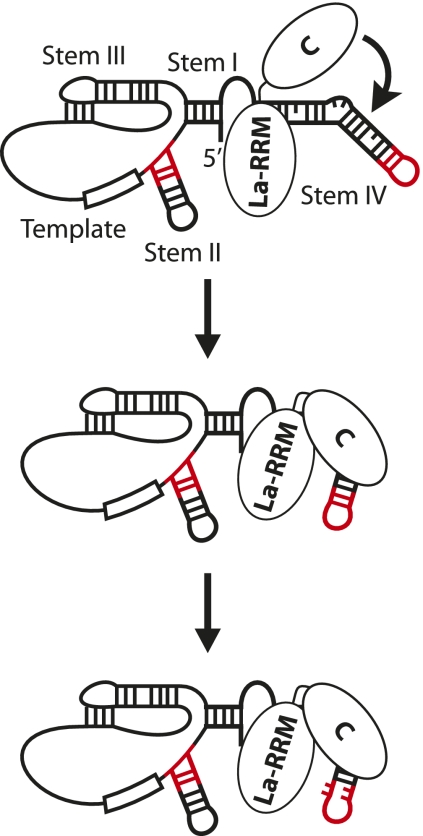
Our results demonstrate that the C-terminal domain of p65 is essential for the RNA conformational change associated with p65 activity. smFRET and RNase protection assays reveal that the C-terminal domain alone induces a related, but not identical, set of conformational rearrangements within TER to those observed with full-length p65. Thus, while the La-RRM domains have no detectable activity with regard to the RNA remodeling activity of p65, they appear to be important for stabilizing the C-terminal domain induced conformational change and helping to improve the affinity of p65 for its RNA substrate. This suggests a model wherein the La-RRM domains bind the poly-U tract of TER, positioning the C-terminal domain for its lower affinity interaction with stem IV and remodeling the RNA for TERT interaction (Fig. 6). Our RNase footprinting results suggest that p65 disrupts contacts within stem–loop IV as part of its activity, in addition to its previously identified bending activity on stem IV. NMR spectroscopy structures of stem–loop IV of Tetrahymena telomerase RNA (Chen et al. 2006; Richards et al. 2006) reveal that, in the absence of p65, stem–loop IV residues are highly ordered, with residues C132 and U138 forming a noncanonical base pair and residues C132, A133, and C134 participating in base stacking interactions. These nucleotides show the greatest deprotection upon addition of p65, suggesting that disruption of these interactions may be responsible for the observed RNase hypersensitivity of these residues. This result is also in accord with the recent report that mutations in stem–loop IV which do not alter p65 affinity for TER can disrupt the ability of p65 to facilitate TERT–TER interactions (Robart et al. 2010).
FIGURE 6.
Model of p65 function. The La-RRM domains and the C-terminal domain of p65 interact with separate sites on telomerase RNA (the 3′ poly-U tract and stem IV, respectively). La-RRM binding of the poly-U tract helps to target the C-terminal domain to stem IV, which, in turn, induces the conformational change associated with p65 activity. This positions TERT-interacting elements (red) in the optimal conformation for assembly. p65 binding also induces a rearrangement in stem–loop IV nucleotides, further facilitating TERT–TER assembly.
Our smFRET assay and RNase footprinting results have not revealed the function of the N-terminal domain of p65. Our results, in combination with previous studies on the effect of N-terminal deletion on the Kd of the TERT–TER interaction (O'Connor and Collins 2006), suggest that the N terminus of p65 is dispensable for the in vitro activity of the protein. Other studies have suggested that p65 may have binding activity outside of stem IV, either in stem I or elsewhere in the RNA (O'Connor and Collins 2006; Berman et al. 2010). It may be that the N-terminal domain helps set up those contacts. An additional possibility is that the N-terminal domain is essential in vivo, perhaps playing a regulatory role such as directing nuclear localization or interaction with other proteins.
MATERIALS AND METHODS
Single-molecule FRET assays
Fluorophore-labeled telomerase RNAs were constructed as previously described (Akiyama and Stone 2009). Briefly, synthetic RNA fragments harboring reactive amine modifications at specific residues (Dharmacon) were labeled with monoreactive Cy3 and Cy5 (GE Life Sciences) and purified by reverse-phase HPLC. A central RNA fragment that did not contain a modification was also generated by targeted RNase H (NEB) cleavage of full-length in vitro transcribed telomerase RNA using targeted cutting oligos (IDT) (Supplemental Table S1). Fragments were assembled by DNA-splinted RNA ligation. Purified fluorophore-labeled RNAs were annealed to the biotin-labeled DNA handle and immobilized on PEGylated quartz slides, and the appropriate p65 construct was flowed over the slide. Slides were imaged on a prism-type total internal fluorescence microscope using an Andor IXON CCD camera with a 100-msec integration time. FRET studies were performed in 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 0.1 mg/mL BSA, 0.1 mg/mL yeast tRNA, 2 mM Trolox, 10% glucose, 10% glycerol, 1 μg/mL catalase, and 1 mg/mL glucose oxidase. FRET is defined as the ratio of IA/(IA+ID), where IA is the acceptor intensity and ID is the donor intensity. Histograms were compiled from the average FRET value obtained from molecules over a 2-sec observation time. Molecules with 0.0 FRET, obtained due to a small fraction of molecules containing bleached acceptor fluorophores, were excluded from histograms by omitting molecules that displayed lower than a minimum threshold value of acceptor intensity.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (O'Connor et al. 2005). Body-labeled TER was transcribed by T7 RNA polymerase (NEB) using α-32P UTP (Perkin-Elmer) and PAGE-purified. 100 pM TER was incubated with various dilutions of the appropriate p65 construct and incubated at 30°C for 20 min. The binding conditions were 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 0.1 mg/mL BSA, 0.1 mg/mL yeast tRNA, 1 mM DTT, and 10% glycerol. The protein-RNA complex was then loaded onto a 5% 37.5:1 acrylamide:bis-acrylamide gel containing 0.5× TBE and 4% glycerol. The gel was run in 0.5× TBE at 200V at 4°C for 3 h, dried, and imaged using an Amersham Phosphor Screen and a GE Typhoon Scanner. Band intensities were quantified using Imagequant and were used to determine Kd values. Percent-bound complexes were plotted in Origin, and Kd values were determined by fitting to the equation F = [(Fmax)(cn)]/[(Kd)n(cn)], where F represents the fraction bound, c represents the concentration, Kd is the dissociation constant (the concentration at which 50% of the RNA is bound), Fmax represents the maximal value of F, and n represents the Hill coefficient.
