The RNA-binding protein Hnrpab is highly expressed in developing nervous tissue and in neurogenic regions of the adult brain, but its role in neural development and function is unknown. The authors developed a mouse that lacks Hnrpab expression to define what role, if any, Hnrpab plays during mouse neural development. They demonstrate that Hnrpab−/− neural stem and progenitor cells undergo altered differentiation patterns in culture, and mature Hnrpab−/− neurons demonstrate increased sensitivity to glutamate-induced excitotoxicity. They also demonstrate that Hnrpab nucleocytoplasmic distribution in primary neurons is regulated by developmental stage.
Keywords: RNA-binding proteins, neural development, neuron survival, regulation of gene expression, mouse genetic knockout
Abstract
The molecular mechanisms that govern the timing and fate of neural stem-cell differentiation toward the distinct neural lineages of the nervous system are not well defined. The contribution of post-transcriptional regulation of gene expression to neural stem-cell maintenance and differentiation, in particular, remains inadequately characterized. The RNA-binding protein Hnrpab is highly expressed in developing nervous tissue and in neurogenic regions of the adult brain, but its role in neural development and function is unknown. We raised a mouse that lacks Hnrpab expression to define what role, if any, Hnrpab plays during mouse neural development. We performed a genome-wide quantitative analysis of protein expression within the hippocampus of newborn mice to demonstrate significantly altered gene expression in mice lacking Hnrpab relative to Hnrpab-expressing littermates. The proteins affected suggested an altered pattern of neural development and also unexpectedly indicated altered glutamate signaling. We demonstrate that Hnrpab−/− neural stem and progenitor cells undergo altered differentiation patterns in culture, and mature Hnrpab−/− neurons demonstrate increased sensitivity to glutamate-induced excitotoxicity. We also demonstrate that Hnrpab nucleocytoplasmic distribution in primary neurons is regulated by developmental stage.
INTRODUCTION
RNA-binding proteins play a central role in regulating gene expression. In the nervous system mutations in RNA-binding proteins lead to misregulation of gene expression that causes neurological diseases such as mental retardation or neurodegenerative disease such as Amyolateral Sclerosis (Penagarikano et al. 2007; Glisovic et al. 2008; Cooper et al. 2009; Da Cruz and Cleveland 2011). The majority of studies involving the function of RNA-binding proteins involve cultured immortalized cell lines to examine targeted Protein–RNA interactions, since they provide access to homogenous and adaptable material for molecular and biochemical studies. However, applying conclusions from such studies to physiological regulation requires caution, since they rely on cells selected for in vitro survival under conditions that are quite different in vivo. To understand the roles that RNA-binding proteins play during mammalian development, genetic manipulation of mice currently provides the best opportunity to ablate gene function and assess accompanying changes in cellular function that depend on this gene's activity.
The Hnrpab protein has long been linked to the regulation of gene expression, primarily from biochemical studies using cell lines. However, an in vivo role for the protein in mammalian development has not been identified. Hnrpab orthologs are defined by the presence of a unique peptide domain in the N terminus that is evolutionarily conserved (Akindahunsi et al. 2005; Czaplinski et al. 2005; Kroll et al. 2009). Also, Hnrpab contains two RNA recognition motifs (RRMs) and an alternatively spliced C terminus. These domains are paralogous in two other conserved genes, AUF1/hnRNPD and JKTBP/hnRNPDL. The alternative splicing gives rise to two isoforms called Hnrpab1 and Hnrpab2. Hnrpab protein was first purified as an activity from 40S hnRNP particles that disrupted RNA secondary structure, and bound both RNA and DNA (Kumar et al. 1987). The cDNA encoding Hnrpab2 was cloned using this peptide sequence, and the corresponding protein was named hnRNP A/B at the time because of the similarity of its domain arrangement to the emerging hnRNP A and hnRNP B family of RNA-binding proteins (Khan et al. 1991). Contrary to its name, the hnRNP A/B gene (HNRPAB or HNRNPAB) is not a member of what is often referred to as the hnRNP A/B family (namely hnRNP A1, hnRNP A2/B1, and hnRNP A3) (Khan et al. 1991). Hnrpab has been independently linked to transcription, splicing, mRNA editing, translation, mRNA stability, and RNA localization through numerous biochemical studies from different laboratories, and different isoforms have been give numerous different names (Smidt et al. 1995; Lau et al. 1997; Bemark et al. 1998; Wang and Parrish 1999; Mikheev et al. 2000; Yabuki et al. 2001; Dean et al. 2002; Fomenkov et al. 2003; Gao et al. 2004; Khateb et al. 2007; Raju et al. 2008).
Regardless of the steps in gene expression that Hnrpab regulates at the cellular level, many other studies have suggested that Hnrpab might have important roles in the development and function of the vertebrate nervous system. An anxiety-associated strain polymorphism in the promoter of the rat arginine-vasopressin gene affected binding of Hnrpab to this region of DNA (Murgatroyd et al. 2004). Hnrpab was also cloned in a screen for genes differentially expressed in the striatum, and its mRNA was induced in rodent brain by treatment with a dopamine receptor agonist (Rushlow et al. 1999, 2000). Two independent screens for defects in Xenopus nervous-system development demonstrated a role for the Hnrpab ortholog in the developing nervous system, where overexpression leads to anterior developmental defects and cell-autonomous inhibition of neural crest cell migration (Dichmann et al. 2008; Yan et al. 2009). The Zebrafish database contains numerous images of the Hnrpab ortholog's mRNA strongly expressed in the nervous system throughout development (Rauch et al. 2003). A directed in situ hybridization screen of RNA-binding protein expression in newborn mouse heads demonstrated enrichment of Hnrpab expression in neural tissue of newborn mice (McKee et al. 2005). An Hnrpab promoter-driven GFP BAC transgene strongly labels neurons within developing mouse brains (Gong et al. 2003). In adult mice, broad expression in the mature brain is observed using in situ hybridization, with regionally elevated mRNA levels observed in the granule cell layers of the hippocampus, dentate gyrus, and cerebellum, as well as in the subventricular zone, rostral migratory stream, and olfactory bulb (Rushlow et al. 1999; Lein et al. 2007).
There are clearly numerous studies consistent with a role for Hnrpab in regulating gene expression in the brain. But, despite the extensive cache of information about this long-known nucleic acid-binding protein, no data relating these independent studies to in vivo function have ever been published. As a first step toward understanding the function of Hnrpab in vivo we raised an Hnrpab knockout mouse and quantify global protein expression changes in the newborn hippocampus using a novel quantitative proteomic approach. Our results demonstrate that Hnrpab plays a role in neural stem cell maintenance and differentiation as well as cell survival after activation of glutamate signaling pathways. Moreover, we found changes in the subcellular distribution of Hnrpab isoforms during neuronal maturation, suggesting that Hnrpab's role in regulating gene expression may change during neuronal development.
RESULTS
Construction of an Hnrpab−/− mouse line
To study the function of Hnrpab in the nervous system, we sought a mouse with an Hnrpab null alelle. A publicly available mouse ES-cell collection contained numerous lines with Hnrpab gene-traps, and the 5′ most of these was putatively located within exon 5 of Hnrpab (Fig. 1A). AV0426 ES-cells were used to create chimeric mice, and offspring from these were screened for germ-line transmission. PCR primers from exon 4 and reverse primers in the gene trap cassette amplified a band specifically from heterozygous mice (Fig. 1B). DNA sequencing of this band confirmed that the gene trap inserted within intron 5 at nucleotide 2524 of the Hnrpab gene (data not shown). These mice are heterozygous for the HnrpabGt(AV0462)Wtsi allele and were mated to generate mice that were homozygous (Fig. 1B). Hnrpab+/Gt(AV0462)Wtsi and HnrpabGt(AV0462)Wtsi/Gt(AV0462)Wtsi mice are not distinguishable from wild-type littermates based on either physical appearance or behavioral differences, although subtle differences in either of these measures cannot be ruled out at this time.
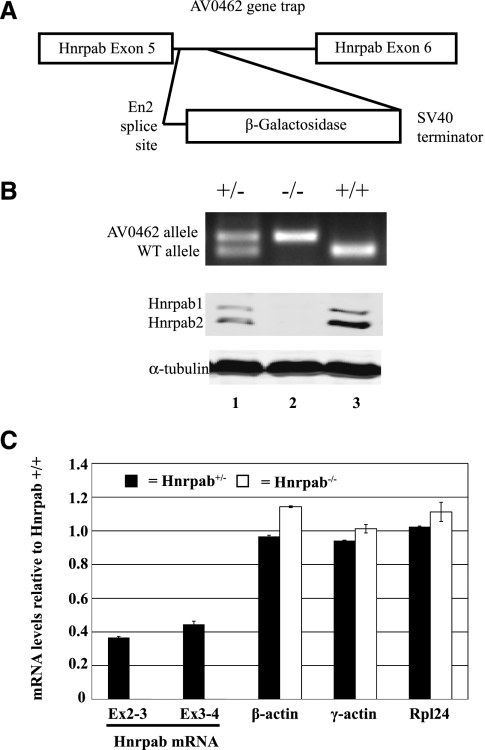
FIGURE 1.
AV0462 ES cell gene trap disrupts Hnrpab expression. (A) A diagram of the region of the Hnrpab gene showing the location of the gene trap insertion. (B, top) Typical PCR genotyping results for AV0462 gene trap heterozygous (lane 1), homozygous (lane 2), and wild-type mice (lane 3). (Middle) Western blots of protein lysates from cerebral cortex of P0 mice using antiserum raised against the N terminus of Hnrpab that recognizes both isoforms of the protein. (Bottom) The α-tubulin loading control to show equal protein loaded in all lanes. (C) The levels of mRNA in heterozygous (black box) or homozygous (white box) P0 cortex relative to wild type were determined using reverse-transcription real-time-PCR (RT–RT–PCR) and plotted above. The error bars represent variation of multiple PCR measurements. Hnrpab exon 2-3 and exon 3-4 were detected in Hnrpab−/− samples, but their decrease was so great that that they are essentially not visible on the chart.
