MALAT-1 noncoding RNA is localized to nuclear speckles despite its mRNA-like characteristics. Here, the authors report the identification of several factors that promote the localization of MALAT-1 to nuclear speckles and also provide evidence that MALAT-1 is involved in the regulation of gene expression. The results suggest that RNPS1, SRm160, and IBP160 contribute to the localization of MALAT-1 to nuclear speckles, where MALAT-1 could be involved in regulating gene expression.
Keywords: MALAT-1, noncoding RNA, nuclear domain, nuclear speckle
Abstract
MALAT-1 noncoding RNA is localized to nuclear speckles despite its mRNA-like characteristics. Here, we report the identification of several key factors that promote the localization of MALAT-1 to nuclear speckles and also provide evidence that MALAT-1 is involved in the regulation of gene expression. Heterokaryon assays revealed that MALAT-1 does not shuttle between the nucleus and cytoplasm. RNAi-mediated repression of the nuclear speckle proteins, RNPS1, SRm160, or IBP160, which are well-known mRNA processing factors, resulted in the diffusion of MALAT-1 to the nucleoplasm. We demonstrated that MALAT-1 contains two distinct elements directing transcripts to nuclear speckles, which were also capable of binding to RNPS1 in vitro. Depletion of MALAT-1 represses the expression of several genes. Taken together, our results suggest that RNPS1, SRm160, and IBP160 contribute to the localization of MALAT-1 to nuclear speckles, where MALAT-1 could be involved in regulating gene expression.
INTRODUCTION
The nuclei of higher eukaryotes are highly compartmentalized and consist of structurally and functionally distinct domains that contain special components involved in transcription, RNA processing, or ribonucleoprotein (RNP) assembly (Misteli 2005). These nuclear domains include the nucleolus, nuclear speckles (also known as the interchromatin granule cluster [IGC]), paraspeckles, gems, PML bodies, and Cajal bodies (Lamond and Earnshaw 1998; Spector 2001; Handwerger and Gall 2006). Several lines of evidence suggest that nuclear speckles function as storage, assembly, and modification compartments, which supply splicing and transcription factors to active transcription sites (Lamond and Spector 2003). Other findings have described a model in which nuclear speckles act as hubs that facilitate efficient integration of distinct steps during gene expression, from transcription to mRNA export (Hall et al. 2006; Han et al. 2011). The function of nuclear speckles is still debatable; therefore, analysis of the structural and functional components of nuclear speckles is an important step toward improving our understanding of nuclear speckle functions.
Recent large-scale transcriptome analyses have revealed a large number of transcripts with low protein-coding potential, known as noncoding RNAs (ncRNAs) (Birney et al. 2007; Yasuda and Hayashizaki 2008). Furthermore, genome-wide transcriptional profiling in many organisms, from Caenorhabditis elegans to humans, has revealed that the majority of genomes, including intergenic regions, are highly transcribed and produce noncoding transcripts along with classical mRNAs (Kapranov et al. 2007). Recently, genome-wide surveillance has identified more than 9000 ncRNAs that interact with the polycomb repressive complex 2 (PRC2) (Zhao et al. 2010). This complex catalyzes trimethylation of histone H3-lysine27 (H3-K27me3). Thousands of transcripts associated with the enhancer region (eRNAs) (Kim et al. 2010; Wang et al. 2011), transcription start-site-associated RNAs (TSSa) (Seila et al. 2008), promoter-associated short RNAs (PASR), promoter-associated long RNAs (PALR) (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009), and long intergenic noncoding RNAs (lincRNAs) (Guttman et al. 2009) have been identified. The ncRNAs can be roughly classified into two groups: small transcripts, such as microRNAs and piwi-interacting RNAs (piRNAs), and long transcripts. Although the biological importance of small ncRNAs has been documented in recent years, the physiological functions of long ncRNAs are poorly understood. Recent studies have revealed that several long ncRNAs are involved in the regulation of genome organization and gene expression, or that they are structural components of functional domains in the nucleus, such as XIST, Air, Kcnq1ot1, and MEN ɛ/β (or NEAT1) (Hannon et al. 2006; Prasanth and Spector 2007; Mercer et al. 2009).
These nuclear ncRNAs exhibit characteristic subcellular localizations. A key factor in the dosage compensation of the mammalian X-chromosome, XIST ncRNA, is distributed along the target X-chromosome, where it silences gene expression by changing chromatin structure (Heard and Disteche 2006; Erwin and Lee 2008; Payer and Lee 2008). The Air transcript targets the paternal chromosome and is involved in triggering the spreading of inactive chromosomal structures associated with imprinting, although the actual mechanism of how this occurs remains to be elucidated (Braidotti et al. 2004). Kcnq1ot1/Lit1, which interacts with histone methyltransferases G9a and the PRC2 complex to bidirectionally silence the genes, localizes to the Kcnq1 domain (Murakami et al. 2007; Pandey et al. 2008). MEN ɛ/β localizes to nuclear paraspeckles, where it acts as an essential structural component (Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009). It is evident that the localization of ncRNAs within the nucleus is important for achieving their structural and biological functions. In many cases, the localization mechanisms are closely associated with structural and functional entities.
Metastasis associated in lung adenocarcinoma transcript 1 (MALAT-1; also known as NEAT2) was originally identified as a transcript showing significant expression in individuals at high risk for the metastasis of nonsmall-cell lung tumors. It is a highly conserved mRNA-like long ncRNA of >8000 nt (Ji et al. 2003), exhibiting different patterns of expression across various tissues. This suggests that MALAT-1 has undefined but potentially important functional roles in mammalian cells (Ji et al. 2003; Hutchinson et al. 2007). The fact that MALAT-1 localizes to nuclear speckles (Hutchinson et al. 2007) led us to formulate the hypothesis that MALAT-1 has a function associated with nuclear speckles, such as pre-mRNA metabolism or transcriptional regulation. Factors that determine the distribution of RNA are linked to ncRNA functional entities, and therefore it is important to identify the cis- and trans-acting factors that influence the localization of MALAT-1 to nuclear speckles.