Protein expression and purification
Proteins were expressed from a pET28 vector behind a 6×-His tag. p65ΔN contained residues 109–542 of p65, p65ΔC contained residues 1–340, and p65CTD contained residues 302–542. All p65 constructs were expressed in Escherichia coli (BL21 DE3). Cells were lysed using a cell disrupter and cell debris pelleted by centrifugation. p65 was purified from lysate on nickel affinity resin (GE Life Sciences) by means of an N-terminal 6×–His tag. The eluant was further purified by ion exchange chromatography on a Hi-trap Q column (GE Life Sciences) and by size-exclusion chromatography on a Superdex 200 column (GE Life Sciences). All proteins were eluted into a final buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 10% glycerol, flash frozen in liquid nitrogen, and stored at −70°C for future use.
RNase protection assays
Stem I-IV TER and full-length TER were synthesized by T7 RNA polymerase (NEB), treated with Turbo DNase (Ambion) and Calf Intestinal Phosphotase (NEB), and PAGE-purified. Purified RNA was end-labeled using phosphonucleotide kinase (NEB) and 32P-labeled γ-ATP (Perkin-Elmer) and PAGE-purified. Purified p65 construct proteins were incubated with 5.0 ng/μl 5′-end-labeled TER construct for 15 min at room temperature in the presence of 20 mM Tris ph 8.0, 100 mM NaCl, 1 mM MgCl2, 0.1 mg/mL yeast tRNA, 1 mM DTT, and 10% glycerol. RNase ONE (Promega) was added at a final concentration of 0.00067 U/μl and incubated at room temperature. After 15 min, a recovery control was added, and the reaction was immediately quenched by phenol:chloroform extraction and ethanol precipitation. RNase T1 (Ambion) digestion was used to generate a reference ladder. The precipitated RNA was fractionated on a 10% 19:1 acrylamide:bis-acrylamide denaturing sequencing PAGE gel. The gel was imaged using an Amersham Phosphor Screen and a GE Typhoon Scanner.
RNase ONE footprinting quantification
RNase footprinting gels were quantified with semi-automated footprinting analysis (SAFA) (Das et al. 2005). Nucleotide bands were assigned using a T1 reference ladder and SAFA's user interface. SAFA fit each lane's intensity profile as the sum of a series of Lorentzian distributions and quantified the relative intensity of each band as the area under each distribution. Band intensities from each lane were then normalized to the amount of material observed in the recovery control. Normalized band intensities from each quantified band from the low protein (LP) control lane were compared against the high protein (HP) lane. For protected residues, the extent of protection at each band was quantified as 1−(HP/LP). For deprotected residues, the extent of hypersensitivity was defined as (HP/LP)−1. This factor was used to scale the sizes of the shapes in Figure 5. Lane intensity profiles for full-length TER (Supplemental Fig. S6) were generated in Imagequant and plotted in Origin.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Kathleen Collins (UC Berkeley) for providing the plasmid constructs used in this study and for useful discussions about this work. We thank Dr. Al Zahler for comments on the manuscript. This work was supported by a National Institutes of Health grant (GM095850) to M.D.S. and a National Institutes of Health training grant (T32 GM8646) to B.M.A.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031377.111.
REFERENCES
- Aigner S, Postberg J, Lipps HJ, Cech TR 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42: 5736–5747 [DOI] [PubMed] [Google Scholar]
- Akiyama BM, Stone MD 2009. Assembly of complex RNAs by splinted ligation. Methods Enzymol 469: 27–46 [DOI] [PubMed] [Google Scholar]
- Axelrod D, Thompson NL, Burghardt TP 1983. Total internal inflection fluorescent microscopy. J Microsc 129: 19–28 [DOI] [PubMed] [Google Scholar]
- Bayfield MA, Yang R, Maraia RJ 2010. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 1799: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Gooding AR, Cech TR 2010. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol 30: 4965–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K 2010. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G 2006. Structure of stem–loop IV of Tetrahymena telomerase RNA. EMBO J 25: 3156–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V 2007. How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell 11: 1673–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci 101: 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- O'Connor CM, Collins K 2006. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol 26: 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Lai CK, Collins K 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem 280: 17533–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O'Connor CM, Collins K 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 12: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Wu H, Trantirek L, O'Connor CM, Collins K, Feigon J 2006. Structural study of elements of Tetrahymena telomerase RNA stem–loop IV domain important for function. RNA 12: 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, O'Connor CM, Collins K 2010. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy TJ, Dokal I 2008. Dyskeratosis congenita: The diverse clinical presentation of mutations in the telomerase complex. Biochimie 90: 122–130 [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev 18: 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye AJ, Romero DP 2002. Phylogenetic relationships amongst tetrahymenine ciliates inferred by a comparison of telomerase RNAs. Int J Syst Evol Microbiol 52: 2297–2302 [DOI] [PubMed] [Google Scholar]