The effect of the gene trap on Hnrpab expression in the CNS was analyzed in pups from a heterozygous to heterozygous mating. We used rabbit polyclonal antiserum against a conserved N terminus peptide (Fig. 1B). In protein lysates from P0 cerebral cortex, two Hnrpab isoforms (Hnrpab1 and Hnrpab2) were detected in Western blots of Hnrpab+/+ and Hnrpab+/Gt(AV0462)Wtsi mice. Although the predicted molecular weight of Hnrpab1 and Hnrpab2 are 36 and 30 kD, respectively, these proteins migrated at ∼42 and 40 kD. In extracts from homozygous mice, these two proteins were not detected, nor is an Hnrpab β-Gal fusion protein predicted by the trapped gene's putative mRNA sequence (Fig. 1B, cf. lanes 1 and 3 with lane 2). To determine whether lack of an Hnrpab-β-Gal protein was due to an effect on accumulation of the chimeric RNA, we used real-time PCR on reverse-transcribed P0 cortex total RNA to determine levels of Hnrpab mRNA relative to wild-type mice. We first used primers that amplify the regions of Hnrpab mRNA corresponding to exon 2–3 and exon 3–4 mRNA, both amplicons are upstream of the gene trap. Heterozygous mice contained ∼40% of the mRNA levels of wild-type mice (Fig. 1C). Homozygous mice had an ∼3000-fold decrease in each of these amplicons, with such a large decrease being measurable in this experiment because of abundance of Hnrpab mRNA in P0 cortex (Fig. 1C). Primers that amplified exon 6–8 (downstream from the gene trap) readily amplified two PCR products from either wild-type or heterozygous mice at a low cycle number (because of two amplicons, the fold difference was not readily quantifiable with real time PCR). These two PCR products, which correspond to the alternatively spliced Hnrpab1 and Hnrpab2 mRNAs, were completely undetectable after 40 cycles in mRNA from homozygous mice (data not shown). Rpl24, β-actin, and γ-actin all contained similar levels of mRNA between wild-type, heterozygous, and homozygous mice (Fig. 1C). We conclude that the small amount of exon 2–3 and exon 3–4 RNA detected in the homozygous animals likely represents residual transcription of the gene trap alleles, but that no functional Hnrpab protein is produced from the AV0462 allele, since neither exon 6–8 mRNA nor antibody reactivity are detected. Therefore, HnrpabGt(AV0462)Wtsi homozygous mice are null for Hnrpab protein expression, and we will refer to this HnrpabGt(AV0462)Wtsi allele as Hnrpab− here for simplicity.
Hnrpab disruption alters hippocampal protein expression
To gain insight into the role of Hnrpab in neural development and function, we performed shotgun proteomics analysis to impartially quantify protein expression changes caused by the loss of Hnrpab in the developing hippocampus (Fig. 2). To quantify protein changes on the proteome-wide scale, we used 15N-labeled hippocampal tissues as internal standards. Our approach is similar to the use of SILAM mouse published previously, except that we labeled the mice with 15N amino acids to increase the numbers and the types of peptides we can quantify (Kruger et al. 2008).
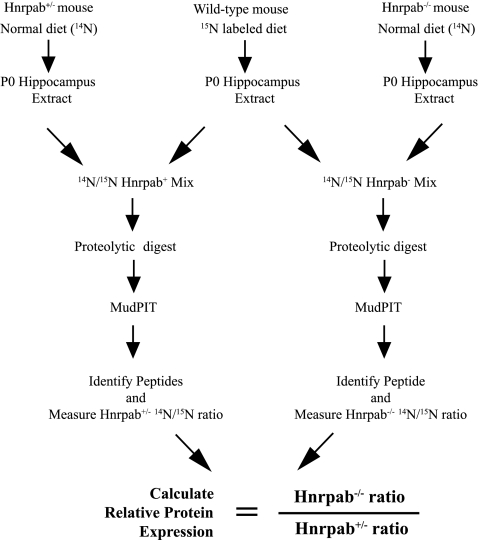
FIGURE 2.
The workflow diagram for quantitative shotgun proteomics of Hnrpab−/− newborn hippocampus. The steps involved in quantifying relative protein levels at the genome-wide scale are diagrammed. Brain tissue from a 15N-labeled littermate was used to determine that incorporation of 15N amino acids was >97% by mass spectrometry. The 15N sample serves as an internal reference by which to compare relative levels of protein between Hnrpab+/− and Hnrpab−/− mice after a Multidimensional Protein Identification Technology analysis (MudPIT).
Protein extract of P0 hippocampus from Hnrpab+/− and Hnrpab−/− (unlabeled, containing 14N amino acids) were fractionated into soluble and insoluble fractions and mixed with equal amount of corresponding 15N-labeled tissue lysates for proteomic analysis (see Materials and Methods). Three littermates of each genotype were prepared and used as biological replicates. SEQUEST was used to identify 14N- and 15N-labeled peptides based on their fragmentation spectra (Eng et al. 1994; Yates et al. 1995). One percent false discovery rate was used to filter protein identifications. CenSus, an algorithm-based quantification software (Park et al. 2008) was used to identify coeluting 14N and 15N peptide peaks from the MS based on MS/MS identifications, generate ratios of coeluting 14N and 15N peptides based on the measured ion intensities, and perform statistical analysis (R2 correlation, ratio distribution of peptides, and others). Only coeluting 14N and 15N peptides with R2 scores >0.5 were used to determine the ratio of a given protein. Protein levels for each animal were expressed as the ratio of unlabeled to 15N-labeled peptides for each protein, and this ratio was averaged over all replicates of the same genotype to represent expression levels for that protein. Differential protein expression was determined by dividing the average Hnrpab−/− ratios (14N Hnrpab−/−/15N standard) by average Hnrpab+/−ratios (14N Hnrpab+/−/15N standard) as illustrated in Figure 2. Soluble and insoluble fractions were analyzed separately.
Volcano plots were used to visualize the changes within the large data sets that we generated from soluble and insoluble fractions (Supplemental Fig. S1; Cui and Churchill 2003). We derived P-values of quantified proteins among biological replicates using the ANOVA test, and plotted significance as the -log10 of P-values with log2 of fold changes (Hnrpab−/−/Hnrpab+/−) on the y- and x-axes, respectively. In the soluble fraction, we found that most of the proteins appeared equidistant from the center, suggesting a normal distribution, with fold changes centered around 1. In the insoluble fraction most changes were centered around 1; however, there was a strong bias toward increased protein in Hnrpab−/− versus Hnrpab+/− hippocampus. The number of these small increases is in contrast to the soluble fraction and it is not likely a random occurrence; we hypothesize that it is a consequence of the lack of Hnrpab expression. The data set also clearly confirmed the lack of Hnrpab peptides in Hnrpab−/− samples.
We used expression ratios of 1.5 for increased proteins or 0.7 for decreased proteins as thresholds to select proteins whose expression was changed in the absence of Hnrpab. Our criteria identified that 349 proteins increased (133 soluble, 216 insoluble; Supplemental Table SI) and 73 proteins decreased (26 soluble, 47 insoluble; Supplemental Table SII) in the Hnrpab−/− hippocampus compared with Hnrpab+/− hippocampus. To understand the biological significance of these differentially regulated proteins we performed pathway-based analysis on our data sets using the Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://ingenuity.com). From the up-regulated protein list, cell death was the most significantly represented cellular function (Supplemental Fig. S2). From the down-regulated protein list, the top 44 entries all had equivalent significance, suggesting that no cellular pathway is predominantly represented (Supplemental Fig. S3).
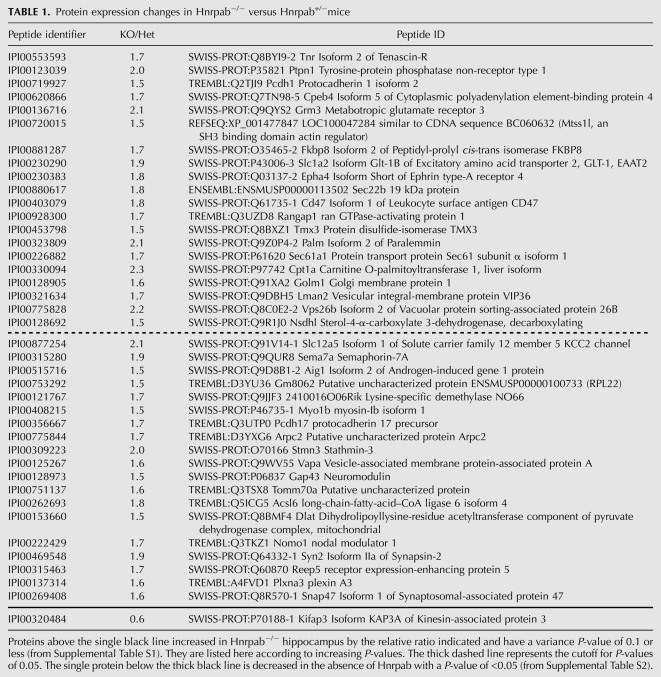
To highlight the most consistent changes, we further restricted the list of affected proteins from our proteomic data to those with ANOVA P-values of 0.1 or lower (Table 1). This list contains 39 increased proteins and one decreased protein. IPA analysis of these increased proteins identified Glutamate Receptor signaling as well as axon guidance signaling as the most significant affected cellular functions (Supplemental Fig. S4). The decreased protein, Kifap3/KAP3, is a kinesin interacting protein involved in anterograde vesicular transport in glutamatergic neurons and is a genetic modifier of ALS in humans (Choi et al. 2008; Orsetti et al. 2011). Results from the proteomic analysis demonstrated that Hnrpab regulates expression of many genes that play important roles in the development of the nervous system.
TABLE 1.