RESULTS
MALAT-1 is stably retained in the nucleus
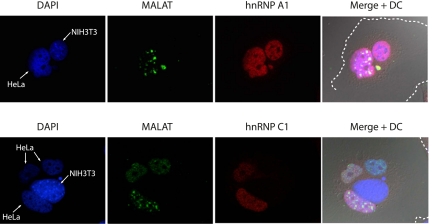
Factors that localize to nuclear speckles, such as subsets of the SR protein and U1, U2, U4, and U5 small nuclear RNAs (snRNAs), shuttle between the nucleus and cytoplasm. This translocation is important for their structural maturation and function (Caceres et al. 1998; Patel and Bellini 2008; Sapra et al. 2009). We used a heterokaryon assay (Pinol-Roma and Dreyfuss 1992) to determine whether MALAT-1 shuttles between the nucleus and cytoplasm. HeLa cells expressing either myc-tagged hnRNP A1 (Michael et al. 1995) or myc-tagged hnRNP C1 (Nakielny and Dreyfuss 1996) were fused with mouse NIH 3T3 cells using polyethylene glycol (PEG). The heterokaryons were then subjected to fluorescent in situ hybridization (FISH) analysis, followed by immunocytochemistry to visualize the localization of MALAT-1 together with that of myc-tagged proteins. Before and after fusion, the cells were treated with cycloheximide to avoid further protein synthesis in the heterokaryons. To distinguish between the human and mouse nuclei, the heterokaryons were stained with 4′,6-diamidino-2-phenylindole (DAPI), which produces a characteristic staining of intranuclear bodies in mouse NIH 3T3 cells. As reported previously, the nucleocytoplasmic shuttling protein, myc-tagged hnRNP A1, originally expressed in HeLa cells, was detected within the mouse nuclei, indicating that successful nucleocytoplasmic shuttling took place under these experimental conditions. A protein that is known to be stably retained in the nucleus, myc-tagged hnRNP C1, was detected only in nuclei derived from HeLa cells. MALAT-1 RNA was detected in HeLa nuclei, but not in mouse NIH 3T3 nuclei (Fig. 1), indicating that MALAT-1 is stably retained in the nucleus after its transcription. Although MALAT-1 was localized to nuclear speckles, some of the MALAT-1 transcript diffused into the nucleoplasm after PEG treatment (Fig. 1). Treatment with PEG did not disrupt the structure of the nuclear speckles, as determined by SRSF2 (SC35) immunostaining; however, PEG treatment may have partially affected the localizing machinery of MALAT-1 in nuclear speckles, but its retention in the nucleus remained stable.
FIGURE 1.
MALAT-1 is stably retained in the nucleus. The localization of human MALAT-1 RNA (green) and Flag-tagged hnRNP-A1/C1 (red) in HeLa-TO cells were examined by fluorescent in situ hybridization and immunocytochemistry, respectively. The nuclei of mouse NIH3T3 cells were easily distinguished from those of human HeLa cells (indicated by arrows) by the characteristic DAPI staining of intranuclear bodies (granules) in the mouse nuclei. Broken white lines in the merged images indicate the heterokaryon's boundaries.
Two independent regions of MALAT-1 direct the localization to nuclear speckles
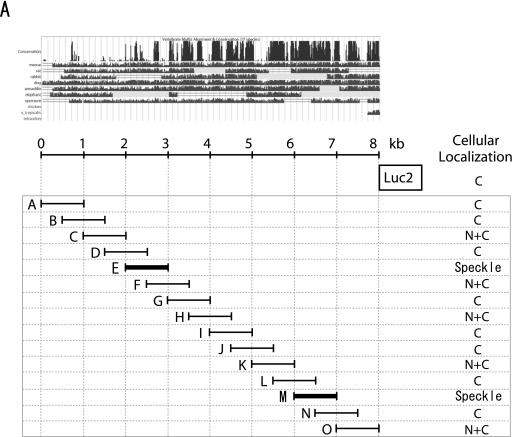
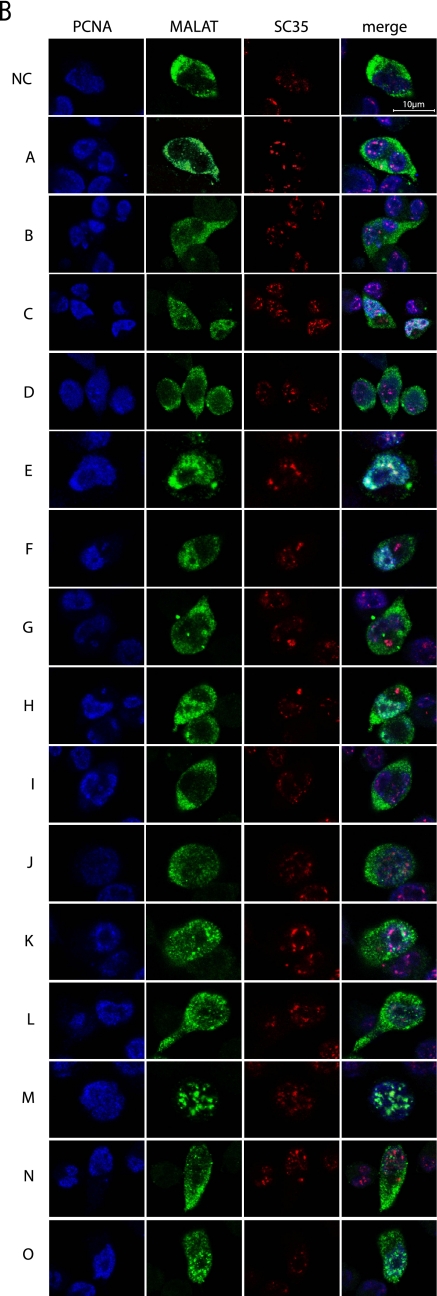
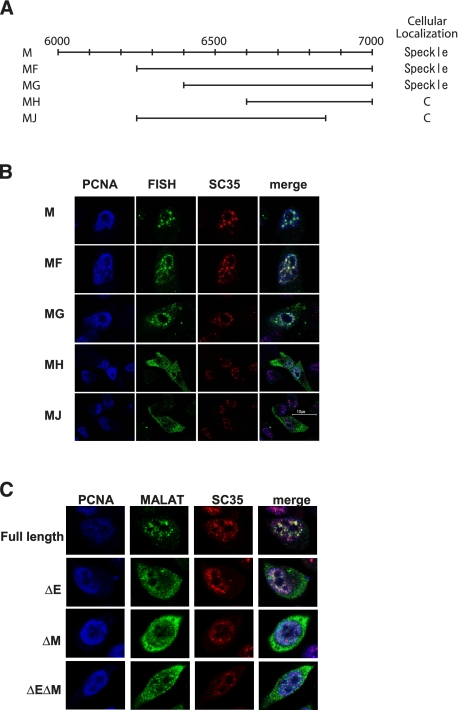
To identify the region in MALAT-1 that is responsible for its localization to nuclear speckles, we constructed a series of plasmids encoding fragments of ∼1 kb of MALAT-1 cDNA fused with luciferase cDNA at the 3′ ends. The transcribed luciferase sequence acts as a tag for detecting and differentiating between endogenously and exogenously expressed MALAT-1 (Fig. 2A). Luciferase mRNA without the MALAT-1 sequence was detected in the cytoplasm (Fig. 2B). In contrast, MALAT-1 fragments fused to luciferase exhibited three different distribution patterns in cells. The RNA signals from region E (1961–3040 nt of the cloned MALAT-1 sequence) and region M (6008–7011 nt of the cloned MALAT-1 sequence) coincided with SRSF2 (SC35) immunostaining (Fig. 2B), although region E showed a slight diffusion away from the nuclear speckles. Regions A, B, D, G, I, J, L, and N of MALAT1 showed exclusive cytoplasmic staining patterns, while the staining for regions C, F, H, K, and O was detected in both the nucleoplasm and cytoplasm. Although the 5′ half of region E overlapped with the D fragment, and the 3′ half of E overlapped with F, RNA signals from regions D and F did not localize to nuclear speckles. Similarly, regions L and N, containing the 5′ and 3′ halves of fragment M, respectively, did not exhibit a nuclear speckle staining pattern. These results suggest that a broad region of fragments E and M were required for localization of MALAT-1 to nuclear speckles. We constructed a series of plasmids encoding serial deletions of the M fragment fused to luciferase cDNA (Fig. 3A) and found that MF (with an ∼250-base deletion from the 5′ end of the M fragment) and MG (with an ∼400-base deletion from the 5′ end of the M fragment) still localized to nuclear speckles (Fig. 3B). In contrast, MH (with an ∼600-base deletion from the 5′ end of the M fragment) and MJ (with an ∼150-base deletion from the 3′ end of the MF fragment) did not localize to nuclear speckles. Taken together, these results indicate that a region covering ∼600 bases, from nucleotides 6387 to 7011, is required for the localization of the M fragment to nuclear speckles. This supports our hypothesis that a broad region of MALAT-1 is required for its localization to nuclear speckles.
FIGURE 2.
Subcellular localization of MALAT-1 fragments. (A) Variants of MALAT-1 RNA used in the experiments. Luc indicates luciferase cDNA. Images from the UCSC Genome Browser (http://genome.ucsc.edu/) represent the data of evolutionary conservations among 17 vertebrates. The human MALAT-1 locus is shown at the top with the graduated sizes. The results of the subcellular distribution analysis are summarized in the right column. C and N+C indicate distribution in the cytoplasm, and nucleoplasm with cytoplasm, respectively. (B) FISH analyses of HeLa-TO cells, transiently transfected with the indicated MALAT-1 variants (green) are displayed, together with immunocytochemistry staining with an anti-PCNA (blue) and anti-SC35 antibody (red). Both images were merged to represent the subcellular localization of the MALAT-1 derivatives. Scale bar, 5 μm.
FIGURE 3.
Subcellular localization of inner fragments derived from the M region of MALAT-1. (A) A map of inner fragments derived from the M region of MALAT-1. (B) FISH analysis of HeLa-TO cells transfected with the indicated inner MALAT-1 fragments (green), together with immunocytochemistry staining of PCNA (blue) and SC-35 (red). The merged images demonstrate the subcellular localization of the inner MALAT-1 fragments. (C) FISH analyses of HeLa-TO cells transfected with the MALAT-1 variants lacking either the E or M regions (ΔE, ΔM), or both (ΔEΔM) (green) are displayed, together with immunocytochemistry staining of PCNA (blue) and SC-35 (red). The merged images demonstrate that these deletion mutants do not localize in nuclear speckles.
We constructed a series of plasmids that express mutant MALAT-1 RNA lacking either regions E or M, or both, to determine whether they were necessary (Fig. 3C). We found that these mutated MALAT-1s did not localize in nuclear speckles, but instead localized within the cytoplasm. These results indicated that the concurrent presence of both regions E and M is essential for the localization of MALAT-1 to nuclear speckles.
A homology search of the E and M regions of MALAT-1 failed to find a conserved sequence that could serve as a common motif responsible for nuclear speckle localization. Any common secondary structure in the E and M regions was not predicted by Mfold or CentroidFold computer tools (Sato et al. 2009). These results suggest that these two regions of MALAT-1 have distinct sequence or secondary structure that directs localization to nuclear speckles.
Depletion of RNPS1, SRm160, or IBP160 disrupts the localization of MALAT-1 to nuclear speckles
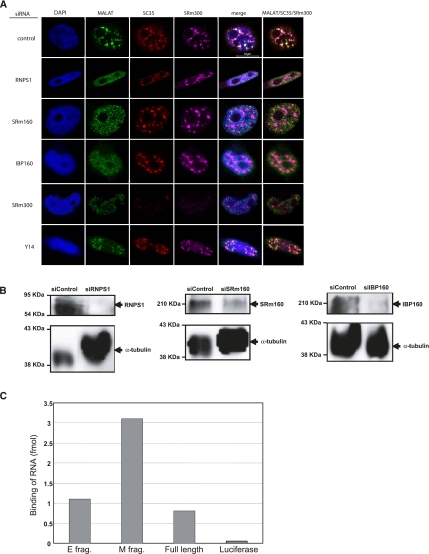
It has been reported that many proteins localize to nuclear speckles, including snRNPs, SR proteins, transcription factors, 3′ end RNA processing factors, structural proteins, kinases, phosphatases, and RNA transport factors (Lamond and Spector 2003; Hall et al. 2006). Recent proteomics analysis has further increased the repertoire of protein factors that localize to nuclear speckles (Saitoh et al. 2004). To test whether speckled proteins contribute to MALAT-1 localization to nuclear speckles, we eliminated the expression of speckled proteins by RNA interference (RNAi), and the localization of MALAT-1 was then observed by FISH analysis.