Protein expression changes in Hnrpab−/− versus Hnrpab+/−mice
Hnrpab disruption alters differentiation of neural stem and progenitor cells
These proteomic analyses are consistent with altered neural development in the Hnrpab−/− mice; to evaluate this we used neurosphere cultures. In the presence of EGF and FGF-2, cells isolated from embryonic cerebral cortex propagate in heterogeneous self-adherent cultures called neurospheres (Doetsch et al. 2002; Marshall et al. 2008). Neurospheres contain self-renewing neural stem cells that give rise to the different neural lineages, as well as several types of lineage-committed progenitor cells. These different cell types within the culture can be clearly distinguished by the expression of lineage-specific marker proteins. Proliferating neurosphere cultures were initiated from E18 Hnrpab+/− and Hnrpab−/− mice; then, after subculturing, were dissociated and plated on coverslips. These were maintained 6 d further in culture with differentiation medium before fixation and immunostaining with neural cell-type marker antibodies.
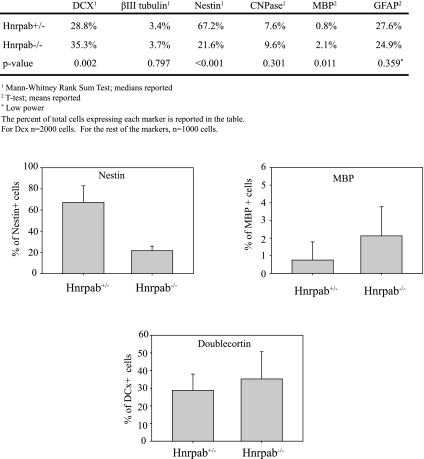
We quantified cells representing different neural lineages within neurosphere cultures and observed changes in several populations. The largest was a decrease in Nestin-expressing (NES) cells in Hnrpab−/− neurosphere cultures (67.2% in Hnrpab+/− vs. 21.6% in Hnrpab−/−) (Fig. 2). Increases in Doublecortin-positive cells (DCX, a neuroblast marker, 28.8% in Hnrpab+/− vs. 35.3% in Hnrpab−/−) (Fig. 3) and Myelin Basic Protein-positive cells (MBP, an oligodendrocyte marker, 0.8% in Hnrpab+/− vs. 2.1% in Hnrpab−/−) had P-values of 0.002 and 0.011, respectively (Fig. 3). Also, a trend toward increased positive cells for an early marker of oligodendrocyte lineage cells, CNPase, was observed (7.6% in Hnrpab+/− vs. 9.6% in Hnrpab−/−, P-value 0.301) (Fig. 3). A trend to slightly fewer Glial Fibrillary Accessory Protein (GFAP)-positive cells was seen (27.6% in Hnrpab+/− vs. 24.9% in Hnrpab−/−, P-value 0.359) (Fig. 3). These data suggest involvement of Hnrpab in neural stem cell maintenance and differentiation of different neural lineages in neurosphere cultures, with increases in expression of some differentiation markers when Hnrpab is disrupted, and a decrease of pluripotent cell types.
FIGURE 3.
Hnrpab disruption affects the differentiation of neural lineage cells in neurosphere cultures. Neurosphere cultures at the second passage were dissociated, plated on coverslips, and grown for 6 d under differentiation conditions, then fixed and immunostained with indicated markers for different neural lineages. For each image, the total number of cells was counted by DAPI staining, and the percentage of those cell that were positive for each marker was calculated. The data represent the average percentage per image field over all the fields imaged (at least 30 fields per marker). This represented a minimum of 1000 cells for all markers, 2000 for Doublecortin. Graphs of the three differences with a P-value of <0.011 are included to illustrate the results. (DCX) Doublecortin; (CNPase) 2′, 3′-cyclic nucleotide 3′-phosphodiesterase; (MBP) Myelin Basic Protein; (GFA) Glial Fibrillary Accessory Protein.
Neurons lacking Hnrpab1 and Hnrpab2 show increased sensitivity to glutamate excitotoxicity and longer neurites
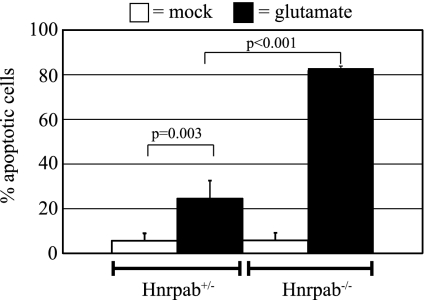
Glutamate receptor signaling and axon guidance signaling topped the lists of affected cellular functions in Hnrpab−/− hippocampus (Supplemental Fig. S4). We considered what consequences these results could have. Glutamate receptor stimulation leads to neuronal depolarization and formation of action potentials. However, this activates cellular stress responses, and excessive glutamate receptor stimulation leads to cell death, a process termed excitotoxicity (Lau and Tymianski 2010). Hypersensitivity to excitotoxicity is believed to underlie many neurodegenerative diseases (Lau and Tymianski 2010). We therefore tested whether Hnrpab−/− neurons would demonstrate an altered sensitivity to cell death after glutamate stimulation. We plated E18 hippocampal neurons from Hnrpab+/− and Hnrpab−/− neurons on coverslips and treated them at 15 d in vitro (DIV) with 50 μM glutamate for 10 min, and allowed them to recover without exogenous glutamate for 6 h prior to fixation. We quantified the percentage of dying neurons in Hnrpab+/− neuron cultures versus Hnrpab−/− cultures in both glutamate-stimulated as well as mock-stimulated cultures to control for the culture manipulations. Very few dying neurons could be found in mock-stimulated Hnrpab+/− or Hnrpab−/− cultures (5.6% and 5.7%, respectively) (Fig. 4). As expected, glutamate treatment increased the number of dying neurons in Hnrpab+/− cultures to 24.5%, consistent with the view that glutamate application has some level of inherent toxicity (Fig. 4). However, 82.8% of the Hnrpab−/− neurons were dying, demonstrating that neurons lacking Hnrpab demonstrate strongly increased sensitivity to glutamate-induced excitotoxicity (Fig. 4).
FIGURE 4.
Hnrpab disruption increases sensitivity of cells to glutamate-stimulated excitotoxicity. At 15DIV, Hnrpab+/− and Hnrpab−/− neurons were treated with 50 μM glutamate for 10 min, or mock treated with the same media changes lacking glutamate, and allowed to recover for 6 h. Cells were then fixed and imaged using 60× magnification, and cell death was scored visually through shrinkage of the nucleus in the DAPI channel and morphology of neurons using DIC microscopy. Error bars represent the standard deviation over three independent experiments. P-values were determined using a two-way ANOVA, followed by the Tukey test for multiple comparisons.
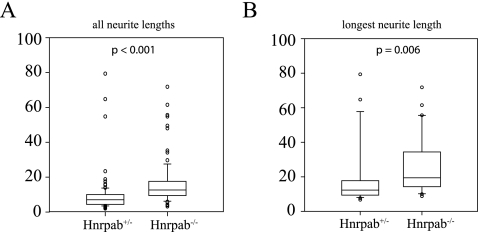
Axon guidance signaling molecules were significantly up-regulated in Hnrpab−/− hippocampus, so we measured the neurite length in Hnrpab+/− and Hnrpab−/− neurons 2 d after plating. Hnrpab−/− neurons had 40% longer neurites than Hnrpab+/− littermate neurons (average length of 11.56 μm for Hnrpab+/− vs. 16.22 μm for Hnrpab−/−) (Fig. 5). Moreover, the average length for the longest neurite of Hnrpab−/− neurons was 32.5% longer than those of Hnrpab+/− littermates (average length of 22.6 μm for Hnrpab+/− vs. 29.9 μm for Hnrpab−/−) (Fig. 5). Longer neurites of cultured neurons is consistent with up-regulation of axon guidance molecules indicated by the proteomic data.
FIGURE 5.
Hnrpab disruption leads to increased neurite length. Primary neurons from Hnrpab+/− and Hnrpab−/− E18 mouse hippocampus were dissociated and plated, then maintained in culture for 2 d prior to fixation. Cells were immunostained for βIII-tubulin, and distance that β-III tubulin extended from the cell body was measured as neurite length. All neurite lengths (A) or longest neurite lengths (B) were plotted with a box-whisker plot and statistics were performed using the Mann-Whitney rank sum test on the median values.
Hnrpab is a nucleocytoplasmic protein in the brain
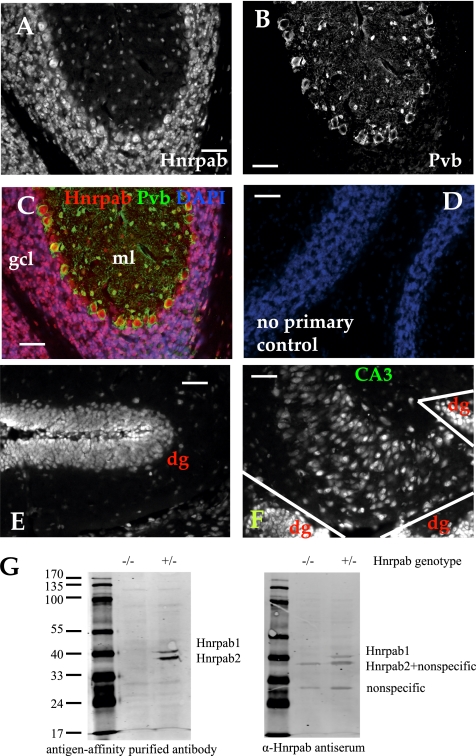
Having established that Hnrpab plays a functional role in the nervous system, we sought to characterize the distribution of Hnrpab within the mature brain. We perfused and sectioned a 55-d-old (postnatal day 55 or P55) mouse brain, then immunostained it using antigen affinity-purified antibody raised against a conserved peptide in the N terminus. This affinity-purified antibody specifically detects Hnrpab1 and Hnrpab2 isoforms in a Western blot (Fig. 6G). Hnrpab staining is visible throughout the brain, and most cells have some level of Hnrpab protein; only occasional cells of unknown identity lacked detectable Hnrpab expression. Overall, we observed agreement of regional Hnrpab protein expression with two previous FISH studies that looked at Hnrpab RNA expression patterns in the brain (Rushlow et al. 1999; Lein et al. 2007). The most prominent Hnrpab staining was observed within the granule cell layers of the hippocampus, dentate gyrus, and cerebellum, so we acquired images within these regions at a higher magnification in order to observe the subcellular distribution of the protein (Fig. 6). DAPI costaining demonstrated that these regions high in Hnrpab expression are also packed with many nuclei. Hnrpab immunoreactivity is enriched in the nuclei of individual cells, although a weaker uniform cytoplasmic distribution throughout the cell soma can often be seen, particularly at the edges of these granule cell layers (Fig. 6A,E). The granule cell layer of the dentate gyrus contained discrete zones of higher Hnrpab expression at the interface between the granule layer and the polymorphic layer (Fig. 6F). The tight packing of the cell bodies within these regions made cytoplasmic signal difficult to characterize in detail; however, in the cerebellum the Purkinje neurons were an exception to this (Fig. 6A–C). Purkinje neurons are GABA-ergic neurons at the interface of the granule cell layer and molecular layers of the cerebellum, easily distinguishable by their size and expression of parvalbumin (PV) in the cytoplasm (Fig. 6B). In Purkinje neurons, Hnrpab staining overlapped with PV, demonstrating cytoplasmic Hnrpab staining (Fig. 6A–C). While nuclear staining was always observed in Hnrpab-expressing cells, cytoplasmic Hnrpab was only sometimes observable in combination with the nuclear staining. Aside from Purkinje neurons, clear examples of the cytoplasmic staining pattern was seen in the cells of the CA3 region in the hippocampus (Fig. 6F). Based on the immunostaining of brain slices, we conclude that at least one isoform of Hnrpab is cytoplasmic during normal neuron function in the brain, although this Hnrpab antibody does not determine whether different isoforms have different subcellular distributions.