SRm300 and SRSF2 (SC35) are core structural components of nuclear speckles (Blencowe et al. 2000; Hall et al. 2006; Hutchinson et al. 2007). We observed that the RNAi repression of SRm300 disrupted the speckle-type staining pattern of SRSF2 (SC35), as well as that of MALAT-1 (Fig. 4A). These results suggest that SRm300 is essential for the core structure formation of nuclear speckles, and also indicate that MALAT-1 requires the nuclear speckle core structure to achieve its speckle-pattern localization within the nucleus. Therefore, we monitored the localization pattern of both SRSF2 (SC35) and SRm300 to assure the stability of the nuclear speckle core structure in cells when other speckle proteins were depleted. The RNAi repression of either RNPS1 or SRm160 resulted in the diffusion of MALAT-1 in the nucleoplasm, although their depletion did not disrupt the core structure of nuclear speckles (Fig. 4A,B). These results suggest that RNPS1 and SRm160 retain MALAT-1 on the nuclear speckles. To evaluate the binding affinity of RNPS1 to MALAT-1 RNA in vitro, we performed a filter-binding assay (Fig. 4C). We observed that purified RNPS1 specifically bound to MALAT-1 RNA, and the binding affinity of RNPS1 to M fragment RNA was much stronger than for the E fragment RNA and full-length MALAT-1. The data correspond with a more intense speckled localization of the M fragment compared with the E fragment (Fig. 2B).
FIGURE 4.
Effects of RNAi-mediated depletion of nuclear proteins on the localization of MALAT-1. (A) FISH analyses of HeLa-TO cells transfected with the indicated siRNAs (endogenous MALAT-1, green) are displayed, together with immunocytochemical staining of SRSF2 (SC35) (red) and SRm300 (cyan). DAPI (blue) was used to visualize nuclei. (B) Western blot analysis of the indicated proteins where expression had been knocked down. (C) Binding activity of RNPS1 to full-length or partial MALAT-1 RNA as measured by a filter-binding assay in vitro.
It was reported that IBP160, one of the speckle-localizing proteins, interacts with RNPS1 and SRm160 (Ideue et al. 2007); therefore, we examined whether IBP160 was also required for nuclear speckle localization of MALAT-1. We found that the RNAi-mediated depletion of IBP160 led to the diffusion of MALAT-1 in the nucleoplasm (Fig. 4A), which is similar to what we observed under the RNAi repression of either RNPS1 or SRm160 (Fig. 4A).
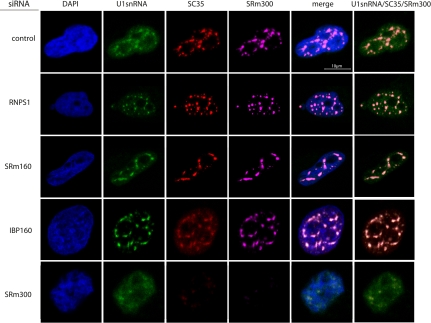
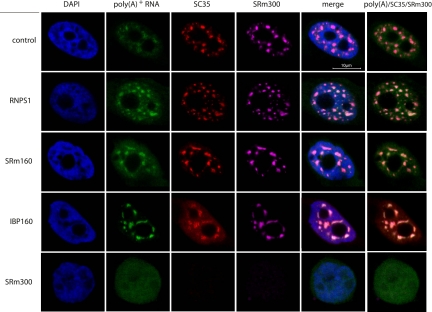
A population of poly(A)+ RNA are localized to nuclear speckles (Carter et al. 1991; Visa et al. 1993), where U snRNAs are also localized (Huang and Spector 1992). Therefore, we next investigated whether the depletion of RNPS1, SRm160, or IBP160 affected the nuclear speckle localization of U1 snRNA and poly(A)+ RNAs. We observed that RNAi repression of RNPS1, SRm160, or IBP160 did not affect the localization of U1 snRNA and poly(A)+ RNA (Figs. 5, 6), suggesting that RNPS1, SRm160, and IBP160 are not involved in the localization of U1 snRNA and poly(A)+ RNAs to nuclear speckles, but they are associated with a retention mechanism for MALAT-1 in the nuclear speckles.
FIGURE 5.
Subcellular localization of U1 snRNA in cells where the expression of nuclear speckle proteins was repressed by RNAi. FISH analyses of HeLa-TO cells transfected with the indicated siRNAs (endogenous U1 snRNA, green) are displayed, together with immunocytochemical staining of SRSF2 (SC35) (red), SRm300 (cyan), and DAPI staining of DNA (blue).
FIGURE 6.
Subcellular distribution of poly(A)+ RNAs in cells where the expression of nuclear speckle proteins was repressed by RNAi. FISH analyses of HeLa-TO cells transfected with the indicated siRNAs (poly(A)+ RNA, green) are displayed, together with immunocytochemistry of SRSF2 (SC35) (red), SRm300 (cyan), and DAPI staining of DNA (blue).
RNPS1 and SRm160 are components of the exon-junction complex (EJC), which play an important role in nonsense-mediated mRNA decay (NMD) and mRNA export (Chang et al. 2007). IBP160, which was originally identified as an intron-binding protein, is required before the formation of the EJC (Hirose et al. 2006; Ideue et al. 2007). Moreover, it has been shown that the depletion of IBP160 causes dysfunction of the NMD pathway, resulting in an increase in the amount of Gas5 ncRNA, an NMD target (Tycowski et al. 1996; Smith and Steitz 1998; Ideue et al. 2007). To rule out the possibility that the delocalization of MALAT-1 is arbitrarily attributed to a vulnerable EJC caused by depletion of RNPS1, SRm160, or IBP160, we examined the effect of depleting Y14, an EJC core factor, on the distribution of MALAT-1. Because the depletion of Y14 did not affect the localization of MALAT-1 (Fig. 4A), the localization of MALAT-1 to nuclear speckles might not be related with the integrity of the EJC. Our observation that the depletion of Magoh, another EJC core factor, did not cause delocalization of MALAT-1 (data not shown) further supports this scenario. We also examined the effect of MALAT-1 depletion on EJC function by monitoring the expression level of Gas5 ncRNA. Depletion of MALAT-1 had no effect on cell viability and did not affect the level of Gas5 mRNA (data not shown). In contrast, the depletion of UPF1, an essential NMD factor, increased Gas5 ncRNA levels as previously reported (Ideue et al. 2007). Together, these results indicate that MALAT-1 does not participate in the NMD pathway.
Effect of MALAT-1 delocalization on gene expression
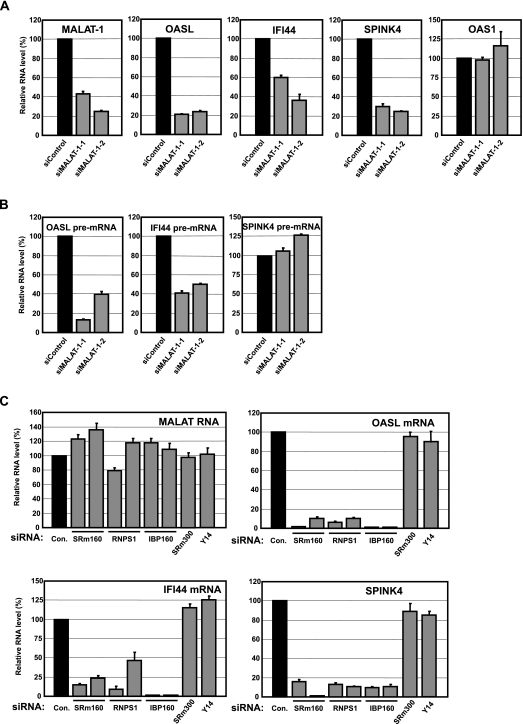
We postulate that MALAT-1 might have as yet undiscovered/novel functions with respect to the transcriptional regulation or pre-mRNA metabolism associated with nuclear speckles. To test this hypothesis, we investigated whether MALAT-1 affects gene expression. We used DNA microarray analysis, covering the whole human genome, to analyze up- and down-regulated genes in two independent MALAT-1 silenced cell lines prepared by RNAi. We found that only 14 genes with significant differences in expression, and quantitative real-time PCR analysis confirmed that the expression of 2′-5′-oligoadenylate synthetase like protein (OASL), interferon-induced protein 44 (IFI44), and serine peptidase inhibitor Kazal type 4 (SPINK4) were reduced in both MALAT-1-silenced cell lines (Fig. 7A). We assume that a loss of MALAT-1 function, either directly or indirectly, results in the down-regulation of these genes in vivo.
FIGURE 7.
Reduced expression of OASL, IFI44, and SPINK4 in cells where the expression of MALAT-1, RNPS1, SRm160, or IBP160 was repressed by RNAi. (A) Total RNAs were prepared from cells transfected with siRNAs against MALAT-1 and then analyzed by qRT–PCR. Using two different siRNAs against MALAT-1 minimizes the possible off-target effects, causing apparent reductions of transcript levels. Each value was normalized to the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Error bars indicate the standard deviation (SD) obtained from three independent experiments. (B) Pre-mRNA levels of the indicated transcripts were measured by qRT–PCR. The primer set for OASL pre-mRNA and the primer set for IFI 44 pre-mRNA amplified the cDNA sequence in the exon 1 and intron 1 junction of each pre-mRNA. The primer set for SPINK4 pre-mRNA amplified the cDNA sequence in the junction between exon 3 and intron 3 of SPINK4. (C) The indicated transcripts were quantified by qRT–PCR analysis using total RNAs prepared from cells transfected with the indicated siRNAs. Each value was normalized to the expression level of GAPDH. Error bars represent the SD obtained from three independent experiments.
To confirm the effect of MALAT-1 on the expression of these three genes, we analyzed their pre-mRNA levels in the MALAT-1 silenced cells (Fig. 7B). The pre-mRNA levels of OASL and IFI44 were reduced, while that of SPINK4 was unaltered between control cells and MALAT-1-silenced cells, suggesting that MALAT-1 can influence gene expression at distinct steps.
We investigated whether the delocalization of MALAT-1 also disrupts its own function toward the downstream genes. We examined the expression levels of OASL, IFI44 and SPINK4 in cells in which MALAT-1 localization was disrupted by RNAi-mediated repression of RNPS1, SRm160, or IBP160. The expression level of MALAT-1 was not changed in those knockdown cells; however, the expression levels of OASL, IFI44, and SPINK4 mRNA were decreased (Fig. 7C), similar to the results from our MALAT-1 depletion experiments (Fig. 7A). In contrast, the repression of Y14, which had no effect upon the localization of MALAT-1, did not influence the levels of OASL, IFI44, and SPINK4 mRNAs (Fig. 7C). These results suggest that the localization of MALAT-1 to nuclear speckles is required for its function. Although SRm300 affects the localization of MALAT-1 (Fig. 4A), depletion of SRm300 did not influence the levels of these genes (Fig. 7C), possibly because of the existence of nuclear speckle subdomains containing MALAT-1 in the SRm300-depleted cells (see Discussion section).
DISCUSSION
In this study, we have shown that the nuclear speckle components, RNPS1, SRm160, and IBP160, are essential factors for the localization of MALAT-1 to nuclear speckles, and that proper localization is controlled by two distinct regions of MALAT-1. We identified three genes, OASL, IFI44, and SPINK4, whose expressions were down-regulated following silencing of MALAT-1, which were also recapitulated by the displacement of MALAT-1 from nuclear speckles.
SRm160, IBP160, and RNPS1 promote nuclear speckle localization of MALAT-1
RNPS1 was originally identified as a pre-mRNA splicing activator and was shown to regulate alternative splicing in vivo (Mayeda et al. 1999; Sakashita et al. 2004). RNPS1, as a component of the EJC, also facilitates the 3′ end processing of mRNA and enhances translational activity (Wiegand et al. 2003; Trembley et al. 2005). SRm160 functions as a splicing coactivator and 3′ end cleavage factor (Blencowe et al. 1998; McCracken et al. 2003), and is capable of regulating alternative splicing in vivo (Cheng and Sharp 2006). Immunoprecipitation experiments have shown that RNPS1 associates with SRm160 (McCracken et al. 2003). IBP160, which can interact with RNPS1 and SRm160, is important for the formation of the EJC, but is dispensable for general splicing events (Ideue et al. 2007). Our results suggest that RNPS1, SRm160, and IBP160 form a complex in vivo, and this complex is required for the localization of MALAT-1 to nuclear speckles.