FIGURE 6.
Immunofluorescence of Hnrpab in the brain of adult mice. (A) Brain sections were immunostained with N-terminal peptide antibody that recognizes both isoforms of Hnrpab. (B) Coimmunostaining with a parvalbumin monoclonal antibody confirms the cytoplasmic localization of Hnrpab in the cytoplasm of cerebellar Purkinje neurons. (C) A merge of these images pseudo-colored together with DAPI. (D) A pseudo-colored, normalized, then merged image from an adjacent brain section that was processed for immunostaining without primary antibodies. (E,F) Hnrpab immunostained fluorescence image from the dentate gyrus (dg) and the CA3 region of the hippocampus, with the adjacent dg regions indicated are shown in E and F, respectively. The polymorphic layer of the dg in E lies in between the two branches of the granule layer that fold back to ∼180° in this image. Scale bar in all images is 50 μm. (G) Hnrpab+/− and Hnrpab−/− protein lysate was separated on a 10% PAGE gel and probed with Hnrpab peptide-affinity purified antibody or antiserum as indicated.
A cytoplasmic pool of Hnrpab increases during neuronal maturation
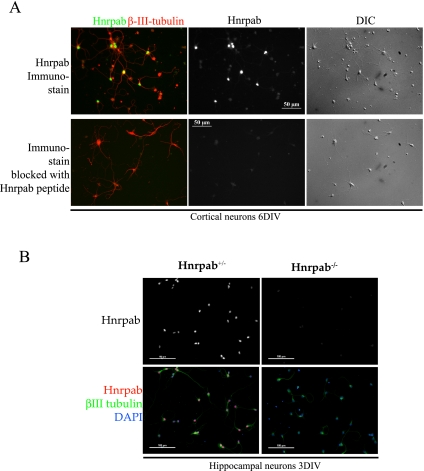
Following the pattern of immunostaining in brain sections, we wanted to understand the requirements for localization of Hnrpab isoforms in neurons, so we first immunostained hippocampal neuron cultures with Hnrpab N terminus peptide antibody and βIII tubulin as a marker for neurons. In neuron cultures 6 DIV, very prominent nuclear staining was detected in all βIII tubulin-positive cells. Weaker, but strictly nuclear signals were also detected in most cells that did not stain with βIII tubulin (Fig. 7A). In these cultures, only very weak cytoplasmic signal was detectable with the N terminus antibody that detects both isoforms (Fig. 7A,B). To determine whether weak cytoplasmic staining was due to Hnrpab immunoreactivity, we blocked the Hnrpab-dependent fluorescence by including excess immunogenic peptide in the staining reaction. This treatment effectively blocked the strong nuclear staining in all cells; however, the intensity of cytoplasmic staining was unaffected by this treatment, suggesting that the very weak cytoplasmic fluorescence in these cells did not reflect a cytoplasmic pool of Hnrpab protein (Fig. 7A). Furthermore, plating neurons from Hnrpab−/− mice demonstrated that the prominent nuclear stain was absent when these neurons were stained with the Hnrpab N terminus peptide antibody, while the same relatively weak cytoplasmic staining remained (Fig. 7B). These results confirm the specificity of our antibody in immunostaining, and suggest that the cytoplasmic appearance of at least one isoform of Hnrpab in neurons is likely to be developmentally regulated, since both isoforms remain primarily nuclear in 6DIV cultured neurons.
FIGURE 7.
Hnrpab localizes to the nucleus in early primary neuron cultures. (A) Primary cortical neurons from an E18 Hnrpab+/− mouse brain were dissociated and plated, and then maintained in culture for 6 d. These were immunostained with N terminus peptide Hnrpab antibody, in the absence (top) or presence (bottom) of the immunogenic peptide. βIII-tubulin was costained as a marker for neurons. Normalized Hnrpab images (middle) are shown together with pseudo-colored Hnrpab/βIII-tubulin merge images (left) and DIC images (right). All images were taken on the same magnification, and the scale bars in the center images are 50 μm. Cell bodies that are βIII-tubulin+ but Hnrpab- in the top panel always coincide with dead neurons in the DIC images. (B) Primary hippocampal neurons from Hnrpab+/− and Hnrpab−/− E18 mouse brains were dissociated and plated, then maintained in culture for 3 d. These were immunostained with N terminus peptide Hnrpab antibody, βIII-tubulin, and DAPI. Normalized images of Hnrpab immunofluorescence (top) and pseudo-colored Hnrpab/βIII-tubulin/DAPI merged images (bottom) are shown. Scale bars in these images represent 100 μm.
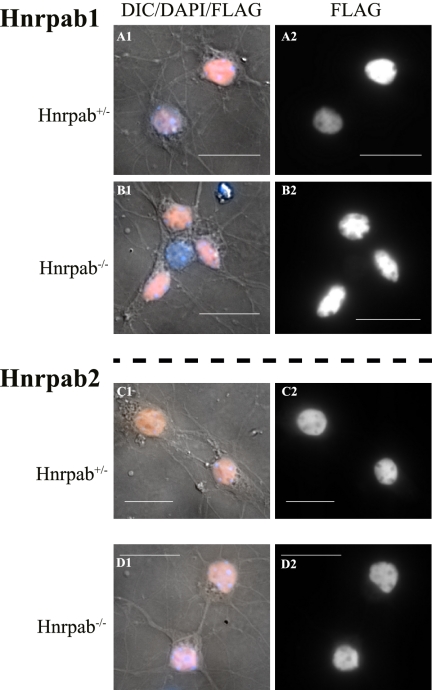
To study the cytoplasmic appearance of the individual isoforms, we expressed recombinant Hnrpab isoforms in cultured neurons. We constructed recombinant lentivirus-like particles (LVPs) designed to express either Hnrpab1 or Hnrpab2, and we incorporated a Flag epitope at the amino terminus to improve our sensitivity of detection. These viruses express only full-length tagged Hnrpab1 or Hnrpab2 by Western blot as expected (data not shown). We plated hippocampal neurons from Hnrpab+/− and Hnrpab−/− neurons on coverslips and first infected these on day 5 with LVPs expressing Hnrpab1 or Hnrpab2. After three more days (8DIV in total), the cells were fixed, immunostained with an anti-Flag antibody and imaged. In Hnrpab+/− cells, Hnrpab1 and Hnrpab2 appear nuclear, albeit a cytoplasmic pool of Hnrpab2 is detectable in the cell body (Fig. 8A,C). Quantifying the ratios of nucleus to cytoplasmic fluorescence shows that Hnrpab−/− cells have the same distribution of either Hnrpab 1 or Hnrpab2 as Hnrpab+/− cells (Supplemental Fig. S5A,B). Uninfected cells do not stain with the anti-Flag antibody and show only weak cytoplasmic fluorescence (autofluorescence) that is not apparent in normalized images (data not shown). We conclude that in immature neurons there is no detectable cytoplasmic Hnrpab1, and only a minor pool of cytoplasmic Hnrpab2 that our peptide antibodies could not clearly detect (cf. Fig. 8, C and D with Fig. 7, A and B).
FIGURE 8.
Hnrpab isoforms are predominantly nuclear in developing neurons. Primary hippocampal neurons from Hnrpab+/− (A,B) and Hnrpab−/− (B,D) E18 mouse brains were dissociated and plated, then maintained in culture for 5 d before infection with recombinant LVPs to express Flag epitope-tagged either Hnrpab1 (A,B) or Hnrpab2 (C,D). These were fixed and processed for immunostaining 3 d later (8DIV), using anti-Flag epitope antibodies and DAPI for the nucleus. Pseudo-colored Flag immunofluorescence (orange) merged with DAPI (blue) and DIC images (gray) (panels labeled 1) are shown next to the Flag immunofluorescence images alone (panels labeled 2). Scale bars represent 20 μm.