RNAi-mediated depletion of RNPS1, SRm160, or IBP160 showed that these factors are not required for nuclear speckle localization of U1 snRNA or poly(A)+ RNA. These data indicate that the role of these proteins leading to MALAT-1 localization to nuclear speckles is highly specific.
Both E and M regions have the localization signal(s) to nuclear speckles (Fig. 2). However, deletion of either E or M abolished the nuclear speckle localization of MALAT-1, even though each of the mutants still contains the other region involved in localization. A possible explanation for this observation is that E and M are more robust isolated than in the context of full-length RNA, as implied by the improved binding of M with RNPS1 (Fig. 4C). The formation of secondary structures in the full-length or isolated fragments of MALAT-1 may affect the robustness of nuclear localization.
MALAT-1 depletion and delocalization from nuclear speckles induces down-regulation of genes
OASL (2′-5′-oligoadenylate synthetase like protein), which is a member of the 2′-5-oligoadenylate synthetases, polymerizes ATP into a series of 2′–5′-linked oligomers of adenosine (2′–5′A) that activate the latent endoribonuclease, RNase L, an antiviral protein (Hovanessian and Justesen 2007). SPINK4 (serine peptidase inhibitor Kazal type 4), which encodes a peptide structurally related to pancreatic secretory trypsin inhibitor, was reported as a candidate gene responsible for celiac disease (Wapenaar et al. 2007). IFI44 (interferon-induced protein 44), which was originally identified to be a gene induced by interferon-α, has been suggested to encode an antiproliferative protein (Hallen et al. 2007).
OASL and IFI44 are both induced by interferons; therefore, MALAT-1 might be involved in the basal expression of the interferon response genes. We thus decided to examine the expression levels of other interferon-inducible genes in MALAT-1 knockdown cells. We found that the expression of 2′,5′-oligoadenylate synthetase 1 (OAS1) and 2′,5′-oligoadenylate synthetase 2 (OAS2), known interferon-inducible genes, was not reduced in MALAT-1 knockdown cells (Fig. 7A; data not shown), suggesting that it does not participate in the basal expression of general interferon response genes. This result also indicates that siRNAs targeting MALAT-1 did not induce an interferon response as a consequence of off-target effects. Although we do not know why altered expression of only three genes was detected, it is possible that MALAT-1 may regulate the expression of inducible genes by responding to certain stress or stimuli. This assumption is very likely because OASL and IFI44 are induced by interferon stimulus. It is possible that MALAT-1 regulates some other aspect of RNA metabolism rather than the absolute level of gene expression. Recently, it has been demonstrated that MALAT-1 is involved in the induction of serum-inducible genes (Yang et al. 2011), which supports our hypothesis that MALAT-1 affects the expression of stimuli-mediated inducible genes. It was also reported that MALAT-1 is involved in alternative splicing of certain transcripts (Tripathi et al. 2010). Therefore, we still cannot rule out the possibility that dysregulation of alternative splicing by MALAT-1 knockdown altered the expression of OASL, IFI44, or SPINK4.
We found that pre-mRNA levels varied between OASL, IFI44, and SPINK4 in the MALAT-1 silenced cells, suggesting that MALAT-1 affects gene expression at the transcriptional and post-transcriptional stages. MALAT-1 may affect either transcriptional activity or RNA processing through interaction with nuclear speckled proteins.
The expression of OASL, IFI44, and SPINK4 was not decreased in the SRm300-silenced cells, even when nuclear speckled localization of MALAT-1 was disrupted. The existence of nuclear speckle subdomains (or subcomplexes) has been previously suggested (Mintz and Spector 2000). Although the depletion of SRm300 disrupts the formation of nuclear speckles, subdomains that contain MALAT-1 might be present in the nucleus, and thus they could participate in the regulation of gene expression. Further investigation of this mechanism will uncover the complete function of MALAT-1 in the nucleus.
MATERIALS AND METHODS
Cell culture and transfection
HeLa-TO cells were grown at 37°C/5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin/streptomycin. Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Plasmid DNA (2 μg) and 4 μL of Lipofectamine 2000 in 500 μL of Opti-MEM I (Invitrogen) was dispensed onto 2 × 105 cells, then incubated for 4–6 h with 37°C/5% CO2. The DNA-Lipofectamine 2000 mixture was replaced with fresh medium and the transfected cells were further incubated for 24 h.
To perform RNAi, cells were transfected with siRNA duplexes synthesized by Sigma or Invitrogen (Table 1) using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's guidelines. Briefly, 12 pmol of siRNA duplex and 1.6 μL of Lipofectamine RNAi MAX (Invitrogen) in 200 μL of Opti-MEM I were layered onto 3 × 104 cells, then incubated for 4–6 h at 37°C/5% CO2. After changing the siRNA-Lipofectamine RNAi MAX complex to the fresh medium, the transfected cells were further incubated for 48 h. Real-time quantitative PCR or Western blot analyses (Fig. 4B) were used to examine whether RNAi achieved efficient reduction of the target sequence.
TABLE 1.
Target sequences for siRNA duplexes
Plasmid constructs
The cDNA encoding human MALAT-1 was amplified using specific primers (5′-GTAGGGCCCTCCATGGCGATTTGCCTTGTGAGCAC-3′ and 5′-GAGCTCGAGGTCCTGAAGACAGATTAGTAGTCAAAG C-3′) with a human cDNA library as a template. The amplified cDNA was then subcloned into the pGEM-T vector (Promega) as instructed by the manufacturer. The resulting cloned human MALAT-1 cDNA spanned nucleotides 493–8443 (8709 nt), corresponding to the sequence reported in GenBank (RefSeq accession no. NR_002819).