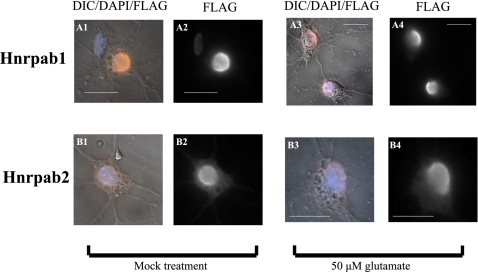
Since immunostaining of brain sections found many neurons with clearly defined cytoplasmic Hnrpab staining, we wanted to know whether neuronal maturation influenced the localization of the Hnrpab isoforms. We plated E18 hippocampal neurons from Hnrpab+/− mice on coverslips as before, but infected these on day 7 with LVPs. At 15DIV the cells were fixed, immunostained with an anti-Flag antibody, and imaged. Hnrpab1 was again largely nuclear, but was now detectable in the cytoplasm (cf. Fig. 9 A1 and A2 with Fig. 8 A1 and A2; Supplemental Fig. S5C). Hnrpab2 was also nuclear, but more clearly detectable in the cytoplasm (cf. Fig. 9 B1 and B2 with Fig. 8, C1 and C2; Supplemental Fig. S5D). Interestingly, Hnrpab2 in the cytoplasm on day 15 is more pronounced than Hnrpab1 (cf. Fig. 9, B1 and B2 with Fig. 9, A1 and A2; cf. ratios in Supplemental Fig. S5, C and D; P-value 0.014 for this comparison). We examined the effect of glutamate excitation on the nucleocytoplasmic appearance of both Hnrpab1 and Hnrpab2. A modest increase in either Hnrpab1 or Hnrpab2 cytoplasmic staining was detected upon glutamate stimulation (Fig. 9, cf. A1 and A2 with A3 and A4, and B1 and B2 with B3 and B4; Supplemental Fig. S5E,F). These results suggest that the localization of Hnrpab to the cytoplasm in brain sections is more associated with a change in stage of neuronal maturation than a state of excitation, and that a larger proportion of the cytoplasmic signal from total Hnrpab staining is due to Hnrpab2.
FIGURE 9.
Hnrpab isoforms appear in the cytoplasm of mature neurons. Primary hippocampal neurons from E18 Hnrpab+/− mouse brains were dissociated and plated on coverslips and then maintained in culture for 8 d before infection with recombinant LVPs to express Flag-epitope tagged either Hnrpab1 (A) or Hnrpab2 (B). On 15DIV cultures were treated with 50 μm of glutamate (panels labeled 3 and 4) or mock treated (panels labeled 1 and 2) for 10 min and then glutamate removed and incubation continued for 6 h. These were fixed and processed for immunostaining using anti-Flag epitope antibodies, and DAPI for the nucleus. Pseudo-colored Flag immunofluorescence (orange) merged with DAPI (blue) and DIC images (panels labeled 1 and 3) are shown next to the Flag immunofluorescence images alone (panels labeled 2 and 4). Scale bars represent 20 μm.
DISCUSSION
A screen of gene expression changes in Hnrpab knockout mice reveals a role for this RNA-binding protein in neural differentiation and excitotoxicity
We raised an Hnrpab knockout mouse from a gene-trapped ES cell collection that lacks detectable expression of both isoforms in late embryonic and early postnatal brains, as well as in any type of cell-cultured cells derived from these brains. We used this Hnrpab null mouse to quantify protein expression changes in the developing hippocampus at the genome-wide level in their in vivo context. The list of the most significant changes reveals that Hnrpab regulates levels of proteins involved in neural development. Consistent with this, neurosphere cultures showed an altered course of differentiation in the absence of Hnrpab expression. Since nestin-positive cells showed the largest decrease in neurosphere cultures, and since we observed an increase in neural progenitor markers, we hypothesize that Hnrpab regulates stem cell maintenance and neural precursor differentiation. We speculate that this role may continue into adulthood, since Hnrpab expression remains high in the neurogenic regions of the brain (subventricular zone and rostral migratory stream), where the adult neural stem cells reside (Rushlow et al. 1999; Lein et al. 2007). Most likely, Hnrpab does not function as a master regulator of neural development, since no class of neural lineage is strongly lost or favored in Hnrpab−/− mice. We think it is more likely that Hnrpab regulates the timing of neural stem cell differentiation, possibly being involved in interpreting environmental signals that influence neural cell fate into changes in gene expression at the transcriptional and/or post-transcriptional levels. Follow-up studies will be required to identify the changes that Hnrpab directly mediates.
The proteomic results also led us to discover that Hnrpab−/− neurons showed increased sensitivity to cell death induced by glutamate stimulation, suggesting that under normal conditions Hnrpab activity prevents cell death that can result from excess neuronal activity. Excitotoxicity results from excessive release of calcium following glutamate receptor simulation, which activates cell stress response and cell death cascades (Wang and Qin 2010; Zhivotovsky and Orrenius 2011). It is also possible that Hnrpab may limit cell death in other tissues where it is expressed. The table of most significantly affected genes includes an increase in Grm3/mGluR3, a G-protein coupled metabotropic glutamate receptor (Table 1). A speculative explanation for glutamate excitotoxicity would be increases in glutamate-stimulated calcium release to toxic levels, due to an increased amount of Grm3 (Hilton et al. 2006). Increases in apoptosis-promoting proteins such as Fkbp8/Fkbp38, a calmodulin-regulated Bcl-I interacting protein, may also exacerbate this phenomenon (Edlich et al. 2006). However, the large number of changes we documented requires future experimentation to determine whether this is the underlying cause for the increase in excitotoxicity. Programmed cell death is very important during neural development; therefore, it is possible that the role of Hnrpab in neural development and excitoxicity both stem from Hnrpab's cellular role in preventing cell death (Martin 2001; Buss et al. 2006). In this scenario, an altered susceptibility to cell death might result in the changes in the neural lineage marker expression that we observe.
The nucleo-cytoplasmic distribution of Hnrpab isoforms suggests a change in the cellular function during neuronal maturation
The localization of Hnrpab protein at first suggests a primarily nuclear function of Hnrpab during development when little or no cytoplasmic signal is detected. This is most consistent with roles in transcription or nuclear mRNA processing such as splicing, editing, or cleavage and polyadenylation. Examples of Hnrpab binding to transcription element containing DNA have been reported; however, the presence of RRMs indicate that it is likely that this protein will play a role in regulating gene expression at the post-transcriptional level (Smidt et al. 1995; Bemark et al. 1998; Mikheev et al. 2000; Yabuki et al. 2001; Gao et al. 2004; Murgatroyd et al. 2004). mRNA localization in the cytoplasm has been demonstrated to require nuclear RNA-binding proteins; however, the precise role of these proteins in the cytoplasmic localization process remains poorly defined (Czaplinski and Singer 2006; Giorgi and Moore 2007). Therefore, nuclear localization does not preclude a protein from playing a role in localizing mRNA in the cytoplasm in immature neurons. In fact, evolutionarily conservation of the Hnrpab protein strongly supports this as cellular role for Hnrpab function. Injecting antibodies into the Xenopus Hnrpab ortholog, 40LoVe, impairs localization of the TGF-β family Vg1 mRNA in the cytoplasm of oocytes, and the Drosophila relative, called Squid, plays a role in localization of different mRNAs during Drosophila oogenesis (Norvell et al. 1999; Czaplinski et al. 2005; Czaplinski and Mattaj 2006; Delanoue et al. 2007). Recombinant Hnrpab2 was suggested to have a detectable preference for an hnRNP A2 responsive element (A2RE), an RNA sequence involved in mRNA transport in oligodendrocytes and neurons, although this particular sequence was demonstrated to bind quite specifically to hnRNP A2 (Hoek et al. 1998; Raju et al. 2008). This observation could be consistent with overlapping roles for Hnrpab2 and hnRNP A2 in mRNA localization in neurons.
Neither bona fide Hnrpab RNA-binding sites nor consensus sites have been identified; however, its preference for G- and/or U-rich RNA or DNA has been reported (Kumar et al. 1987; Sarig et al. 1997; Khateb et al. 2004; Raju et al. 2008). Hnrpab1 specifically associates with AU-rich stability elements (AREs) of some mRNAs, suggesting a role in cell-type and differentiation-regulated mRNA stability for at least some mRNAs. This is interesting because Hnrpab belongs to the AUF1/hnRNP D family of proteins, and AUF1/hnRNP D regulates the stability of many ARE-containing mRNAs (Dean et al. 2002; Czaplinski et al. 2005; Kroll et al. 2009). Under normal conditions, calcium influx upon glutamate stimulation would be expected to impact the stability of many A–U-rich element-containing mRNAs in neurons (Klein et al. 1999). It is therefore appealing to consider that increased sensitivity to excitotoxicity may result from improper regulation of Hnrpab target mRNA stability when Hnrpab is lacking. The larger increase of Hnrpab2 in the cytoplasm after neuronal maturation raises the possibility that cytoplasmically localized Hnrpab2 may take on a role that is distinct from Hnrpab1, such as regulating translation or mRNA stability in this compartment. Similar to Hnrpab, multiple isoforms of the Drosophila Squid protein have distinct roles in mRNA localization and translation (Norvell et al. 1999, 2005; Clouse et al. 2008). It will be worthwhile to delineate the mechanism of how Hnrpab1 and Hnrpab2 are targeted to the nucleus, how the mechanism of localization for each isoform is regulated during neuronal development, and what role Hnrpab1's unique exon 7 plays in regulating this activity.
In contrast to the cytoplasmic immunostaining pattern observed for many other RNA-binding proteins in neurons, we found no evidence for a primarily punctate localization for either Hnrpab isoform in the cytoplasm of neurons in culture or in brain sections. In fact, the localization that we observed contradicts studies using antibodies raised against the unique exon 7 sequence of Hnrpab1, which showed very strong punctate staining throughout the cytoplasm of cells in the brain (Raju et al. 2008, 2011). We never observed punctate staining using affinity-purified antibody preparations from four independent Hnrpab immune-sera (all raised against the Hnrpab N terminus), or with recombinant lentiviral-expressed epitope-tagged Hnrpab1 (Figs. 4–7; data not shown). Our data show that Hnrpab1 becomes detectably cytoplasmic in mature neurons, but is always uniform in its appearance, not strongly granular or punctate.
Changes in gene expression when Hnrpab is disrupted identifies Hnrpab cellular function
We observed hundreds of changes in protein expression when animals develop in the absence of Hnrpab. Classifying these changes allowed us to successfully predict a phenotype for Hnrpab−/− neural cells, although we do not yet know how many of these changes are due directly to the absence of Hnrpab regulation, either transcriptional or post-transcriptional. Hnrpab disruption favors increases in affected proteins at a 5:1 ratio over decreases. This could suggest that Hnrpab has a widespread role in repressing transcription or translation of many transcripts, or for promoting mRNA instability. Alternatively, Hnrpab may directly regulate only one (or a few) regulatory protein that targets many other genes, making many of the changes that we observe indirectly dependent on Hnrpab function. Future experiments will be needed to address the mechanism of these gene expression changes and their role in neural development and neuron activity. With a viable Hnrpab−/− mouse, numerous other experiments to explore neuronal function and survival in living mice are possible and may lead to novel insights into how regulation of gene expression influences neurological disease processes and mouse behavior. Many neurodegenerative diseases are thought to involve excitotoxicity and are largely untreatable. Therefore, understanding the mechanism of Hnrpab−/− sensitivity to excitotoxicity may lead to novel approaches to close this gap.