A series of MALAT-1 cDNA fragments (fragments A to O, and derivatives of the M fragment) were amplified using the cloned human MALAT-1 cDNA as a template and the corresponding primers listed in Table 2. The amplified products were digested with ApaI (Takara Bio), followed by subcloning of the digested cDNA into the ApaI site of pGL CMV MCS Luc2 pA. This plasmid was constructed by ligating an EcoRV–ApaI DNA fragment harboring the CMV promoter derived from pcDNA hygro (Invitrogen), and an EcoRV–ApaI DNA fragment from pGL 4.13 SV40 (Promega) harboring a luciferase (Luc2) cDNA, a poly(A) signal, and a bacterial origin of replication. Detailed construction details of the pGL CMV MCS Luc2 pA plasmid are available upon request. To make the plasmids expressing mutated MALAT-1, the MALAT-del-E, and MALAT-del-M primer sets described in Table 2 were used.
TABLE 2.
Oligonucleotides used to prepare the MALAT-1 fragment by PCR and those used in qRT-PCR analysis
Heterokaryon assay
The heterokaryon assay was performed as described previously (Pinol-Roma and Dreyfuss 1992). HeLa cells were transfected with a plasmid expressing either myc-tagged hnRNP A1 or myc-tagged hnRNP C1 using Lipofectamine 2000 (Invitrogen). The cells were then seeded onto coverslips, followed by coincubation with untransfected mouse NIH 3T3 cells for 3 h in the presence of 75 μg/mL cycloheximide. Cocultured cells were fused with 50% PEG4000 (Sigma) in phosphate-buffered saline for 2 min at room temperature, and the resulting heterokaryons were further incubated for 3 h in media containing 100 μg/mL cycloheximide before fixation. Fixed heterokaryons were processed for FISH analysis, followed by immunocytochemistry, to visualize MALAT-1 and myc-tagged proteins.
FISH and immunocytochemistry
To prepare the template cDNA for the RNA probes, a 3.3-kb fragment of MALAT-1 was amplified by PCR using the full-length MALAT-1 cDNA as a template and specific primers (5′-GGATGTGTAACTGAGGCGGGG-3′ and 5′-CACTACTCCAAGCATTGGGGAACAC-3′). The amplified MALAT-1 fragment was subcloned into pGEM-T (Promega). A digoxigenin (DIG)-labeled MALAT-1 RNA probe was prepared using DIG RNA labeling mix (Roche) and T7 RNA polymerase (Promega) according to the manufacturer's instructions. The transcribed DIG-labeled MALAT-1 RNA probe was partially digested in alkaline buffer (60 mM Na2CO3, 40 mM NaHCO3, at pH 10.2) to enhance the permeability of the probe (Cox et al. 1984). FISH was carried out to visualize MALAT-1 according to a previously described protocol (Sone et al. 2007). To detect the transiently expressed MALAT-1 fragment conjugated to luciferase mRNA, fixed cells were first treated with DNase I to eliminate transfected plasmid DNA prior to hybridization with a DIG-labeled luciferase RNA probe. The hybridized probes were detected using standard immunocytochemical procedures with monoclonal sheep anti-DIG (Roche) and polyclonal goat Alexa Fluor 488-conjugated anti-sheep IgG (Invitrogen) antibodies.
For in situ hybridization of U1 snRNA, cells were prepared according to a previously described method (Huang and Spector 1992). An Alexa Fluor 594-conjugated 2′-O-alkyl antisense oligonucleotide probe (Gene Design, Inc.) was used to visualize U1 snRNA as described previously (Carmo-Fonseca et al. 1991). Visualization of nuclear poly(A) RNA was carried out with an Alexa Fluor 594-conjugated oligonucleotide dT34 probe (Gene Design, Inc.) as previously described (Tokunaga and Tani 2008).
To visualize nuclear speckles by immunocytochemistry, we used the following antibodies: mouse monoclonal anti-SRSF2 (SC35) (Sigma); goat polyclonal Alexa Fluor 594-conjugated anti-mouse IgG (Invitrogen); rabbit polyclonal anti-SRm300 (kindly provided by Dr. Blencowe); goat polyclonal Alexa Fluor 647-conjugated anti-rabbit IgG (Invitrogen); rabbit polyclonal anti-PCNA (FL-261; Santa Cruz); and mouse monoclonal anti-myc (clone 9E10; Santa Cruz). For the detection of DNA, samples were incubated with DAPI (Wako). Fluorescent images were obtained using a confocal microscope TCS SP5 with a 63×/1.4NA HC PL APO CS objective (Leica). Acquired confocal images were pseudocolored and merged using LAS AF image analysis software (Leica). All images shown here are representative of consistent data from repeated experiments.
Filter-binding assay
The 32P-labeled RNA substrates were prepared using a MAXIscript In Vitro Transcription Kit (Ambion) according to the manufacturer's guidelines. Binding reaction mixtures (20 mM HEPES at pH 7.6, 100 mM KCl, 20% [w/v] glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT, 0.5 U/μL RNasin Plus [Promega]) with 7.5 nM recombinant RNPS1 (Mayeda et al. 1999), 0.5 nM 32P-labeled RNA substrate, and 20 μg/mL yeast tRNA were incubated at 37°C for 20 min. The reaction mixtures were filtered through a nitrocellulose membrane (Whatman) at room temperature and washed three times with 50 mL of buffer (20 mM HEPES at pH 7.6, 100 mM KCl, 20% [w/v] glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT, 0.5 U/μL RNasin Plus [Promega]). The radioactivity of nitrocellulose membrane filters was measured using a liquid scintillation counter.
Total RNA extraction and microarray analysis
Total RNA was extracted from HeLa cells transfected with siRNA using Trizol (Invitrogen) according to the manufacturer's instructions, followed by DNase I treatment to eliminate contamination of genomic DNA. The yield and purity of isolated RNA were determined using a spectrophotometer (NanoDrop) and visually checked by formaldehyde agarose gel electrophoresis. Total RNA (800 ng) was labeled with either Cy3 or Cy5 dye using an Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent). Labeled complementary RNAs derived from the total RNA of control cells and from MALAT-1-silenced cells were hybridized to the same microarray slide carrying 60-mer probes from the Whole Human Genome 4x44K Oligo Microarray Kit (Agilent Technologies). A flip labeling (dye-swap or reverse labeling with Cy3 and Cy5 dyes) procedure was followed to nullify the dye bias associated with unequal incorporation of the two Cy dyes into cDNA. The use of a dye-swap approach provides more stringent selective conditions for altered gene expression profiling than the use of a simple single- or two-color approach. Hybridization and washing procedures were performed according to the manufacturer's instructions, and the hybridized microarrays were scanned using an Agilent Microarray scanner G2565BA.