It is also interesting to consider that some of the changes that we observe, in fact represent a protein signature for how cells must compensate for the lack of Hnrpab. Cellular compensation for loss of embryonic expression of many vital genes has been observed. This allows animals to develop in the absence of such genes, demonstrating that functional cellular plasticity is inherent in mammalian development; however, the cellular mechanisms that adjust for this are generally unknown. If Hnrpab indeed plays some essential role that can be compensated for, then our proteomic results indicate that the cells appear to adjust gene expression networks by altering many different pathways slightly, as opposed to strongly up-regulating one pathway.
MATERIALS AND METHODS
Raising Hnrpab−/− mice
Gene trap AV0462 ES cells in the Wellcome Trust Sanger Institute collection harbored a putative insertion of the pGT01xr gene trap vector into intron 5 of the Hnrpab gene. ES cell-line expansion, gene-trap verification, blastocyst injection, and germ-line screening of chimeras were all performed using standard techniques from the Mutant Mouse Regional Resource Center facility at University of California at Davis. We designed a single PCR reaction to genotype, with a sense-strand primer to exon 4 (5′-ggtggcttgtttcttcttg) in combination with two antisense primers that bind either to intron 5 (to amplify the wild-type allele, 5′-gaagagccagctgtttccag), or the En2 intron of pGT01xr (5′-ggctaccggctaaaacttga). The wild-type allele produces a band of 429 nucleotides (nt) and the Gt(AV0462)Wtsi allele produces a band of 745 nt. Fifteen microliter reactions with Sigma Jump Start Taq PCR mix are used with 1 μL of genomic DNA from mouse tail, prepared with DNeasy Blood and tissue kit (Qiagen). A 58° annealing and 1-min extension time is used. Homozygous males and females breed normally on the mixed genetic background of the germ-line transmitted mice, and this colony is maintained inbred. We routinely mate a heterozygous female with a homozygous male to generate a 1:1 ratio of Hnrpab+/− and Hnrpab−/− mice in each litter.
Hnrpab antisera and affinity-purified antibodies
Amino acids 6–24 of mouse Hnrpab (NH2-EEQPMETTGATENGHEAAP-COOH) were used to raise polyclonal rabbit antibodies (ProteinTech Group). This peptide is present in both isoforms of Hnrpab and is 100% conserved in mouse, rat, and most other mammalian Hnrpab orthologs. We also purified recombinant 6his-tagged human Hnrpab N terminus protein (amino acids 1–71) and ProteinTech Group-raised polyclonal antisera to this. To affinity purify antibodies, either the peptide or 6his-Hnrpab1-71 was attached to a NHS-Sepharose to a high concentration as per the manufacturer's instructions. Immune serum was reacted with resin in batch, then poured into a column for washing (PBS-0.5 M NaCl) before the antigen-purified antibodies were eluted with 0.1 M glycine (pH 2.5), 500 mM NaCl, and immediately neutralized with 1/10 vol of 1M Tris (pH 8.0). This was dialyzed against PBS, concentrated in Dialysis tubing (MWCO 3000) covered in PEG powder (MW 30,000), then dialyzed extensively against PBS-20% glycerol in new dialysis tubing with 10,000 MWCO. This antibody (∼100 μg/mL) was aliquoted and stored at −80°C for long-term storage, or at 4°C for short-term use. Both serum and affinity-purified antibody recognized the expected two isoforms of Hnrpab on Western blots.
Immunostaining of brain sections
A 55-d-old mouse was perfused with PBS-4%PFA and the brain dissected, cryo-protected, frozen, and mounted in tissue-mounting medium. Saggital sections were prepared and post-fixed on the slides with PBS-4%PFA and permeabilized with PBS-1%TritonX-100. After washing in PBS, blocking was performed in CAS block (Zymed) for 1 h, and primary antibodies diluted in CAS block. Hnrpab N-terminal peptide affinity-purified antibody was used at 1:50 and Mouse antiparvalbumin (Sigma P3088) was also used 1:50, and these primary antibodies were incubated with sections overnight at 4°C. After washing in PBS, secondary antibodies (anti-mouse IgG FITC and anti-rabbit IgG-Cy3, Jackson Immunoresearch) were diluted to 1:500 in PBS and incubated for 1 h. After several washes, DAPI was included in the final PBS wash at 300–600 nM. Brain sections were mounted in Pro-long Gold anti-fade (molecular probes). Microscopy was performed on a Nikon TiE widefield fluorescence microscope with appropriate fluorescence filters, and images acquired using a Photometrics cool snap HQ2 camera.
Neuron culture and immunofluorescence
Culture methods are based on routine practices for embryonic neuron cultures (Goslin and Banker 1998; Meberg and Miller 2003; Raju et al. 2008). Hnrpab+/Gt(AV0462)Wtsi mothers are mated with Hnrpab Gt(AV0462)Wtsi / Gt(AV0462)Wtsi males and embryonic day 18 (E18) mouse pups are dissected from the pregnant females, transferred, and extensively washed in ice-cold sterile PBS. Brains are dissected in ice-cold Hanks Basal Salt Solution, w/o Mg++ or Ca++ (HBSS) with 5 mM HEPES (pH 7.5), hippocampi are removed without meninges from individual animals, and tails are kept separate and quickly processed for genotyping. Brain tissue is treated with 0.25% trypsin in HBSS at 37° for 20 min. Trypsin is inactivated by adding Ovomucoid Inhibitor to 1 mg/mL and DNaseI added to 0.2 mg/mL, and cells were incubated for 5 min at room temperature. Tissue pieces are rinsed three times with room temperature Hibernate-E (BrainBitsLLC) containing 2 mM Glutamax and 1× B27, and then dissociated carefully by 10 passes through a flame-polished Pasteur pipette. Cultures are filtered with a 0.4-μm mesh and stored on ice in the dark until genotyping is completed (typically within 24 h). Equal numbers of neurons from animals of the same genotype are then pooled prior to plating 75,000 to 100,000 viable cells on 18-mm glass coverslips coated with 50 μg/mL Poly-L-Lysine hydrobromide in boric acid buffer (50 mM Boric Acid, 12.5 mM Sodium Borate, decahydrate). The hippocampal neurons are diluted in in Neurobasal 1xB27 and 2 mM glutamax with 25 μM glutamate for plating. Coverslips are maintained within one well of a 12-well dish. After cells attach to coverslips (typically within 1 h) the media is changed to remove cell debris. Media half changes occur every 4 d using a 1:1 mixture of fresh Neruobasal-B27-2mM Glutamax and glia-conditioned medium (Meberg and Miller 2003). Cells were maintained at 37°C with 5% CO2 in a humidified incubator.
For glutamate stimulation, complete medium change to fresh Neurobasal-B27-1mM Glutamax was very carefully performed, and after 10 min, cells were rinsed once in plain Neurobasal and then changed back to fresh Neurobasal-B27-Glutamax, and incubation continued for 6 h. All media for glutamate stimulation was equilibrated in 5% CO2 at 37°C prior to application to the cells. Fixation was performed by three serial threefold dilutions of culture medium with PBS-4% PFA, followed by a last complete change with PBS-4% PFA. After 20 min, coverslips were changed into PBS-0.1 M Glycine for 20 min. For storage, cells were changed into 80% methanol and kept at −20. Stored coverslips were rehydrated by six twofold serial dilutions of the storage solution with PBS, followed by a final wash with PBS. These were permeabilized with PBS-0.5% IGEPAL-60 for 5 min. Coverslips were rinsed in PBS and blocked for 30 min with CAS block (Zymed). Hnrpab antibody was diluted 1:50 in CAS block for immunostaining, and anti-Flag M2 monoclonal mouse antibody was diluted into TBST (50 mM Tris at pH 8.0, 150 mM NaCl, 0.1% Tween 20) at a concentration of 10–20 μg/mL. Primary antibodies were incubated with samples overnight at 4°C in a humidified chamber. Coverslips were washed for 1 h with four changes of PBS. Coverslips were incubated with secondary antibody at 1:500 in CAS block for 1 h at room temperature (anti-rabbit IgG -Cy3 or anti-mouse IgG-Alexa 546 for Hnrpab and FlagM2, respectively). Coverslips were washed for 1 h with four changes of PBS, 300 nM DAPI included in the last wash. Coverslips were mounted and imaged as described for brain sections above, with the exception that a QuantEM camera with 512 × 512 pixel chip with the multiplier off was used for image acquisition of the virus-infected neurons.
For neurite length measurements, embryonic day 18 cortical neurons were prepared as described above and allowed to grow for 2 d in vitro. The cells were then fixed with 4% PFA and permeabilized with NP40. The cells were immunostained using β−III tubulin (Sigma T8660) in the presence of CAS block (Invitrogen 00-8120) to block nonspecific interactions. A total of 26 Heterozygous and 30 KO cells with 117 and 99 neurites (respectively) were analyzed using the Nikon NIS Elements Software. A neurite was defined as a β−III tubulin-positive extension from the cell body. The lengths of individual neurites per cell were recorded and the longest neurite from each cell determined. Statistics were performed using a Mann-Whitney Rank Sum Test on the median values of the lengths of each neurite and the longest neurite per cell.
To quantify Nuclear/Cytoplasmic Ratios of Flag-tagged Hnrpab isoforms, at least 12 cells per condition were analyzed using the Nikon NIS Elements Software. The sum intensity Flag staining in the nucleus was determined along with the sum intensity of the total cell soma. The cytoplasmic distribution of the Hnrpab isoforms was calculated by subtracting the nuclear intensity from the total intensity. The ratio of nuclear signal to cytoplasmic signal was then calculated. Statistical analyses of the nuclear to cytoplasmic ratios were performed using the Mann-Whitney Sum Rank Test or students t-test, controlling for multiple comparisons with the Bonferroni adjustment.
Hnrpab expressing virus-like particle (LVP) production
Triple-Flag-tagged Hnrpab1 and Hnrpab2 were cloned by add-on PCR into a Tat and Rev-dependent lentiviral vector (pHAGE-UbC-GIR) (Mostoslavsky et al. 2006). After LVP infection, target cells use the human Ubiquitin C promoter to transcribe an mRNA that will translate three tandem Flag epitopes at the N terminus of either Hnrpab1 or Hnrpab 2, and a Internal Ribosomal Entry Site (IRES), followed by Green Fluorescent Protein from Zoanthus species as a marker for infection of living cells. LVPs are produced by cotransfection of these vectors individually into HEK 293-T cells with Tat, Rev, Gag-pol, and VSV-G envelope protein-expressing vectors as described (Mostoslavsky et al. 2006). Virus-containing culture supernatant is harvested on the first, second, and third days after transfection. Debris is cleared from the virus containing supernatant at 3000g for 10 min. This supernatant from the first and second day is stored on ice until the third day post-transfection. On the third day, all 3 d of debris-cleared supernatant are combined and filtered with a 0.4-μm PES-syringe filter. LVPs are concentrated by ultracentrifugation at 100,000g for 2 h. After removal of the supernatant, the pellet is resuspended in 1.2 mL of DMEM (no serum), aliquoted into 100-μL aliquots, and stored at −80°C. For infection, one aliquot is thawed on ice and infection performed in primary neuron cultures by addition of the appropriate amount of viral stock solutions directly to neuronal culture medium. One aliquot of LVPs prepared as described can effectively infect up to four wells of neurons in a 12-well dish when plated at densities described above.
Proteomic methods
Materials
Invitrosol was purchased from Invitrogen. Trypsin (modified, sequencing grade) was obtained from Promega. Other laboratory reagents were purchased from Sigma-Aldrich and Thermo Fisher Scientific unless noted otherwise.
Sample preparation
An entire litter from Hnrpab+/− and Hnrpab−/− animals was dissected within several hours of birth. Hippocampi from each mouse were combined and fractionated into soluble fractions and insoluble fractions, and the corresponding tails used to genotype the samples. Hippocampus tissues were homogenized in the first mass spectrometry compatible lysis buffer (50 mM Ammonium Bicarbonate,, 0.5× invitrosol, protease, and phosphatase inhibitors [Roche]) using the Precellys 24 tissue homogenizer (Bertin Technologies). After homogenization, tissue lysates were cleared by centrifugation. The cleared supernatant was collected as the soluble fraction. The remaining pellet was solubilized in the second mass spectrometry compatible lysis buffer (50 mM Ammonium Bicarbonate, 8M Urea, 1× invitrosol, protease and phosphatase inhibitors) and collected as the insoluble fraction. The protein concentration from each fraction was determined using the EZQ protein assay.
For quantitative global protein analysis, we used brain lysates from metabolically labeled mice as internal protein standards. C57BL/6 mice were labeled metabolically using stable isotope-labeled (15N) amino acids (SILAM) according to the feeding regimen established in the Chen laboratory. The isotopic incorporation was to >97% of 15N amino acids into proteins in the brain tissue as determined by LC-MS/MS. Age-matched 15N-labeled hippocampus tissues were homogenized and fractionated using the same method described above, and the protein concentration was determined using the EZQ protein assay.
Trypsin digestion
A total of 30 μg of unlabeled soluble or insoluble hippocampus lysates were mixed with 30 μg of corresponding 15N-labeled hippocampus lysates and diluted in 50 mM Ammonium Bicarbonate for trypsin digestion. Trypsin was added to each sample at a ratio of 1:30 enzyme/protein along with 2 mM CaCl2 and incubated for 16 h at 37°C. Following digestion, all reactions were acidified with 90% formic acid (2% final) to stop proteolysis. Then, samples were centrifuged for 30 min at 14,000 rpm to remove insoluble material. The soluble peptide mixtures were collected for LC-MS/MS analysis.
Multidimensional chromatography and tandem mass spectrometry
Peptide mixtures were pressure loaded onto a 250-μm inner diameter (i.d.) fused-silica capillary packed first with 3 cm of 5 μm strong cation exchange material (Partisphere SCX, Whatman), followed by 3 cm of 10 μm of C18 reverse-phase (RP) particles (Aqua). Loaded and washed microcapillaries were connected via a 2-μm filtered union (UpChurch Scientific) to a 100-μm i.d. column that had been pulled to a 5-μm i.d. tip using a P-2000 CO2 laser puller (Sutter Instruments), then packed with 13 cm of 3 μm of C18 reverse-phase (RP) particles (Aqua) and equilibrated in 5% acetonitrile, 0.1% formic acid (Buffer A). This split-column was then installed in-line with a NanoLC Eskigent HPLC pump. The flow rate of channel 2 was set at 300 nL/min for the organic gradient. The flow rate of channel 1 was set to 0.5 μL/min for the salt pulse. Fully automated 11-step chromatography runs were carried out. Three different elution buffers were used: 5% acetonitrile, 0.1% formic acid (Buffer A); 98% acetonitrile, 0.1% formic acid (Buffer B); and 0.5 M ammonium acetate, 5% acetonitrile, 0.1% formic acid (Buffer C). In such sequences of chromatographic events, peptides are sequentially eluted from the SCX resin to the RP resin by increasing salt steps (increase in Buffer C concentration), followed by organic gradients (increase in Buffer B concentration). The last chromatography step consists of a high-salt wash with 100% Buffer C, followed by acetonitrile gradient. The application of a 2.5-kV distal voltage electrosprayed the eluting peptides directly into a LTQ-Orbitrap XL mass spectrometer equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Full MS spectra were recorded on the peptides over a 400–2000-m/z range by the Orbitrap, followed by five tandem mass (MS/MS) events sequentially generated by LTQ in a data-dependent manner on the first, second, third, and fourth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan).
Database search and interpretation of MS/MS data sets
Tandem mass spectra were extracted from raw files, and a binary classifier—previously trained on a manually validated data set—was used to remove the low quality MS/MS spectra. The remaining spectra were searched against a mouse protein database containing 56,871 protein sequences downloaded as FASTA-formatted sequences from EBI-IPI (database version 3.75, released on August, 19, 2010) (Kersey et al. 2004) and 124 common contaminant proteins, for a total of 56,995 target database sequences. To calculate confidence levels and false-positive rates, we used a decoy database containing the reverse sequences of 56,995 proteins appended to the target database (Elias and Gygi 2007) and the SEQUEST algorithm (Eng et al. 1994; Yates et al. 1995) to find the best matching sequences from the combined database.
SEQUEST searches were done using the Integrated Proteomics Pipeline (IP2, Integrated Proteomics Inc.) on Intel Xeon X5450 X/3.0 PROC processor clusters running under the Linux operating system. The peptide mass search tolerance was set to 50 ppm. No differential modifications were considered. No enzymatic cleavage conditions were imposed on the database search, so the search space included all candidate peptides whose theoretical mass fell within the 50-ppm mass tolerance window, despite their tryptic status.
The validity of peptide/spectrum matches was assessed in DTASelect2 (Tabb et al. 2002) using SEQUEST-defined parameters, the cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, and +3) and tryptic status (fully tryptic, half-tryptic, and nontryptic), resulting in nine distinct subgroups. In each one of the subgroups, the distribution of XCorr and DeltaCN values for (1) direct and (2) decoy database hits was obtained, and the two subsets were separated by quadratic discriminant analysis. Outlier points in the two distributions (for example, matches with very low Xcorr, but very high DeltaCN, were discarded. Full separation of the direct and decoy subsets is not generally possible; therefore, the discriminant score was set such that a false-positive rate of 1% was determined based on the number of accepted decoy database peptides. This procedure was independently performed on each data subset, resulting in a false-positive rate independent of tryptic status or charge state.
In addition, a minimum sequence length of 7 amino acid residues was required, and each protein on the final list was supported by at least two independent peptide identifications unless specified. These additional requirements—especially the latter—resulted in the elimination of most decoy database and false-positive hits, as these tended to be overwhelmingly present as proteins identified by single peptide matches. After this last filtering step, the false identification rate was reduced to below 1%.
Quantitative global protein analysis
SEQUEST identified 14N- and 15N-labeled peptides based on their fragmentation spectra. CenSus, an algorithm-based quantification software (Park et al. 2008) was used to identify coeluting 14N and 15N peptide peaks from the MS based on MS/MS identifications, generate ratios of coeluting 14N and 15N peptides based on the measured ion intensities, and perform statistical analysis (R2 correlation, ratio distribution of peptides, etc). Only coeluting 14N and 15N peptides with R2 scores >0.5 were used for protein quantification. Relative expression level between 14N-labeled (Hnrpab+/− or Hnrpab−/−) and 15N-labeled (wild type) for each protein was calculated by averaging the ratio of 14N- to 15N-labeled peptides among animals of a the same genotype. Differential protein expression between Hnrpab+/− or Hnrpab−/− hippocampus lysates was calculated by dividing the Hnrpab−/− 14N/15N ratios by Hnrpab+/− 14N/15N ratios.
Functional analysis of an entire data set
Network analysis using ingenuity pathway analysis software can organize gene-expression changes into groups of genes, which highly influence one another governing specific biological functions. Proteins whose expression was changed by Hnrpab disruption were uploaded into ingenuity software (http://www.ingenuity.com) to perform network analysis. The software scans the input gene-expression data to provide networks by using the Ingenuity Pathway Knowledge Base, which is a database created from data mining for expression and functional relationships between molecules extracted from previously published peer-reviewed papers found in NCBI PubMed, Medline, and several other databases. These proteins were associated with biological pathways using Ingenuity's Knowledge Base. Right-tailed Fisher's exact test with the Benjamini-Hochberg multiple correction to control for false positives was used to calculate a P-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
K.C. is supported by funds from the Stony Brook School of Medicine. This work was supported by grants to S.N. and E.C. from the Susan Komen Catalyst Research Award, and the mass spectrometer used in this study was funded by the shared instrument grant (NIH/NCRR 1 S10 RR023680-1).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030742.111.