To detect the genes that showed a significant differential expression between control and MALAT-1 knockdown samples, each slide image was processed using Agilent Feature Extraction software (version 9.5.3.1). This program measures the Cy3 and Cy5 signal intensities of whole probes. Normalization was performed by locally weighted linear regression, which calculates the log ratio of dye-normalized Cy3- and Cy5-signals, as well as the final error in the log ratio. The P-values were based on the propagated error and universal error models. In this analysis, the threshold for genes showing significant levels of differential expression was less than 0.01. Additionally, generated artifactual data were eliminated before analysis with the software. The list of differentially expressed genes was generated and annotated using GeneSpring version GX 7.3.1 (Agilent). The data set (accession no. GSE20506) is available at the Gene Expression Omnibus (GEO) database at NCBI.
Quantitative Real Time Reverse Transcription–PCR (qRT–PCR)
Total RNA was prepared from cells using Trizol (Invitrogen) and was reverse transcribed to produce cDNA with a Quantitect Reverse Transcription Kit (Qiagen). Aliquots of cDNA were subjected to PCR using Syber premix II reaction mixture (Takara Bio) and the primer sets listed in Table 2 according to the manufacturer's instructions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an endogenous normalization control.
ACKNOWLEDGMENTS
We thank Dr. S. Nakagawa (Riken) for instruction in FISH analysis, Dr. B. Blencowe (University of Toronto) for providing the rabbit polyclonal anti-SRm300 antibody, and Dr. N. Kataoka (Kyoto University) for providing the plasmids expressing myc-tagged hnRNP A1 and myc-tagged hnRNP C1 together with instructions for the heterokaryon assay. We also thank Drs. K. Prasanth and S. Lang for critically reading our manuscript. This work was supported by the Naito Foundation and Takeda Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028639.111.
REFERENCES
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028−1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA 1998. A coactivator of pre-mRNA splicing. Genes Dev 12: 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Bauren G, Eldridge AG, Issner R, Nickerson JA, Rosonina E, Sharp PA 2000. The SRm160/300 splicing coactivator subunits. RNA 6: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidotti G, Baubec T, Pauler F, Seidl C, Smrzka O, Stricker S, Yotova I, Barlow DP 2004. The Air noncoding RNA: An imprinted cis-silencing transcript. Cold Spring Harb Symp Quant Biol 69: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev 12: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI 1991. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J 10: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB 1991. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol 115: 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74 [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Sharp PA 2006. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol 26: 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, DeLeon DV, Angerer LM, Angerer RC 1984. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol 101: 485–502 [DOI] [PubMed] [Google Scholar]
- Erwin JA, Lee JT 2008. New twists in X-chromosome inactivation. Curr Opin Cell Biol 20: 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB 2006. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen LC, Burki Y, Ebeling M, Broger C, Siegrist F, Oroszlan-Szovik K, Bohrmann B, Certa U, Foser S 2007. Antiproliferative activity of the human IFN-α-inducible protein IFI44. J Interferon Cytokine Res 27: 675–680 [DOI] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD 2011. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol 21: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG 2006. Subnuclear organelles: New insights into form and function. Trends Cell Biol 16: 19–26 [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA 2006. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol 71: 551–564 [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA 2006. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol Cell 23: 673–684 [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Justesen J 2007. The human 2′-5′oligoadenylate synthetase family: Unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 89: 779–788 [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL 1992. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci 89: 305–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39 doi: 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T 2007. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev 21: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC 1998. Structure and function in the nucleus. Science 280: 547–553 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR 1999. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J 18: 4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Longman D, Johnstone IL, Caceres JF, Blencowe BJ 2003. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J Biol Chem 278: 44153–44160 [DOI] [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G 1995. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83: 415–422 [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Spector DL 2000. Compartmentalization of RNA processing factors within nuclear speckles. J Struct Biol 129: 241–251 [DOI] [PubMed] [Google Scholar]
- Misteli T 2005. Concepts in nuclear architechture. Bioessays 27: 477–487 [DOI] [PubMed] [Google Scholar]
- Murakami K, Oshimura M, Kugoh H 2007. Suggestive evidence for chromosomal localization of non-coding RNA from imprinted LIT1. J Hum Genet 52: 926–933 [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol 134: 1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Patel SB, Bellini M 2008. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 36: 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355: 730–732 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL 2007. Eukaryotic regulatory RNAs: an answer to the “genome complexity” conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL 2004. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 15: 3876–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A 2004. Human RNPS1 and its associated factors: A versatile alternative pre-mRNA splicing regulator in vivo. Mol Cell Biol 24: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM 2009. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell 34: 179–190 [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T 2009. MENɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T 2009. CentroidFold: A web server for RNA secondary structure prediction. Nucleic Acids Res 37: W277–W2 80 doi: 10.1093/nar/gkp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18: 6897–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Spector DL 2001. Nuclear domains. J Cell Sci 114: 2891–2893 [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K, Tani T 2008. Monitoring mRNA export. Curr Protoc Cell Biol Chapter 22. Unit 22: 13 doi: 10.1002/047113030.cb2213s41 [DOI] [PubMed] [Google Scholar]
- Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y 2004. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell 16: 101–111 [DOI] [PubMed] [Google Scholar]
- Trembley JH, Tatsumi S, Sakashita E, Loyer P, Slaughter CA, Suzuki H, Endo H, Kidd VJ, Mayeda A 2005. Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol Cell Biol 25: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379: 464–466 [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E 1993. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res 208: 19–34 [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapenaar MC, Monsuur AJ, Poell J, van ‘t Slot R, Meijer JM, Meijer GA, Mulder CJ, Mearin ML, Wijmenga C 2007. The SPINK gene family and celiac disease susceptibility. Immunogenetics 59: 349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci 100: 11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Hayashizaki Y 2008. The RNA continent. Adv Cancer Res 99: 77–112 [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of Polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]