REFERENCES
- Akindahunsi AA, Bandiera A, Manzini G 2005. Vertebrate 2xRBD hnRNP proteins: a comparative analysis of genome, mRNA and protein sequences. Comput Biol Chem 29: 13–23 [DOI] [PubMed] [Google Scholar]
- Bemark M, Olsson H, Heinegard D, Leanderson T 1998. Purification and characterization of a protein binding to the SP6 κ promoter. A potential role for CArG-box binding factor-A in κ transcription. J Biol Chem 273: 18881–18890 [DOI] [PubMed] [Google Scholar]
- Buss RR, Sun W, Oppenheim RW 2006. Adaptive roles of programmed cell death during nervous system development. Annu Rev Neurosci 29: 1–35 [DOI] [PubMed] [Google Scholar]
- Choi J, Ha CM, Choi EJ, Jeong CS, Park JW, Baik JH, Park JY, Costa ME, Ojeda SR, Lee BJ 2008. Kinesin superfamily-associated protein 3 is preferentially expressed in glutamatergic neurons and contributes to the excitatory control of female puberty. Endocrinology 149: 6146–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse KN, Ferguson SB, Schupbach T 2008. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev Biol 313: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Churchill GA 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4: 210 doi: 10.1186/gb-2003-4-4-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Mattaj IW 2006. 40LoVe interacts with Vg1RBP/Vera and hnRNP I in binding the Vg1-localization element. RNA 12: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Singer RH 2006. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci 31: 687–693 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Kocher T, Schelder M, Segref A, Wilm M, Mattaj IW 2005. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev Cell 8: 505–515 [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW 2011. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol doi: 10.1016/j.conb.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JL, Sully G, Wait R, Rawlinson L, Clark AR, Saklatvala J 2002. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem J 366: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C 2007. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell 13: 523–538 [DOI] [PubMed] [Google Scholar]
- Dichmann DS, Fletcher RB, Harland RM 2008. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev Dyn 237: 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A 2002. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron 36: 1021–1034 [DOI] [PubMed] [Google Scholar]
- Edlich F, Weiwad M, Wildemann D, Jarczowski F, Kilka S, Moutty MC, Jahreis G, Lucke C, Schmidt W, Striggow F, et al. 2006. The specific FKBP38 inhibitor N-(N′,N′-dimethylcarboxamidomethyl)cycloheximide has potent neuroprotective and neurotrophic properties in brain ischemia. J Biol Chem 281: 14961–14970 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Fomenkov A, Huang YP, Topaloglu O, Brechman A, Osada M, Fomenkova T, Yuriditsky E, Trink B, Sidransky D, Ratovitski E 2003. P63 α mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J Biol Chem 278: 23906–23914 [DOI] [PubMed] [Google Scholar]
- Gao C, Guo H, Wei J, Mi Z, Wai P, Kuo PC 2004. S-nitrosylation of heterogeneous nuclear ribonucleoprotein A/B regulates osteopontin transcription in endotoxin-stimulated murine macrophages. J Biol Chem 279: 11236–11243 [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ 2007. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol 18: 186–193 [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925 [DOI] [PubMed] [Google Scholar]
- Goslin K, Banker G 1998. Rat hippocampal neurons in low-density culture. In Culturing nerve cells, (ed. G. Banker and K. Goslin), pp. 339–370. MIT press, Cambridge, MA [Google Scholar]
- Hilton GD, Nunez JL, Bambrick L, Thompson SM, McCarthy MM 2006. Glutamate-mediated excitotoxicity in neonatal hippocampal neurons is mediated by mGluR-induced release of Ca++ from intracellular stores and is prevented by estradiol. Eur J Neurosci 24: 3008–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Kidd GJ, Carson JH, Smith R 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry 37: 7021–7029 [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4: 1985–1988 [DOI] [PubMed] [Google Scholar]
- Khan FA, Jaiswal AK, Szer W 1991. Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett 290: 159–161 [DOI] [PubMed] [Google Scholar]
- Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M 2004. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res 32: 4145–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M 2007. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res 35: 5775–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein N, Curatola AM, Schneider RJ 1999. Calcium-induced stabilization of AU-rich short-lived mRNAs is a common default response. Gene Expr 7: 357–365 [PMC free article] [PubMed] [Google Scholar]
- Kroll TT, Swenson LB, Hartland EI, Snedden DD, Goodson HV, Huber PW 2009. Interactions of 40LoVe within the ribonucleoprotein complex that forms on the localization element of Xenopus Vg1 mRNA. Mech Dev 126: 523–538 [DOI] [PubMed] [Google Scholar]
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M 2008. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134: 353–364 [DOI] [PubMed] [Google Scholar]
- Kumar A, Sierakowska H, Szer W 1987. Purification and RNA binding properties of a C-type hnRNP protein from HeLa cells. J Biol Chem 262: 17126–17137 [PubMed] [Google Scholar]
- Lau A, Tymianski M 2010. Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch 460: 525–542 [DOI] [PubMed] [Google Scholar]
- Lau PP, Zhu HJ, Nakamuta M, Chan L 1997. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem 272: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176 [DOI] [PubMed] [Google Scholar]
- Marshall GP 2nd, Ross HH, Suslov O, Zheng T, Steindler DA, Laywell ED 2008. Production of neurospheres from CNS tissue. Methods Mol Biol 438: 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ 2001. Neuronal cell death in nervous system development, disease, and injury (Review). Int J Mol Med 7: 455–478 [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA 2005. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol 5: 14 doi: 10.1186/1471-213X-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg PJ, Miller MW 2003. Culturing hippocampal and cortical neurons. Methods Cell Biol 71: 111–127 [DOI] [PubMed] [Google Scholar]
- Mikheev AM, Mikheev SA, Zhang Y, Aebersold R, Zarbl H 2000. CArG binding factor A (CBF-A) is involved in transcriptional regulation of the rat Ha-ras promoter. Nucleic Acids Res 28: 3762–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC 2006. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci 103: 16406–16411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Wigger A, Frank E, Singewald N, Bunck M, Holsboer F, Landgraf R, Spengler D 2004. Impaired repression at a vasopressin promoter polymorphism underlies overexpression of vasopressin in a rat model of trait anxiety. J Neurosci 24: 7762–7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev 13: 864–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvell A, Debec A, Finch D, Gibson L, Thoma B 2005. Squid is required for efficient posterior localization of oskar mRNA during Drosophila oogenesis. Dev Genes Evol 215: 340–349 [DOI] [PubMed] [Google Scholar]
- Orsetti V, Pegoraro E, Cima V, D'Ascenzo C, Palmieri A, Querin G, Volpe M, Ermani M, Angelini C, Soraru G 2011. Genetic variation in KIFAP3 is associated with an upper motor neuron-predominant phenotype in amyotrophic lateral sclerosis. Neurodegener Dis 8: 491–495 [DOI] [PubMed] [Google Scholar]
- Park SK, Venable JD, Xu T, Yates JR 3rd 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods 5: 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST 2007. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet 8: 109–129 [DOI] [PubMed] [Google Scholar]
- Raju CS, Goritz C, Nord Y, Hermanson O, Lopez-Iglesias C, Visa N, Castelo-Branco G, Percipalle P 2008. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol Biol Cell 19: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju CS, Fukuda N, Lopez-Iglesias C, Goritz C, Visa N, Percipalle P 2011. In neurons, activity-dependent association of dendritically transported mRNA transcripts with the transacting factor CBF-A is mediated by A2RE/RTS elements. Mol Biol Cell 22: 1864–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch GJ, Lyons DA, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot WS 2003. Submission and curation of gene expression data. ZFIN Direct Data Submission (http://zfin.org)
- Rushlow WJ, Rajakumar N, Flumerfelt BA, Naus CC 1999. Characterization of CArG-binding protein A initially identified by differential display. Neuroscience 94: 637–649 [DOI] [PubMed] [Google Scholar]
- Rushlow WJ, Rajakumar B, Flumerfelt BA, Naus CC, Rajakumar N 2000. Changes in CArG-binding protein A expression levels following injection(s) of the D1-dopamine agonist SKF-82958 in the intact and 6-hydroxydopamine-lesioned rat. Neuroscience 98: 69–78 [DOI] [PubMed] [Google Scholar]
- Sarig G, Weisman-Shomer P, Fry M 1997. Telomeric and tetraplex DNA binding properties of qTBP42: a homologue of the CArG box binding protein CBF-A. Biochem Biophys Res Commun 237: 617–623 [DOI] [PubMed] [Google Scholar]
- Smidt MP, Russchen B, Snippe L, Wijnholds J, Ab G 1995. Cloning and characterisation of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res 23: 2389–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR 3rd 2002. DTASelect and contrast: Tools for assembling and comparing protein identification from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Parrish CR 1999. A heterogeneous nuclear ribonucleoprotein A/B-related protein binds to single-stranded DNA near the 5′ end or within the genome of feline parvovirus and can modify virus replication. J Virol 73: 7761–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin ZH 2010. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis 15: 1382–1402 [DOI] [PubMed] [Google Scholar]
- Yabuki T, Miyagi S, Ueda H, Saitoh Y, Tsutsumi K 2001. A novel growth-related nuclear protein binds and inhibits rat aldolase B gene promoter. Gene 264: 123–129 [DOI] [PubMed] [Google Scholar]
- Yan CY, Skourides P, Chang C, Brivanlou A 2009. Samba, a Xenopus hnRNP expressed in neural and neural crest tissues. Dev Dyn 238: 204–209 [DOI] [PubMed] [Google Scholar]
- Yates JR 3rd, Eng JK, McCormack AL, Schieltz D 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67: 1426–1436 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S 2011. Calcium and cell death mechanisms: A perspective from the cell death community. Cell Calcium 50: 211–221 [DOI] [PubMed] [Google Scholar]